Abstract
The interactions between the subgroup A avian leukosis virus [ALV(A)] envelope glycoproteins and soluble forms of the ALV(A) receptor Tva were analyzed both in vitro and in vivo by quantitating the ability of the soluble Tva proteins to inhibit ALV(A) entry into susceptible cells. Two soluble Tva proteins were tested: the 83-amino-acid Tva extracellular region fused to two epitope tags (sTva) or fused to the constant region of the mouse immunoglobulin G heavy chain (sTva-mIgG). Replication-competent ALV-based retroviral vectors with subgroup B or C env were used to deliver and express the two soluble tv-a (stva) genes in avian cells. In vitro, chicken embryo fibroblasts or DF-1 cells expressing sTva or sTva-mIgG proteins were much more resistant to infection by ALV(A) (∼200-fold) than were control cells infected by only the vector. The antiviral effect was specific for ALV(A), which is consistent with a receptor interference mechanism. The antiviral effect of sTva-mIgG was positively correlated with the amount of sTva-mIgG protein. In vivo, the stva genes were delivered and expressed in line 0 chicken embryos by the ALV(B)-based vector RCASBP(B). Viremic chickens expressed relatively high levels of stva and stva-mIgG RNA in a broad range of tissues. High levels of sTva-mIgG protein were detected in the sera of chickens infected with RCASBP(B)stva-mIgG. Viremic chickens infected with RCASBP(B) alone, RCASBP(B)stva, or RCASBP(B)stva-mIgG were challenged separately with ALV(A) and ALV(C). Both sTva and sTva-mIgG significantly inhibited infection by ALV(A) (95 and 100% respectively) but had no measurable effect on ALV(C) infection. The results of this study indicate that a soluble receptor can effectively block infection of at least some retroviruses and demonstrates the utility of the ALV experimental system in characterizing the mechanism(s) of viral entry.
The first step in retrovirus infections involves an interaction between the viral envelope glycoprotein and a specific receptor on the surface of the host cell. A variety of cell surface proteins, including type I surface proteins, which span the membrane once, and polytropic surface proteins, which span the membrane multiple times, have been identified as receptors for retroviruses (30). The avian leukosis-sarcoma virus (ALV) group of retroviruses provides a powerful model system for studying envelope-receptor interactions; different members of these closely related viruses use distinct cellular receptors to gain entry into cells. The members of the ALV group are classified into a number of envelope subgroups, designated A to J, on the basis of host range, cross-interference of infection, and neutralization by antibodies (49). The susceptibility of chicken cells to ALV envelope subgroups A to E is determined by differences at three genetic loci designated tv-a, tv-b, and tv-c. The tv-a receptor controls susceptibility to subgroup A ALV, the tv-c receptor controls susceptibility to subgroup C, and the tv-b receptor controls susceptibility to subgroups B, D, and E. Susceptibility or resistance to viral infection is conferred by distinct alleles at each locus. There are two ways that resistance to retroviral infection can occur at the cell surface: (i) the cell is genetically resistant, i.e., a functional version of the specific receptor is not present on the surface of the cell; and (ii) the receptors are saturated with viral envelope glycoproteins that physically block the receptor, a phenomenon known as receptor interference (30, 48, 49).
Cells and animals that express retroviral envelope glycoproteins, due to a naturally occurring or genetically engineered endogenous virus, are highly resistant to retroviruses using the same receptor and have less virus-associated pathogenesis. Based on the example of the resistance of chicken lines that express endogenous subgroup E envelope glycoproteins to ALV(E) infection (38), Crittenden and colleagues demonstrated that insertion of the ALV(A) envelope gene into the germ line of chickens and its subsequent expression provided resistance to infection by ALV(A) strains by receptor interference (12, 18, 41, 42). However, the general utility of receptor interference as an antiviral strategy may be limited for retroviruses that express cytotoxic envelope glycoproteins (17, 21).
Three cell surface proteins have been identified as ALV receptors: Tva, the receptor for ALV(A) (5, 6, 50); CAR1, the receptor for ALV(B) and ALV(D) (10, 45); and SEAR, the receptor for ALV(E) (1). The normal cellular function of the Tva receptor is currently unknown; however, the extracellular domain contains a 40-amino-acid region which is homologous to the ligand-binding region of the low-density lipoprotein receptor (7, 39, 40). To aid in the characterization of the interactions between Tva and the ALV(A) envelope glycoproteins, soluble forms of the 83-amino-acid extracellular domain of the Tva receptor protein (sTva) were constructed by Connolly et al. (11), who reported that preincubation of the sTva proteins with different envelope subgroup ALVs caused a specific block to infection of susceptible chicken cells by ALV(A), but had no effect on ALV(B) or ALV(C) infection. In a recent study, sTva produced and purified with a baculovirus expression system blocked infection of turkey cells by ALV(A) (4).
In this study, the interactions between the ALV(A) envelope glycoproteins and soluble forms of the receptor were analyzed in chicken cells in vitro and in vivo. To determine if cells and chickens expressing sTva proteins are resistant to ALV(A) infection, ALV-based replication-competent retroviral vectors were used to efficiently deliver and express stva genes (20). The vectors are available with five different envelope subgroups (A to E), which enables multiple genes to be delivered and expressed in virtually every cell. We have found that two genes encoding sTva proteins, stva and stva-mIgG, were efficiently delivered and broadly expressed by ALV-based retroviral vectors both in cultured cells and in chickens. Both the sTva and sTva-mouse immunoglobulin G (sTva-mIgG) proteins significantly inhibited ALV(A) infection in vitro and in vivo. The antiviral effect was specific for ALV(A), consistent with a receptor interference mechanism.
MATERIALS AND METHODS
Soluble receptor and retroviral vector constructs.
The two soluble receptor gene constructs, contained in the plasmids pLC126 and pKZ457, were gifts of John A. T. Young (Harvard Medical School). The stva gene, encoding the 83-amino-acid Tva extracellular domain fused to a 9-amino-acid antibody epitope tag derived from influenza virus hemagglutinin and followed by six histidine residues, was isolated from pLC126 (11) as a NcoI-PstI fragment and cloned into the NcoI and PstI sites of the CLA12NCO adapter plasmid (20, 29). The stva-mIgG gene, encoding the 83-amino-acid Tva extracellular domain fused to the constant region of the mouse IgG heavy chain (nucleotides 353 to 1072) (47), was isolated from pKZ457 as a NcoI-BlpI fragment. The BlpI site was made blunt, and the modified fragment was cloned into the NcoI and SmaI sites of CLA12NCO. Both the stva and stva-mIgG genes had been modified to contain NcoI sites at their initiator ATGs. The soluble receptor gene cassettes were isolated as ClaI fragments from the adapter plasmids and cloned into the unique ClaI site of the RCASBP, RCAS, and RCOSBP retroviral vectors with subgroup B and subgroup C envelope genes. The RCAS family of replication-competent retroviral vectors have been described previously (19, 20, 29, 36, 37).
The stva-mIgG gene isolated as a ClaI fragment from the CLA12NCO adapter plasmid was subcloned into the TFANEO expression vector (17). TFANEO is a companion expression vector to the RCAS family of retroviral vectors. The expression cassette of TFANEO consists of two long terminal repeats derived from the RCAS vector that provide strong promoter, enhancer, and polyadenylation sites flanking a unique ClaI insertion site. The TFANEO plasmid also contains a neo resistance gene expressed under the control of the chicken β-actin promoter and an ampicillin resistance gene for selection in E. coli.
The RCASBP(A)AP, RCASBP(B)AP, and RCASBP(C)AP retroviral vectors which contain the heat-stable human placental alkaline phosphatase gene (AP) have been described previously (20, 22, 23). The AP gene, contained on a SalI fragment, was cloned into the SalI site of the CLA12 adapter plasmid and then subcloned into the RCASBP vectors as a ClaI fragment (gift of Constance Cepko).
Cell culture and virus propagation.
Chicken embryo fibroblasts (CEFs) derived from 10-day, line 0 embryos (C/E) (2) were grown in Dulbecco’s modified Eagle’s medium (GIBCO/BRL) supplemented with 10% tryptose phosphate broth (GIBCO/BRL), 5% fetal bovine serum (GIBCO/BRL), 5% newborn calf serum (GIBCO/BRL), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Quality Biological, Inc., Gaithersburg, Md.) as previously described (20). DF-1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (28, 44). Both CEF and DF-1 cultures were passaged 1:3 when confluent.
Virus propagation was initiated by calcium phosphate transfection of plasmid DNA that contained the retroviral vector in proviral form (20). In standard transfections, 5 μg of purified plasmid DNA was introduced into DF-1 cells or early-passage chicken embryo fibroblasts (CEF) by the calcium phosphate precipitation method (31). Viral spread was monitored by assaying culture supernatants for ALV capsid protein by either Western transfer analysis or enzyme-linked immunosorbent assay (ELISA) (46). Virus stocks were generated from the cell supernatants. The supernatants were cleared of cellular debris by centrifugation at 2,000 × g for 10 min at 4°C and stored in aliquots at −80°C. DF-1 cells transfected with the TFANEO plasmid were grown in 500 μg of G418 (GIBCO/BRL) per ml to select for neomycin-resistant cells. Clones were isolated by using cloning cylinders (Bellco Glass Inc., Vineland, N.J.), expanded, and maintained with standard medium supplemented with 250 μg of G418 per ml.
ALV AP challenge assay.
In a direct AP challenge assay, CEF or DF-1 cell cultures (∼30% confluent) were incubated with 10-fold serial dilutions of the RCASBP/AP virus stocks for 36 to 48 h at 39°C. In a preabsorption AP challenge assay, the 10-fold viral serial dilutions were first mixed for 3 h at 4°C with 2 ml of supernatant containing sTva-mIgG and then assayed as above. The assay for AP activity was modified from procedures of Cepko and coworkers (16, 22, 23). Cells were fixed in 4% paraformaldehyde in Dulbecco’s phosphate-buffered saline (PBS) for 30 min at 25°C, washed twice in PBS for 5 min each, and incubated for 1 h at 65°C to inactivate endogenous AP activity. The cells were then washed twice with AP detection buffer (100 mM Tris · Cl [pH 9.5], 100 mM NaCl, 50 mM MgCl2) for 10 min and exposed to the AP chromogenic substrates nitroblue tetrazolium (330 μg/ml) and 5-bromo-4-chloro-3-indolyl phosphate (170 μg/ml) (GIBCO/BRL). Enzymatically active AP produces an insoluble purple precipitate. The reaction was stopped by the addition of 20 mM EDTA (pH 8.0) in PBS.
Immunoprecipitation and Western transfer analysis of sTva-mIgG proteins.
A 500-μl aliquot of culture supernatant or serum was incubated with 50 μl of anti-mouse IgG-agarose beads (Sigma) for ≥1 h at 4°C. The sTva-mIgG agarose bead complexes were collected by centrifugation and washed twice in dilution buffer (50 mM Tris-buffered saline [TBS], 1% Triton X-100, 1 mg of bovine serum albumin per ml), once in 50 mM TBS, and once in 0.05 M Tris · Cl (pH 6.8). The washed complexes were collected by centrifugation, resuspended in 50 μl of 1× Laemmli buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.05 M Tris · Cl [pH 6.8], 0.1% bromophenol blue) without β-mercaptoethanol, and heated for 5 min at 100°C. The agarose in the samples was collected by centrifugation for 2 min, and the supernatants were transferred to new tubes. Prior to gel electrophoresis, 1.0 μl of β-mercaptoethanol was added to each 50-μl sample and the samples were heated for 5 min at 100°C. The denatured immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide) and transferred to a nitrocellulose membrane. The filters were blocked with 10% nonfat dry milk (NFDM) in PBS, probed with 0.05 μg of peroxidase-conjugated goat anti-mouse IgG antibodies (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) per ml in rinse buffer (100 mM NaCl, 10 mM Tris · Cl [pH 8], 1 mM EDTA, 0.1% Tween 20) plus 1% NFDM, and washed in rinse buffer. Protein-antibody complexes were detected with the Western blot chemiluminescence reagent (NEN) as specified by the manufacturer. The immunoblot was then exposed to Kodak X-Omat film.
Mouse IgG ELISA.
Immulon I 96-well plates (Dynatech Labs, Alexandria, Va.) were coated with 2.4 μg of goat anti-mouse IgG Fc fragment (Pierce, Rockford, Ill.) per ml (100 μl per well) in coating buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate [pH 9.5]) and incubated overnight at 4°C. The plate was washed three times with wash buffer (0.1% Tween 80 in PBS) and incubated with blocking buffer (5% NFDM in PBS) for 1 h at 37°C. The standard control protein, ImmunoPure mouse IgG Fc fragment (Pierce), and the sTva-mIgG proteins in culture supernatant or chicken serum were serially diluted in blocking buffer. The blocking buffer was removed from the wells, the diluted samples (100 μl/well) were added, and the wells were incubated for 1 h at 37°C. The wells were then washed three times with wash buffer and incubated for 1 h at 37°C with 0.8 μg of goat anti-mouse IgG Fc fragment antibody conjugated to horseradish peroxidase (Pierce) per ml in blocking buffer. The wells were washed three times with wash buffer and incubated with substrate (1.0 mg of 5-aminosalicylic acid [Sigma, St. Louis, Mo.] per ml, 18 mM potassium phosphate monobasic, 2 mM sodium phosphate dibasic [pH 6.0], 0.01% hydrogen peroxide) for 1 h at room temperature in subdued light. The absorbance at 490 nm was read with a microtiter plate reader. The linear range for a standard experiment was between 0.5 and 50 ng of ImmunoPure mouse IgG Fc fragment per ml.
In vivo ALV challenge assay.
Line 0 embryos were somatically infected with RCASBP(B), RCASBP(B)stva, or RCASBP(B)stva-mIgG by injecting unincubated eggs near the blastoderm with 100 μl containing 106 CEF or DF-1 cells producing the virus. Line 0 is a White Leghorn line that is genetically susceptible to all ALV subgroups except subgroup E and is free of endogenous proviruses that are closely related to ALV (2). Viremic chicks were identified at hatch by ELISA for the ALV capsid protein p27. Viremic and uninfected control chickens were infected intra-abdominally with 105 infectious units of either RAV-1 [an ALV(A) isolate] or RAV-49 [an ALV(C) isolate]. Blood was collected at 2, 4, or 9 weeks postchallenge, and the serum was assayed for infectious subgroup A, B, or C ALV by the in vitro ALV assay (see below). Statistical analyses of the data were done by the Fisher’s exact test (two tailed).
In vitro ALV assay.
The presence of infectious ALV in chickens was determined by assaying the serum samples on a panel of cell lines with different ALV envelope subgroup susceptibilities. The panel of indicator cell lines included line 0 CEF (C/E), which supports ALV(A), ALV(B), and ALV(C) replication; line alv6 CEF (C/A), which supports ALV(B) and ALV(C) replication; line RP30 B-cell line (C/B) which supports ALV(A) and ALV(C) replication; and line 15.C-12 CEF (C/C), which supports ALV(A) and ALV(B) replication. Serum samples (100 μl) were added to the cells, and the cells were incubated for 9 days in medium (containing 5% serum) to allow ALV to spread. The medium was changed after 3 days to avoid detection of ALV proteins in the original serum sample. The cells were then solubilized by two rapid freeze-thaw cycles to release ALV Gag antigens. The ALV capsid protein was detected by ELISA. A positive sample was defined as having an optical density reading of >0.200. The in vitro ALV assay can detect infectious ALV titers as low as 10 IFU/ml.
RNase protection assay.
Total RNA was isolated from cells in culture or from homogenized tissues of experimental birds by the RNazol B method (Tel-Test, Inc., Friendswood, Tex.). Sequence-specific RNA probes were cloned into pBluescript KS as follows. The RAV-1 envelope sequences were cloned as an XhoI-XbaI fragment (GenBank accession no. M19113; nucleotides 248 to 676) (9); the RAV-2 envelope sequences were cloned as a BamHI-SalI fragment (GenBank accession no. M14902; nucleotides 612 to 1080) (8); the stva probe was generated from an EcoRI-PstI fragment from plasmid pLC126 (GenBank accession no. L22752; nucleotides −9 to 386) (6, 11); and the stva-mIgG probe was generated from a ClaI-BamHI fragment derived from the adapter plasmid construct which contains the 5′ ClaI site and transcription leader from the CLA12NCO adapter plasmid and a synthetic sequence encoding the 83-amino acid Tva extracellular domain from pKZ457 that is different from the stva gene. A fragment of the chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (GenBank accession no. K01458; nucleotides 163 to 361) (35) was used as a control for the quantity and quality of the RNA. The constructs were linearized by restriction endonuclease digestion and gel purified. 32P-labeled antisense RNA probes were synthesized with the RNA transcription kit (Stratagene, La Jolla, Calif.). The probes were hybridized with 20 μg of total RNA in 20 μl of hybridization solution (80% formamide, 10 mM sodium citrate [pH 6.4], 300 mM sodium acetate [pH 6.4], 1 mM EDTA) overnight at 42°C. RNase protection assays were performed with the RPA II RNase protection kit (Ambion, Austin, Tex.). The RNA samples were digested with the RNase A-T1 mixture diluted 1:75. The protected RNA probe fragments were separated on a 6% acrylamide–7.6 M urea gel and exposed to Kodak X-Omat film.
PCR assays.
DNA was isolated from cells in culture or tissues of experimental birds by using the QIAamp tissue kit (Qiagen). Each PCR mixture contained 1.25 μl of 10× PCR buffer (final concentrations, 50 mM Tris · Cl [pH 8.3], 50 mM KCl, 7 mM MgCl2, and 1.1 mM β-mercaptoethanol), 1.25 μl of 1.7-mg/ml bovine serum albumin, 0.5 μl of each deoxynucleoside triphosphate at 25 mM, 0.5 μl of each primer (A260 = 5), 6.0 μl of H2O, and 1.0 μl of DNA (genomic DNA, ∼100 ng/μl; plasmid DNA, ∼2 ng/μl). The reaction mixtures were heated to 90°C for 1 min, and the reactions were initiated by the addition of 1.5 μl of Taq DNA polymerase (Promega, Madison, Wis.) diluted 1:10 (vol/vol) (0.75 U). Thirty cycles of PCR were carried out as follows: 90°C for 40 s and then 59°C for 80 s. Diagnostic primers used to detect ALV(A) env (9) were 5′-GGGACGAGGTTATGCCGCTG-3′ (∼50 bp upstream of KpnI site) and 5′-GGGCGTGCGCGCATTACCAC-3′ (nucleotides 871 to 851), yielding a 937-bp fragment. The PCR extension temperature was increased to 62°C for amplification of ALV(A) env. Diagnostic primers used to detect ALV(B) env (8) were 5′-GACCGACCCAGGGAACAATC-3′ (nucleotides 713 to 732) and 5′-ATGAGGAAAATTGCGGGTGG-3′ (nucleotides 1141 to 1122), yielding a 429-bp fragment. Diagnostic primers used to detect stva (6, 11) were 5′-GGAATGTGACTGGTAATGGA-3′ (nucleotides 56 to 75) and 5′-GCCTTAGTGATGGTGATGGT-3′ (nucleotides 369 to 350), yielding a 314-bp fragment. Diagnostic primers used to detect stva-mIgG were 5′-CCATCCGTCTTCATCTTCCCT-3′ (nucleotides 974 to 994) and 5′-TGGTGCGGTGTCCTTGTAGTT-3′ (nucleotides 1562 to 1542), yielding a 589-bp fragment of the mouse IgG gene (47). The amplified DNA fragments were separated on 0.8% agarose gels and visualized with ethidium bromide.
RESULTS
Experimental approach.
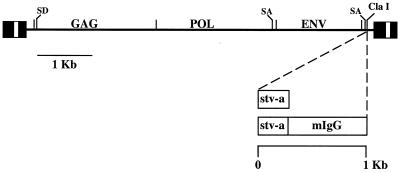
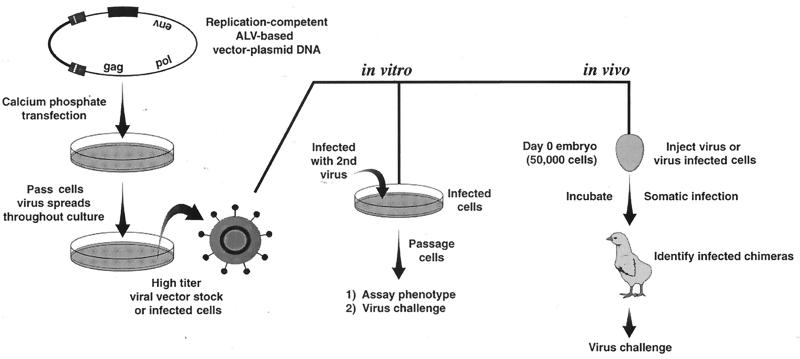
The stva and stva-mIgG receptor gene fusions were subcloned into the CLA12NCO adapter plasmid, which contains a transcriptional leader sequence and has a consensus ATG start site contained in a NcoI site. These sequences work very efficiently with the promoter-enhancer elements of the ALV-based retroviral vectors to express experimental genes at high levels (29). The RCAS family of retroviral vectors were derived from the Schmidt-Ruppin A strain of Rous sarcoma virus and are present in proviral form on pBR-based plasmids (20). Experimental genes are inserted into the vectors in the unique ClaI site (which replaces the src gene in RSV) and are translated from a spliced mRNA. Retroviral vectors that carry and express the stva and stva-mIgG genes are shown schematically in Fig. 1. Virus propagation was initiated by transfection of plasmid DNA containing the retroviral vector into avian cells (Fig. 2). The culture was then passaged until a maximum viral titer was achieved (6 to 10 cell passages depending on the vector) (19). Because vectors that use different receptors are available, this system can be used to deliver multiple genes to virtually all cells in the culture (25). Cell cultures that express sTva or sTva-mIgG from a subgroup B or C vector were subsequently challenged with ALV(A) to quantitate the antiviral effect of the sTva proteins.
FIG. 1.
Schematic of the sTva antiviral gene constructs and the ALV-based replication-competent retroviral vectors.
FIG. 2.
General procedure for using the replication-competent ALV-based retroviral vector system in vitro and in vivo. Reprinted, in part, from reference 20 with permission.
Antiviral effect of sTva in vitro and in vivo.
The initial experiments testing the effects of soluble receptors on viral replication were done with the sTva protein. The stva gene was introduced into the RCASBP vector, which produces the highest-titer viral stocks and the highest level of expression of an experimental protein. To quantitate the antiviral effect of sTva on ALV(A) infection, CEF fully infected with the vector alone [either RCASBP(B) or RCASBP(C)], CEF infected with vectors that express the stva gene [RCASBP(B)stva or RCASBP(C)stva], or uninfected CEF were challenged with RCASBP(A)AP, the subgroup A RCASBP vector containing the human placental AP reporter gene. The results of three assays are shown in Table 1. CEF cultures producing sTva, either from RCASBP(B) or from RCASBP(C), were >100-fold more resistant to infection by RCASBP(A)AP than were cultures infected with the vector alone. CEF cultures infected by the RCASBP(B) or RCASBP(C) vector alone were ∼threefold less susceptible to RCASBP(A)AP infection. The antiviral effect of sTva was specific for RCASBP(A)AP infection, since infection by viruses with other envelope subgroups were not inhibited (Table 1).
TABLE 1.
Relative resistance of CEF expressing sTva or sTva-mIgG to ALV infection
| Vector | Titer (mean ± SD) and resistance to infection with challenge virusa:
|
||
|---|---|---|---|
| RCASBP(A)AP | RCASBP(B)AP | RCASBP(C)AP | |
| Uninfected CEF | (1.5 ± 0.1) × 106 | (2.6 ± 0.5) × 105 | (8.1 ± 0.4) × 104 |
| RCASBP(B) | (4.3 ± 0.1) × 105 (3.5) | NDb | (2.0 ± 0.1) × 104 (4.1) |
| RCASBP(B)stva | (2.8 ± 0.1) × 103 (536) | ND | (1.9 ± 0.3) × 104 (4.3) |
| RCASBP(C) | (6.0 ± 0.1) × 105 (2.5) | (8.5 ± 0.2) × 104 (3.1) | ND |
| RCASBP(C)stva | (3.9 ± 0.2) × 103 (385) | (1.3 ± 0.1) × 105 (2.0) | ND |
| RCASBP(C)stva-mIgG | (2.8 ± 1.3) × 103 (536) | (7.3 ± 0.8) × 104 (3.6) | ND |
The resistance of the cells to ALV(A) infection was determined by dividing the mean titer obtained on the control uninfected DF-1 cells by the mean titer obtained for each experimental group.
ND, not done.
The CEF cultures infected with RCASBP(B) and RCASBP(B)stva were used to inoculate unincubated line 0 eggs to produce chickens viremic with RCASBP(B) or RCASBP(B)stva. Viremic chickens produced in this manner are tolerant to most ALV antigens, since the early embryo was infected. The chickens were challenged with 105 infectious units of RAV-1, an aggressive ALV(A) strain, to quantitate the antiviral effect of sTva. Blood samples were collected from representative birds in each group at 2 weeks postchallenge and from all birds 9 weeks postchallenge. The sera were assayed for ALV(A) and ALV(B) by the in vitro ALV assay (see Materials and Methods). The results of the challenges are summarized in Table 2. Of the birds infected with the RCASBP(B) vector alone and then challenged with RAV-1, 96% produced ALV(A) at both experimental time points as expected. However, 95% of the birds infected with RCASBP(B)stva did not produce detectable levels of ALV(A). These results demonstrate that sTva has a strong antiviral effect on ALV(A) infection both in vitro and in vivo. These results also demonstrate the utility of using vectors with different subgroups in vivo since experimental birds could be infected with both the RCASBP(B) vector and RAV-1. However, the level of sTva expression could not be quantitated, since neither the hemagglutinin nor the histidine epitope tags included on the sTva protein allowed efficient immunoprecipitation of the protein.
TABLE 2.
Chickens expressing sTva are resistant to ALV(A) infection
| Vector | No. of birds in which virus subgroup detected/total no. testeda
|
|||||
|---|---|---|---|---|---|---|
| 2 wk
|
9 wk
|
|||||
| A and B | B | A | A and B | B | A | |
| Uninfected | 0/10 | 0/10 | 0/10 | 0/9 | 0/9 | 0/9 |
| RCASBP(B) | 9/9 | 9/9 | 0/9 | 6/6 | 6/6 | 0/6 |
| RCASBP(B)+RAV-1 | 10/10 | 10/10 | 9/10b | 15/15 | 15/15 | 15/15b |
| RCASBP(B)stva | 10/10 | 10/10 | 1/10 | 11/12 | 11/12 | 0/12 |
| RCASBP(B)stva+RAV-1 | 10/10 | 10/10 | 0/10b | 20/20 | 20/20 | 1/20b |
All of the birds in each group were not necessarily assayed at both time points.
Statistical difference of P < 0.0001 within a column.
Antiviral effect of sTva-mIgG in vitro.
Although the tagged version of sTva could not be immunoprecipitated efficiently, we used an sTva immunoadhesin, sTva-mIgG, consisting of the 83-amino-acid Tva extracellular domain fused to the constant region of the mouse IgG heavy chain that could be immunoprecipitated and quantitated. The stva-mIgG gene was introduced into the RCASBP(C) vector. To quantitate the antiviral effect of sTva-mIgG, CEF cultures infected with RCASBP(C)stva-mIgG, RCASBP(C)stva, or RCASBP(C) were challenged with either RCASBP(A)AP or RCASBP(B)AP. CEF expressing sTva-mIgG were ∼200-fold more resistant to RCASBP(A)AP infection than were cells infected with the vector alone (Table 1). There was no statistical difference in the antiviral effect produced by sTva and sTva-mIgG in these experiments. The antiviral effect was specific for ALV(A), since no significant change in susceptibility was observed when the cultures were challenged with RCASBP(B)AP.
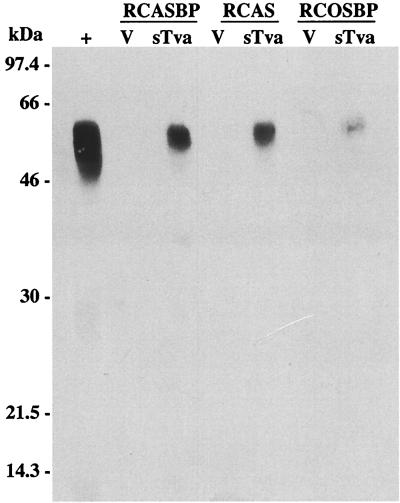
We and others have recently described ALV replication in a permanent, nontransformed cell line derived from line 0 CEF called DF-1 (28, 44). ALV and ALV-based retroviral vectors replicate and express inserted genes in DF-1 cells at levels similar to CEF, and DF-1 can be used to generate clonal cell lines. The antiviral effect of sTva-mIgG produced in DF-1 cultures infected with RCASBP(C)stva-mIgG (Table 3) was similar to that seen in CEF cultures (Table 1). The sTva-mIgG protein was immunoprecipitated from cell culture supernatants with anti-mouse IgG antibody conjugated to agarose beads and analyzed by Western immunoblotting of SDS-PAGE gels (Fig. 3). The immunoprecipitated sTva-mIgG protein migrated as a broad band (50 to 60 kDa) due to posttranslational modification and as a minor ∼38-kDa band (also see Fig. 4). The ∼38-kDa band is probably a degradation product of sTva-mIgG, since both bands appeared after immunoprecipitation with an ALV(A) surface glycoprotein immunoadhesin and the amount of the ∼38-kDa band increased after repeated freeze-thaw cycles of the viral supernatants (data not shown). Stable clonal DF-1 cell lines that express different levels of sTva-mIgG under the control of the TFANEO expression vector were generated (data not shown). These cell lines do not produce infectious ALV and are resistant to RCASBP(A)AP infection at levels similar to those of cultures expressing sTva-mIgG from the retroviral vectors (data not shown). Therefore, chronic ALV infection does not make a major contribution to the antiviral effect obtained.
TABLE 3.
Relative resistance of DF-1 cells expressing sTva-mIgG to ALV(A) infection
| Vector | RCASBP(A)AP titer (mean ± SD) | Resistancea | Concn of sTva-mIgG (ng/ml)b |
|---|---|---|---|
| Uninfected DF-1 | (3.1 ± 0.1) × 106 | ||
| RCASBP(C) | (1.2 ± 0.1) × 106 | 2.6 | |
| RCASBP(C)stva-mIgG | (5.4 ± 0.2) × 103 | 574 (221) | 1714 ± 97 (48 nM) |
| RCAS(C) | (6.2 ± 0.2) × 105 | 5 | |
| RCAS(C)stva-mIgG | (5.7 ± 0.4) × 103 | 544 (109) | 789 ± 26 (22 nM) |
| RCOSBP(C) | (9.0 ± 0.9) × 105 | 3.4 | |
| RCOSBP(C)stva-mIgG | (5.2 ± 0.8) × 104 | 60 (18) | 394 ± 26 (11 nM) |
The resistance of the cells to ALV(A) infection was determined by dividing the mean titer obtained on the control uninfected DF-1 cells by the mean titer obtained for each experimental group. The resistance of the cells to ALV(A) infection relative to the vector alone is given in parentheses.
Concentration of sTva-mIgG protein in the supernatants as quantitated by ELISA for the mIgG tag. The molar concentration of the sTva-mIgG protein is given in parentheses. The apparent molecular weight of sTva-mIgG was calculated from the amino acid sequence and found to be 35,911.
FIG. 3.
sTva-mIgG receptor expression levels in DF-1 cells. The sTva-mIgG protein was immunoprecipitated with goat anti-mouse IgG agarose beads from supernatants (500 μl) of DF-1 cultures infected with the RCASBP(C), RCAS(C), or RCOSBP(C) vector alone (V) or containing the stva-mIgG gene (sTva). The immunoprecipitates were denatured, separated by SDS-PAGE (12% polyacrylamide), and analyzed by Western transfer. The filter was probed with peroxidase-conjugated goat anti-mouse IgG, and the bound protein-antibody complexes were visualized by chemiluminescence on Kodak X-Omat film. sTva-mIgG protein expressed transiently in human embryonic kidney 293 cells was included as a positive control (+).
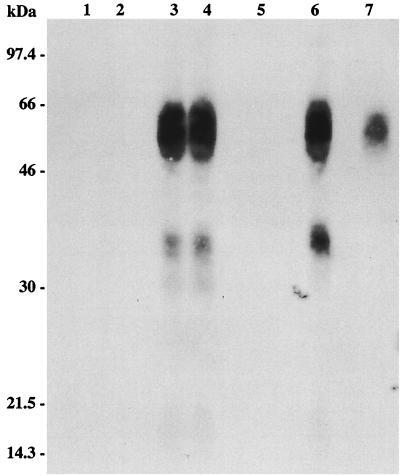
FIG. 4.
sTva-mIgG expression in sera of chickens infected with RCASBP vectors. The sTva-mIgG protein was immunoprecipitated from chicken serum (500 μl) and analyzed as described in the legend to Fig. 3. Lanes: 1, uninfected control; 2, RCASBP(B) vector-alone-infected bird; 3 and 4, two RCASBP(B)stva-mIgG-infected birds; 5, RCASBP(C) vector-alone-infected bird; 6, RCASBP(C)stva-mIgG-infected bird; 7, RCASBP(B)stva-mIgG-infected DF-1 cells as a positive control.
Relationship between sTva-mIgG expression level and the antiviral effect.
We have previously reported that the RCAS and RCOSBP vectors replicate to lower titers and produce lower levels of protein from an inserted gene compared to RCASBP (16, 20, 44). RCAS contains a different pol gene from RCASBP, and RCOSBP lacks the strong transcriptional enhancer contained in the LTRs of RCAS and RCASBP. To express sTva-mIgG at different levels in cells, the stva-mIgG gene was subcloned into the RCAS(C) and RCOSBP(C) retroviral vectors. DF-1 cultures infected with RCASBP(C)stva-mIgG, RCAS(C)stva-mIgG, or RCOSBP(C)stva-mIgG were challenged with RCASBP(A)AP to determine the antiviral effect of different levels of sTva-mIgG on ALV(A) infection. The level of sTva-mIgG produced by these DF-1 cultures was quantitated by ELISA for the mouse IgG tag (Table 3) and visualized by immunoprecipitation of sTva-mIgG followed by Western immunoblot analysis (Fig. 3). As expected, cultures infected with RCASBP produced the highest level of sTva-mIgG and the greatest antiviral effect, ∼200-fold compared to the vector alone control. Cultures infected with RCAS produced slightly lower levels of sTva-mIgG protein [2-fold lower than RCASBP(C)] and a lower antiviral effect (∼100-fold). While RCASBP(A)AP infected both the RCASBP(C)stva-mIgG and RCAS(C)stva-mIgG cultures with equal efficiency, the RCAS(C) vector-alone control culture was less susceptible to RCASBP(A)AP infection than was RCASBP(C) vector alone. Therefore, we believe that the twofold difference between the antiviral effect of RCASBP(C)stva-mIgG and RCAS(C)stva-mIgG is significant. Finally, cultures infected with RCOSBP produced the lowest level of sTva-mIgG protein [∼4-fold lower than RCASBP(C)] and a modest antiviral effect (∼15-fold) compared to the vector alone control.
The antiviral effect of sTva and sTva-mIgG on ALV(A) infection may represent the minimum antiviral effect attainable in vitro as measured by the direct ALV AP challenge assay. The assays were done on subconfluent cell cultures (30%), where the levels of the soluble receptor protein had not accumulated to the levels expressed by a confluent culture. To determine the antiviral effect of higher levels of sTva-mIgG, RCASBP(A)AP was pretreated with supernatants collected from confluent DF-1 cultures infected with RCASBP(B), RCASBP(B)stva-mIgG, RCOSBP(B), or RCOSBP(B)stva-mIgG and then assayed as before. Preabsorption of RCASBP(A)AP with high levels of sTva-mIgG significantly increased the antiviral effect compared to a direct assay: RCASBP(B) stva-mIgG pretreatment increased the antiviral effect of the direct assay ∼30-fold, and RCOSBP(B)stva-mIgG pretreatment increased the direct antiviral effect ∼60-fold (data not shown).
Delivery and expression of sTva and sTva-mIgG in vivo.
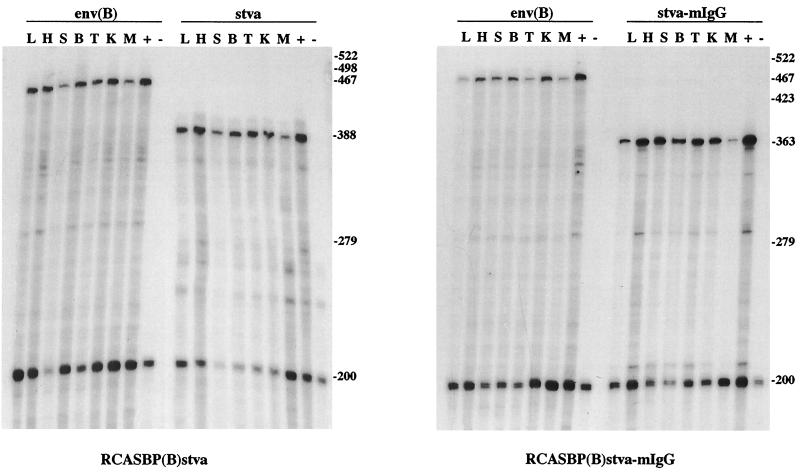
To characterize the efficiency of RCASBP delivery and expression of the sTva proteins in chickens, unincubated line 0 eggs were injected with CEF producing the RCASBP(B) or RCASBP(C) vectors alone, the vectors with the stva gene, or the vectors with the stva-mIgG gene. Viremic chickens were identified on the day of hatching by an ELISA for ALV capsid protein. The sTva-mIgG protein was immunoprecipitated from serum samples of both RCASBP(B)stva-mIgG- and RCASBP(C)stva-mIgG-infected birds and visualized by Western immunoblot analysis (Fig. 4). The sera from eight representative birds [five infected with RCASBP(B)stva-mIgG and three infected with RCASBP(C)stva-mIgG] contained 18.8 ± 2.2 μg/ml as quantitated by ELISA for the mouse IgG tag. The stva, stva-mIgG, and RCASBP(B) env RNA expression levels in liver, heart, spleen, bursa, thymus, kidney, and muscle tissues of infected birds were analyzed by the RNase protection assay. Results of RNase protection analyses of a representative bird infected with RCASBP(B)stva and a representative bird infected with RCASBP(B)stva-mIgG are shown in Fig. 5. Relatively high levels of the stva or stva-mIgG and ALV(B) env RNAs were detected in all tissues assayed, indicating that the inserted genes were delivered and expressed efficiently by the RCASBP(B) vector.
FIG. 5.
Analysis of viral and soluble receptor RNA levels in tissues of chickens infected with RCASBP(B). The figure shows autoradiograms of 6% polyacrylamide–7.6 M urea gels used to separate the protected RNA probe fragments produced in an RNase protection assay with RNA from a bird infected with RCASBP(B)stva or RCASBP(B)stva-mIgG. RNA was prepared from liver (L), heart (H), spleen (S), bursa (B), thymus (T), kidney (K), and muscle (M) tissues of each bird. RNA from DF-1 cells infected with the appropriate virus was included as a positive control (+). RNA from the bursa of an uninfected bird was the negative control (−). ALV(B) env RNA protects a 467-nucleotide fragment from the 522-nucleotide 32P-labeled full-length probe [env(B)]; stva RNA protects a 388-nucleotide fragment from the 498-nucleotide probe (stva); and stva-mIgG RNA protects a 363-nucleotide fragment from the 423-nucleotide probe (stva-mIgG). Each assay mixture contained a chicken GAPDH probe as a control for RNA quality and quantity. GAPDH RNA protects a 200-nucleotide fragment from the 279-nucleotide GAPDH probe.
Antiviral effect of sTva-mIgG in vivo.
Chickens infected with the RCASBP(B) vector alone, RCASBP(B)stva, or RCASBP(B)stva-mIgG were split into two groups and challenged with 105 infectious units of either RAV-1 (subgroup A) or RAV-49 (subgroup C). Blood was collected from each bird 4 weeks after challenge, and the serum was assayed for ALV(A), ALV(B), and ALV(C) by the in vitro ALV assay (Table 4). As expected, ALV(B) was detected in virtually all of the birds since the RCASBP(B) vector was used for gene delivery. ALV(A) was not detected in the sera of RAV-1-challenged birds containing the stva or the stva-mIgG genes. However, ALV(A) was detected in the sera of birds infected with the RCASBP(B) vector alone and challenged with RAV-1. In contrast, birds in all three experimental groups were equally susceptible to RAV-49 challenge, as shown by the presence of ALV(C) in the majority of the birds. Since 19% of the birds challenged with RAV-49 did not produce detectable levels of ALV(C), the titer of the RAV-49 stock may have been lower than expected. The antiviral effect of sTva and sTva-mIgG was specific for ALV(A), consistent with the proposed mechanism of antiviral action, receptor interference.
TABLE 4.
Chickens expressing either sTva or sTva-mIgG are resistant to ALV(A) infection but not to ALV(C) infection
| Vector | No. of birds in which virus subgroup detected/total no. testeda
|
||
|---|---|---|---|
| B | A | C | |
| RCASBP(B)+RAV-1 | 8/8 | 8/8b | |
| RCASBP(B)stva+RAV-1 | 5/5 | 0/5bd | |
| RCASBP(B)stva-mIgG+RAV-1 | 19/19 | 0/19be | |
| RCASBP(B)+RAV-49 | 4/4 | 3/4c | |
| RCASBP(B)stva+RAV-49 | 4/4 | 4/4cd | |
| RCASBP(B)stva-mIgG+RAV-49 | 7/8 | 6/8ce | |
Assays were done 4 weeks after challenge.
The 0/5 and 0/19 results are statistically different from 8/8 (P < 0.0001).
These results are not statistically different (P > 0.5).
Statistical difference of P < 0.008.
Statistical difference of P < 0.0001.
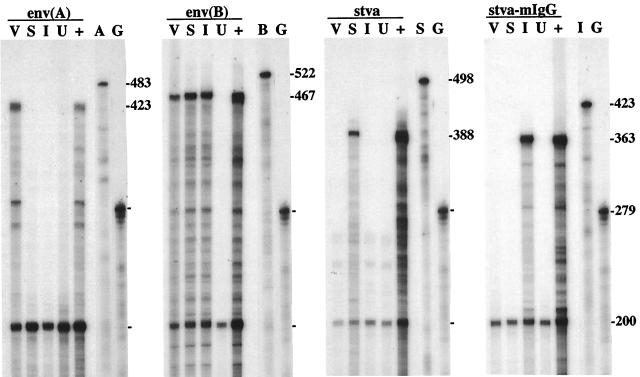
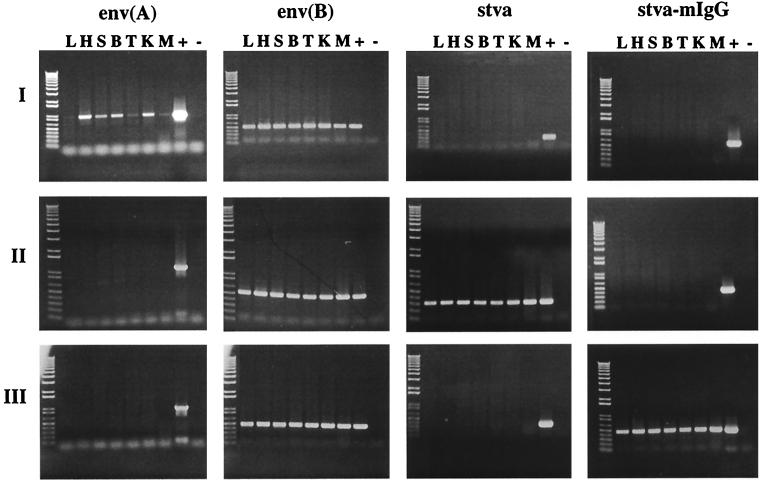
Representative birds from each RAV-1-challenged experimental group were analyzed for the presence of ALV(A) and ALV(B) env, stva, and stva-mIgG sequences in RNA and DNA isolated from a variety of tissue samples from each bird. We were concerned that a low level of RAV-1 infection and replication in a subset of tissues may go undetected by the in vitro ALV assay due to virus inactivation by the sTva or sTva-mIgG proteins in the serum. Antisense riboprobes and primer sets were developed to specifically detect each target sequence by the RNase protection assay and PCR. RNA and DNA were isolated from liver, heart, spleen, bursa, thymus, kidney, and muscle tissue of each bird and analyzed by RNase protection assay and PCR assay. Results of a representative RNase protection assay of RNA of one tissue (bursa) from a bird in each experimental group and an uninfected control bird are shown in Fig. 6. Results of representative PCR analysis of DNA isolated from tissues of a RAV-1 challenged bird from each experimental group are shown in Fig. 7. RAV-1 RNA and DNA were detected only in tissues of birds infected with the RCASBP(B) vector challenged with RAV-1. Therefore, we conclude that the expression of sTva and sTva-mIgG significantly reduces, if not eliminates, infection by the ALV(A) strain RAV-1 in chickens.
FIG. 6.
Analysis of viral and soluble receptor RNA levels in the bursas of birds infected with RCASBP(B) vectors. The figure shows an autoradiogram of a 6% polyacrylamide–7.6 M urea gel used to separate the protected RNA probe fragments produced in an RNase protection assay with RNA from the bursas of birds infected with RCASBP(B) alone (V), RCASBP(B)stva (S), or RCASBP(B)stva-mIgG (I) or uninfected (U). RNA from DF-1 cells infected with the appropriate virus was included as a positive control (+). ALV(A) env RNA protects a 423-nucleotide fragment from the 483-nucleotide 32P-labeled full-length probe [env(A)]; ALV(B) env RNA protects a 467-nucleotide fragment from the 522-nucleotide probe [env(B)]; stva RNA protects a 388-nucleotide fragment from the 498-nucleotide probe (stva); and stva-mIgG RNA protects a 363-nucleotide fragment from the 423-nucleotide probe (stva-mIgG). Each assay mixture contained a chicken GAPDH probe as a control for RNA quality and quantity. GAPDH RNA protects a 200-nucleotide fragment from the 279-nucleotide GAPDH probe. Full-length probes not treated with RNase are shown in the two right lanes of each panel: lane A, env(A); lane B, env(B); lane S, stva; lane I, stva-mIgG; lane G, GAPDH.
FIG. 7.
Analysis of viral and soluble receptor DNA in tissues of RCASBP(B)-infected birds and challenged with RAV-1. Genomic DNA was isolated from liver (L), heart (H), spleen (S), bursa (B), thymus (T), kidney (K), and muscle (M) samples from birds infected with RCASBP(B) alone (I), RCASBP(B)stva (II), and RCASBP(B)stva-mIgG (III) and challenged with RAV-1. DNA isolated from DF-1 cells infected with the appropriate virus was used as a positive control (+). DNA isolated from the bursa of an uninfected control bird was used as the negative control (−). DNA sequences were detected by PCR with specific primer pairs: RAV-1 challenge virus DNA [env(A)] was detected with primers specific for ALV(A) env, yielding a 937-bp fragment; RCASBP(B) vector DNA [env(B)] was detected with primers specific for ALV(B) env, yielding a 429-bp fragment; stva DNA (stva) and stva-mIgG DNA (stva-mIgG) were detected with specific primers yielding fragments of 314 bp and 589 bp, respectively. The amplified DNA fragments were separated on 0.8% agarose gels and visualized with ethidium bromide. The left lane of each panel contains 1-kb plus DNA ladder molecular size markers (GIBCO/BRL).
DISCUSSION
Cells expressing the sTva proteins showed significant resistance to ALV(A) infection, presumably due to the secreted receptor proteins binding the glycoproteins of the invading virion, thus blocking the interactions of the virus and the membrane-bound Tva, a form of receptor interference. Tva has been hypothesized to be necessary and sufficient to mediate ALV(A) entry (3, 6, 50). Several possible mechanisms could account for the sTva inhibition of ALV(A) entry. sTva binding of an ALV(A) surface glycoprotein may lead to an irreversible conformational change in SU and TM. Several studies have shown that sTva binding to purified ALV(A) envelope glycoproteins induces a temperature-dependent conformational change in the glycoproteins and appears to convert the envelope glycoproteins to a membrane-binding state (4, 14, 15, 24, 27). We suggest that the binding of sTva or sTva-mIgG to the envelope glycoproteins on the surface of the virus induces a conformational change in both SU and TM similar to the events leading to fusion of the viral and host cell membranes and converts SU and TM into a form that is unable to bind Tva on the surface of the cell. A conformational change may also lead to the loss of some of the SU subunits (34). Finally, sTva may inhibit ALV(A) entry by simply binding to SU and physically blocking the access of membrane-bound Tva to the virion. By whatever mechanism(s), the sTva proteins block the entry of ALV(A) into cultured cells and cells and tissues of chickens.
The replication-competent ALV-based retroviral vector experimental system enabled the efficient delivery and expression of the stva and stva-mIgG genes both in cultured cells and in virtually all the cells and tissues of the chicken. We have previously infected cultured cells with RCASBP(A) and RCASBP(B) vectors that express different proteins (25). It is now clear that other combinations of ALV retroviral vectors [ALV(B) followed by ALV(A); ALV(C) followed by ALV(A)] can be used in CEF and DF-1 cells in vitro and in vivo. As reported previously, the replication of some RCASBP(B) viruses in CEF and DF-1 cells and of RCASBP(C) viruses on DF-1 cells was somewhat cytopathic (28, 44). The cytopathic effect manifests itself as a pause in the growth (2 to 6 days), after which the cells recover, divide at a normal rate, and express the viral and experimental proteins. Although we have no direct evidence, we believe that only the cells that produce high levels of the RCASBP(B) or RCASBP(C) envelope glycoproteins show the cytopathic effects. The replication of subgroup B and C RCAS and RCOSBP viruses, which replicate to lower titers than RCASBP, does not cause detectable cytopathic effects. We did find that chronic infection of CEF and DF-1 cells with an ALV vector resulted in a lower level of resistance to infection by other ALV env subgroup vectors (two- to fivefold) compared to uninfected cells. This may indicate that while the ALV glycoproteins specifically and efficiently interact with the appropriate receptor, resulting in receptor interference, the high-level expression of one type of envelope glycoprotein on the cell surface may interfere either directly or indirectly with the ability of other ALVs to interact with their host receptors.
Both the RCASBP(B) and RCASBP(C) retroviral vectors were efficient in generating viremic chickens without detectable pathologic effects in short term infections, and the infected chickens expressed relatively high levels of sTva-mIgG protein in their serum. Chickens infected with RCASBP(B) were also efficiently infected with ALV(A). The RCASBP(B) vector efficiently delivered and expressed the stv-a and stva-mIgG genes in all tissues tested and resulted in a significant antiviral effect on ALV(A) infection and replication. By delivering the stva genes in the early embryo, the immune system of the chicken can be evaded. The birds are tolerized to the sTva and sTva-mIgG proteins and to most of the ALV antigens, since the viral vector used to deliver stva and stva-mIgG and the challenge viruses are virtually identical except for regions of SU. The expression of the sTva and sTva-mIgG proteins in vivo allowed us to test the effects of the viral glycoprotein-soluble receptor interactions in a wide variety of cells. One RAV-1-challenged, RCASBP(B)stva-infected bird did contain infectious ALV(A) in its serum. Unfortunately, the bird died of nonviral causes before tissues could be obtained. We are currently analyzing the ALVs present in the serum for ALV(A) to see if it is possible to obtain mutants that are resistant to the sTva-mIgG antiviral effect.
CD4, an important cell surface protein of T lymphocytes, is the primary receptor for human immunodeficiency virus type 1 (HIV-1). Several groups developed and expressed soluble forms of CD4 (sCD4) and demonstrated that recombinant sCD4 proteins could bind specifically to HIV-1 envelope glycoproteins and inhibit HIV-1 infection in vitro (13, 26, 32, 34, 43, 49). However, injecting recombinant sCD4 protein in animal and human trials had little, if any, antiviral effect. It is unclear whether one should expect an exogenously injected recombinant antiviral protein to be effective against cell-to-cell transmission of the virus. For this type of interference to be successful, it may be necessary to use a gene therapy approach in which target cells actively express the soluble receptor. The gene therapy approach has been tested for HIV-1: an sCD4 gene construct was expressed by a murine leukemia virus-based retroviral vector in human T-cell lines and in primary peripheral blood lymphocytes (33). In cell culture populations engineered to express sCD4 (30 to 50% of the cells contained the sCD4 gene), HIV-1 replication was inhibited 50 to 70%, indicating that an sCD4 antiviral approach against HIV-1 might be more effective under the right conditions and in the right environment. Since the initial sCD4 studies were published, the chemokine receptors have been identified as coreceptors necessary for efficient HIV-1 entry into cells (30). Since both CD4 and a chemokine receptor are required for efficient HIV-1 entry into cells, sCD4 alone may not be an effective inhibitor of HIV-1 entry. The ALV receptors do not appear to require coreceptors for the efficient entry of ALV into cells.
The results of this study clearly indicate that a soluble receptor interference antiviral strategy can effectively block the replication of at least some retroviruses and that this approach may be applicable to other virus groups that require specific viral glycoprotein-host receptor interactions for entry into the cell. The application of this strategy in the protection of animals against specific viral diseases is relatively straightforward, since transgenic technology can be used to introduce genes into animals and the transgenes will produce the desired protein without provoking an immune response. However, both the efficient delivery and expression of genes and the tolerance of the foreign proteins by the immune system are problems that remain to be solved if this type of antiviral approach is to be applied to humans.
ACKNOWLEDGMENTS
We thank Kurt Zingler and John A. T. Young (Harvard Medical School) for the pLC126 and pKZ457 plasmids, and we thank V. Shane Pankratz (Department of Health Sciences Research Section of Biostatistics at the Mayo Clinic) for the statistical analysis of the data.
This work was supported in part by the USDA NRI Competitive Grants Program (96-35204-3787), the Siebens Foundation under the Harold W. Siebens Research Scholar Program, and the Mayo Foundation (M.J.F.); by the Mayo Graduate School and the National Cancer Institute predoctoral training grant (T32CA75926) (S.L.H.); and by the National Cancer Institute, DHHS, under contract to ABL (S.H.H.).
REFERENCES
- 1.Adkins H B, Brojatsch J, Naughton J, Rolls M M, Pesola J M, Young J A T. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc Natl Acad Sci USA. 1997;94:11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astrin S M, Buss E G, Hayward W S. Endogenous viral genes are non-essential in the chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 3.Balliet J W, Bates P. Efficient infection mediated by viral receptors incorporated into retroviral particles. J Virol. 1998;72:671–676. doi: 10.1128/jvi.72.1.671-676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balliet J W, Berson J, D’Cruz C M, Huang J, Crane J, Gilbert J M, Bates P. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1999;73:3054–3061. doi: 10.1128/jvi.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates P, Rong L, Varmus H E, Young J A T, Crittenden L B. Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus. J Virol. 1998;72:2505–2508. doi: 10.1128/jvi.72.3.2505-2508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 7.Belanger C, Zingler K, Young J A. Importance of cysteines in the LDLR-related domain of the subgroup A avian leukosis and sarcoma virus receptor for viral entry. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bova C A, Manfredi J P, Swanstrom R. Env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 9.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A T. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 11.Connolly L, Zingler K, Young J A T. A soluble form of a receptor for subgroup A avian leukosis and sarcoma viruses (ALSV-A) blocks infection and binds directly to ALSV-A. J Virol. 1994;68:2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crittenden L B, Salter D W, Federspiel M J. Segregation, viral phenotype, and proviral structure of 23 avian leukosis viruses inserted in the germ line of chickens. Theor Appl Genet. 1989;77:505–515. doi: 10.1007/BF00274271. [DOI] [PubMed] [Google Scholar]
- 13.Daar E S, Li X L, Moudgil T, Ho D D. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damico R, Rong L, Bates P. Substitutions in the receptor-binding domain of the avian sarcoma and leukosis virus envelope uncouple receptor-triggered structural rearrangements in the surface and transmembrane subunits. J Virol. 1999;73:3087–3094. doi: 10.1128/jvi.73.4.3087-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federspiel M J, Bates P, Young J A T, Varmus H E, Hughes S H. A system for tissue-specific gene targeting: Transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci USA. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federspiel M J, Crittenden L B, Hughes S H. Expression of avian reticuloendotheliosis virus envelope confers host resistance. Virology. 1989;173:167–177. doi: 10.1016/0042-6822(89)90232-8. [DOI] [PubMed] [Google Scholar]
- 18.Federspiel M J, Crittenden L B, Provencher L P, Hughes S H. Experimentally introduced defective endogenous proviruses are highly expressed in chickens. J Virol. 1991;65:313–319. doi: 10.1128/jvi.65.1.313-319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Federspiel M J, Hughes S H. Effects of the gag region on genome stability: avian retroviral vectors that contain sequences from the Bryan strain of Rous sarcoma virus. Virology. 1994;203:211–220. doi: 10.1006/viro.1994.1478. [DOI] [PubMed] [Google Scholar]
- 20.Federspiel M J, Hughes S H. Retroviral gene delivery. Methods Cell Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- 21.Federspiel, M. J., and S. H. Hughes. Unpublished data.
- 22.Fekete D M, Cepko C L. Retroviral infection coupled with tissue transplantation limits gene transfer in the chicken embryo. Proc Natl Acad Sci USA. 1993;90:2350–2354. doi: 10.1073/pnas.90.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields-Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Givol I, Tsarfaty I, Resau J, Rulong S, da Silva P P, Nasioulas G, DuHadaway J, Hughes S H, Ewert D L. Bcl-2 expressed using a retroviral vector is localized primarily in the nuclear membrane and the endoplasmic reticulum of chicken embryo fibroblasts. Cell Growth Differ. 1994;5:419–429. [PubMed] [Google Scholar]
- 26.Harbison M A, Gillis J M, Pinkston P, Byrn R A, Rose R M, Hammer S M. Effects of recombinant soluble CD4 (rCD4) on HIV-1 infection of monocyte/macrophages. J Infect Dis. 1990;161:1–6. doi: 10.1093/infdis/161.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Himly M, Foster D N, Bottoli I, Iacovoni J S, Vogt P K. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 29.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. Adapter plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 31.Kingston R E, Chen C A, Okayama H. Introduction of DNA into eukaryotic cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 911–919. [Google Scholar]
- 32.Klasse P J, McKeating J A. Soluble CD4 and CD4 immunoglobulin-selected HIV-1 variants: a phenotypic characterization. AIDS Res Hum Retroviruses. 1993;9:595–604. doi: 10.1089/aid.1993.9.595. [DOI] [PubMed] [Google Scholar]
- 33.Morgan R A, Baler-Bitterlich G, Ragheb J A, Wong-Staal F, Gallo R C, Anderson W F. Further evaluation of soluble CD4 as an anti-HIV type 1 gene therapy: demonstration of protection of primary human peripheral blood lymphocytes from infection by HIV type 1. AIDS Res Hum Retroviruses. 1994;10:1507–1515. doi: 10.1089/aid.1994.10.1507. [DOI] [PubMed] [Google Scholar]
- 34.Orloff S L, Kennedy M S, Belperron A A, Maddon P J, McDougal J S. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolated with reduced sensitivity to sCD4. J Virol. 1993;67:1461–1471. doi: 10.1128/jvi.67.3.1461-1471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panabieres F, Piechaczyk M, Rainer B, Dani C, Fort P, Riaad S, Marty L, Imbach J L, Jeanteur P, Blanchard J. Complete nucleotide sequence of the messenger RNA coding for chicken muscle glyceraldehyde-3-phosphate dehydrogenase. Biochem Biophys Res Commun. 1984;118:767–773. doi: 10.1016/0006-291x(84)91461-x. [DOI] [PubMed] [Google Scholar]
- 36.Petropoulos C J, Hughes S H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petropoulos C J, Payne W, Salter D W, Hughes S H. Appropriate in vivo expression of a muscle-specific promoter by using avian retroviral vectors for gene transfer. J Virol. 1992;66:3391–3397. doi: 10.1128/jvi.66.6.3391-3397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson H L, Astrin S M, Senior A M, Salazar F H. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol. 1981;40:745–751. doi: 10.1128/jvi.40.3.745-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rong L, Bates P. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J Virol. 1995;69:4847–4853. doi: 10.1128/jvi.69.8.4847-4853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong L, Gendron K, Bates P. Conversion of a human low-density lipoprotein receptor ligand-binding repeat to a virus receptor: identification of residues important for ligand specificity. Proc Natl Acad Sci USA. 1998;95:8467–8472. doi: 10.1073/pnas.95.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salter D W, Crittenden L B. Artificial insertion of a dominant gene for resistance to avian leukosis virus into the germ line of the chicken. Theor Appl Genet. 1989;77:457–461. doi: 10.1007/BF00274263. [DOI] [PubMed] [Google Scholar]
- 42.Salter D W, Crittenden L B. Insertion of a disease resistance gene into the chicken germline. Bio/Technology. 1991;16:125–131. [PubMed] [Google Scholar]
- 43.Schacker T, Collier A C, Coombs R, Unadkat J D, Fox I, Alam J, Wang J-P, Eggert E, Corey L. Phase I study of high-dose, intravenous rsCD4 in subjects with advanced HIV-1 infection. J Acquired Immune Defic Syndr. 1995;9:145–152. [PubMed] [Google Scholar]
- 44.Schaefer-Klein J, Givol I, Barsov E V, Whitcomb J M, VanBrocklin M, Foster D N, Federspiel M J, Hughes S H. The EV-0-derived cell line DF-1 supports efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 45.Smith E J, Brojatsch J, Naughton J, Young J A T. The CAR1 gene encoding a cellular receptor specific for the subgroup B and D avian leukosis viruses maps to the chicken tvb locus. J Virol. 1998;72:3501–3503. doi: 10.1128/jvi.72.4.3501-3503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith E J, Fadly A M, Okazaki W. An enzyme-linked immunoabsorbant assay for detecting avian leukosis sarcoma viruses. Avian Dis. 1979;23:698–707. [PubMed] [Google Scholar]
- 47.Tucker P W, Marcu K B, Slightom J L, Blattner F R. Structure of the cloned gene for the constant region of murine gamma 2b immunoglobulin heavy chain. Science. 1979;206:1299–1306. doi: 10.1126/science.117549. [DOI] [PubMed] [Google Scholar]
- 48.Weiss R. Experimental biology and assay of RNA tumor viruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 209–260. [Google Scholar]
- 49.Weiss R A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J A, editor. The retroviruses. Vol. 2. New York, N.Y: Plenum Press; 1992. pp. 1–108. [Google Scholar]
- 50.Young J A T, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]