Abstract
Brome mosaic virus (BMV) RNA replication is directed by two virus-encoded proteins, 1a and 2a. The amino-terminal half of 1a is a distant homolog of alphavirus nonstructural protein nsP1, which has been implicated in capping viral RNAs. In this study, we examined the enzymatic activities of BMV 1a expressed in yeast, where the protein is fully functional in RNA replication. 1a methylated GTP, dGTP, and the cap analogs GpppG and GpppA, using S-adenosylmethionine (AdoMet) as the methyl donor. Product analysis by nuclear magnetic resonance spectroscopy showed that 1a methylation was specific for guanine position 7. Additionally, 1a interacted with GTP to form a covalent 1a-m7GMP complex. This reaction was specific for GTP, required AdoMet, and was accompanied by transfer of 3H-methyl from AdoMet to the covalent 1a-guanylate complex. The covalent complex could be immunoprecipitated by 1a antibodies. The 1a-m7GMP complex was inhibited in catalyzing further methyltransferase reactions. Mutation of conserved amino acids in the N-terminal half of 1a reduced both methyltransferase and covalent complex formation activities to very low or undetectable levels. Covalent 1a-guanylate complex formation took place in similar, AdoMet-dependent fashion in extracts of BMV-infected barley protoplasts. These results show that BMV 1a has activities similar to those of alphavirus nsP1, demonstrating conservation of these putative capping functions across a wide span of sequence divergence within the alphavirus-like superfamily. Conservation of this unusual combination of functions also supports the inference that the superfamily caps viral RNAs by an unusual pathway proceeding via a m7GMP intermediate.
The methylated cap structure at the 5′ end of eukaryotic mRNAs is crucial for mRNA stability and efficient initiation of translation. RNA capping in eukaryotic cells proceeds by a series of three reactions, which have been conserved from yeast to mammals (40). mRNA triphosphatase removes the 5′ γ-phosphate of nascent mRNA; then mRNA guanylyltransferase adds a GMP moiety so that a 5′-5′ triphosphate bridge is formed. This reaction proceeds via a covalent enzyme-GMP intermediate. Finally, the capping guanosine moiety is methylated by mRNA guanine-7-methyltransferase to yield the cap structure m7G(5′)ppp(5′)N1pN2p…, which is in some cases further modified by mRNA ribose-O-methyltransferase, acting on the 2′ position of the first two bases. Since capping of cellular mRNAs is a nuclear function closely associated with RNA polymerase II transcription (8, 27), it is not accessible to cytoplasmic viruses. Therefore, many of these encode their own capping systems, some of which conserve the cellular capping reactions. However, various groups of RNA viruses have evolved diverse alternate pathways for cap acquisition (42).
The brome mosaic virus (BMV) genome consists of three positive-sense RNA molecules. RNA1 and RNA2 encode proteins 1a and 2a, which are required for RNA replication (25, 45), whereas the 3a protein encoded by the 5′ half of RNA3 is needed for cell-to-cell movement within infected plants (5, 29). A subgenomic RNA4 representing the 3′ end of RNA3 is translated to yield the capsid protein (reviewed in references 1 and 43). All positive-sense BMV RNAs have a 5′ cap structure, m7GpppG (11). BMV RNA replication occurs in 1a- and 2a-containing complexes associated with the endoplasmic reticulum of infected cells (33). The ability of BMV to direct virus-specific RNA replication in the yeast Saccharomyces cerevisiae (22) facilitates studies of multiple aspects of its replication (19, 32, 44).
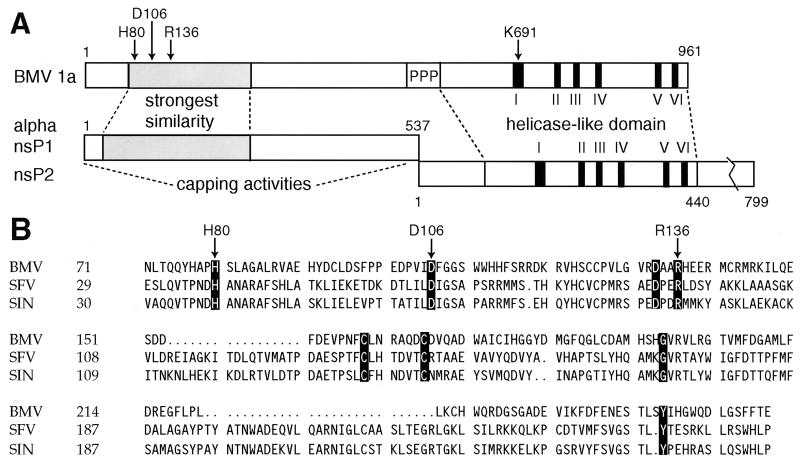
BMV belongs to the large alphavirus-like superfamily of RNA viruses, which in addition to the animal alphaviruses includes the plant bromo-, tobra-, tobamo-, tymo-, carla-, and potexviruses and other groups (2, 15, 23). The hallmark of the alphavirus-like superfamily is conservation of three domains in the RNA replication proteins encoded by these viruses. BMV protein 2a contains one of these domains, which is conserved with RNA-dependent RNA polymerases (17). The other two domains are found in BMV protein 1a (Fig. 1A) (2). These are a DEAD box helicase-related domain comprising the C-terminal half of 1a (16) and a domain implicated in RNA capping at the N terminus of 1a (4, 35). Separating these two domains in 1a is a proline-rich protease-sensitive region (31). In alphaviruses, domains corresponding to the N-terminal and C-terminal halves of BMV 1a are present in nonstructural proteins nsP1 and nsP2, respectively, which are cleaved from a larger precursor by a viral proteinase (Fig. 1A).
FIG. 1.
Comparison of BMV 1a with alphavirus replicase proteins nsP1 and nsP2. (A) Schematic showing BMV 1a and alphavirus nsP1 and nsP2. The region of most significant similarity between alphavirus nsP1s and the N-terminal half of BMV 1a is shaded (2, 4, 35). The C-proximal BMV 1a and alphavirus nsP2 helicase-like domains have six highly conserved motifs, numbered I to VI. In 1a, the nsP1 and nsP2-related domains are separated by a proline-rich linker region, marked PPP. The BMV 1a residues mutated in this study are marked at the top with arrows. (B) Sequence alignment of BMV 1a and the nsP1 proteins of the alphaviruses Semliki Forest virus (SFV) and Sindbis virus (SIN) in the region of their strongest similarity (shown shaded in Fig. 1A). The most strongly conserved residues in the alphavirus-like superfamily are highlighted, and residues mutated in this study are marked with arrows.
Two enzymatic activities have been described for alphavirus nsP1. First, guanine-7-methyltransferase, which is able to methylate GTP and dGTP, as well as some 5′-5′ dinucleotides containing guanosine (26, 38). Mutations in Sindbis virus (SIN) nsP1 affecting the methyltransferase activity were found within the nsP1 domain conserved with the N-terminal portion of BMV 1a (28). Based on secondary structure predictions, a large portion of this conserved domain is structurally related to cellular S-adenosylmethionine (AdoMet)-dependent methyltransferases (4). Second, nsP1 forms a covalent complex with m7GMP (3). This complex corresponds to the covalent enzyme-GMP intermediate formed by cellular mRNA guanylyltransferases but is distinct from them in that it contains a methyl group at position 7 of the guanosine ring. Therefore, the covalent m7GMP-nsP1 complex has been proposed to represent an intermediate in a novel pathway for cap formation, where methylation of the capping guanosine precedes its transfer to the 5′ end of mRNA (3).
As the domain corresponding to alphavirus nsP1 is only weakly conserved within the superfamily, and in particular between BMV 1a and alphaviruses (Fig. 1B), the degree to which such functions might be conserved in BMV or other alphavirus-like viruses has been uncertain. To explore the possible conservation of these activities within the superfamily and to advance understanding of BMV as a model for positive-strand RNA virus replication, we have tested for similar functions in BMV 1a. Here we show that BMV 1a has guanine-7-methyltransferase and covalent m7GMP binding activities that are similar to those of alphavirus nsP1, though distinct in some particulars.
MATERIALS AND METHODS
Plasmids and plasmid construction.
BMV 1a was expressed in S. cerevisiae from pB1CT19 (22), a 2μm plasmid containing a HIS3 selectable marker and the 1a open reading frame flanked by constitutive ADH1 promoter and ADH1 polyadenylation sequences. pRS423 (9) has the same selectable marker and was used as a negative control plasmid.
Point mutations were constructed in pB1CT19 with the unique site elimination method (Chameleon kit; Stratagene). The oligonucleotides used were as follows (the nucleotides differing from the parent plasmid are shown boldface): d(CAGTATCATGCGCCCGCTAGCCTGGCTGGTGC) for the H80A mutation, d(GAAGACCCCGTTATAGCGTTCGGAGGGTCTTGG) for D106A, d(GTTAGAGACGCTGCCGCTCATGAGGAGAGGATG) for R136A, and d(GTTGCGGGATGCGGTGCTACCACTGCCATAAAAG) for K691A. The selection primer d(GATCTTTCGAACAGGCCATATGCAGTTGTCGAAC) destroys a unique BsiWI site within the HIS3 gene but does not lead to a coding change. Restriction fragments SapI-MluI, PflMI-MluI, PflMI-MluI, and NcoI-PmeI of the mutant plasmids, respectively, were used to replace the corresponding fragment in wild-type pB1CT19 backbone, and the area of the transferred fragment was sequenced in each case to verify the presence of the desired mutation and the absence of additional mutations.
1a expression and membrane isolation.
S. cerevisiae YPH500 (Matα ura 3-52 lys2-801 ade2-101 tyr1-Δ63 his3-Δ200 leu2-Δ1) was transformed by the lithium acetate method (20) with pB1CT19 or its derivatives or with pRS423. Yeast cultures were grown at 30°C in a defined synthetic medium containing 2% glucose (6). Histidine was omitted to maintain plasmid selection.
Cells from 250 ml of culture at an optical density at 600 nm of 0.5 to 0.8 were collected, and the cell wall was removed with lyticase (36). Pelleted spheroplasts were stored at −80°C and thawed on ice. The following procedures were performed at 4°C. The spheroplasts were resuspended with 1 ml of lysis buffer (50 mM HEPES, pH 7.2, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 μg each of aprotinin, leupeptin, and pepstatin per ml); 250 μl of acid-washed glass beads was added, and the cells were mechanically broken in a Mini-Beadbeater-8 (Biospec Products) for 1.5 min at maximal setting. The total lysate (without glass beads) was mixed with 5 ml of 67% sucrose (wt/wt) in HN buffer (50 mM HEPES [pH 7.5], 100 mM NaCl) containing the above protease inhibitors. The suspension was poured into an SW41 Ti ultracentrifuge tube (Beckman) and overlaid by 5 ml of 50% (wt/wt) and 1 ml of 10% sucrose in HN buffer. The tubes were centrifuged overnight (ca. 16 h) at 35,000 rpm. The top 3 ml, containing flocculent membranous material, was collected and diluted with 9 ml of HN buffer. This suspension was centrifuged for 1 h at 35,000 rpm in an SW41 Ti rotor, the supernatant was discarded, and the pellet was carefully resuspended with 300 μl of lysis buffer. The membrane preparations were stored in aliquots at −80°C. One microliter of a typical, BMV 1a-containing preparation catalyzed the formation of approximately 2.5 pmol of m7GTP under the standard methyltransferase reaction conditions specified below.
Barley protoplasts were prepared, infected with BMV or mock infected, and incubated for 23 h as described elsewhere (24). A total of 7 × 106 protoplasts were broken in 1 ml of lysis buffer with 30 strokes in a tight-fitting Dounce homogenizer. Membranes were prepared by flotation and repelleting exactly as described above.
Enzyme assays.
The methyltransferase was assayed in a 25-μl final volume containing 50 mM HEPES (pH 7.2), 2 mM MgCl2, 2 mM dithiothreitol, 1.2% n-octyl-β-d-glucopyranoside, 10 mM GTP, 10 μM AdoMet, 0.75 μCi of Ado[methyl-3H]Met (80 Ci/mmol; Amersham), and 2 to 3 μl of the enzyme preparation. In testing the substrate specificity, the various methyl acceptors, including GTP, were present at 3 mM. The reaction mixtures were incubated for 50 min at 30°C and then transferred onto ice; 1 ml of 10 mM ammonium acetate (pH 8.5) was added. The labeled reaction products were isolated by ion exchange in 1 ml DEAE-Sephadex columns prepared in pasteur pipettes, which were washed with 100 mM NaCl in the same buffer. Elution of the nucleotides was achieved by 500 mM NaCl in the same buffer, and the incorporated label was measured by liquid scintillation (26).
For product identification, a large-scale reaction (total volume, 600 μl) was performed in the above buffer with 5 mM GpppG and 5 mM AdoMet as substrates. It contained 200 μl of a more concentrated enzyme preparation (obtained by resuspending the final yeast membrane pellets with one-third of the usual volume of buffer) and was incubated for 16 h at 30°C. The reaction products were isolated by ion exchange as described above except that 200-μl fractions were collected when eluting with 500 mM NaCl. The first five fractions containing UV-absorbing material were pooled, dried extensively in vacuo, and redissolved in 600 μl of D2O. The 500-MHz 1H nuclear magnetic resonance (NMR) spectra of this sample as well as of 0.5 mM solutions of pure GpppG and 7-methyl-GpppG (New England Biolabs) in D2O were obtained at the National Magnetic Resonance Facility, Madison, Wis.
Covalent guanylate binding reactions (3) were performed in a 30-μl final volume with 50 mM HEPES (pH 7.2), 10 mM KCl, 2 mM MgCl2, 5 mM dithiothreitol, 1.2% n-octyl-β-d-glucopyranoside, 100 μM AdoMet, and 10 μCi of [α-32P]GTP (800 Ci/mmol; New England Nuclear). In experiments where the complex was labeled with AdoMet-derived 3H, the substrates used were 100 μM GTP and 5 μCi of Ado[methyl-3H]Met (80 Ci/mmol). The reaction mixtures were incubated for 20 min at 30°C, and reactions were stopped by addition of sodium dodecyl sulfate (SDS) to 2% (final concentration) followed by boiling for 3 min. The samples were analyzed in SDS-polyacrylamide gels and visualized by autoradiographic film exposure or by a Molecular Dynamics PhosphorImager model 425 imaging system. In some reactions, vaccinia virus capping enzyme (Gibco BRL) was used as a control and [β,γ-32P]GTP (25 Ci/mmol; ICN) or [8-3H]GTP (8 Ci/mmol; Amersham) was used to determine the groups transferred to the covalent complex with 1a.
Immunoprecipitation and Western blotting.
For immunoprecipitation, SDS-denatured samples were diluted with 20 volumes of TET buffer (1% Triton X-100, 50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA). Polyclonal rabbit antiserum reactive against the N-terminal half of BMV 1a (amino acids 1 to 502) (33) or against BMV 2a was added to 1:200 dilution, followed by 10 μl of protein A-agarose beads (Boehringer Mannheim). The reaction mixtures were incubated overnight with gentle mixing. The beads were then washed three times with TET buffer, followed by boiling in SDS and polyacrylamide gel electrophoresis (PAGE) analysis.
For Western blotting, proteins separated in SDS-PAGE were transferred to polyvinylidene fluoride membrane (Immobilon P; Millipore). After blocking with 5% nonfat dry milk, and treatment with anti-1a antiserum (1:6,000), detection was performed with Immun-Star kit (Bio-Rad) and a Lumi-Imager luminescence imager (Boehringer Mannheim).
RESULTS
Enrichment of 1a by membrane isolation.
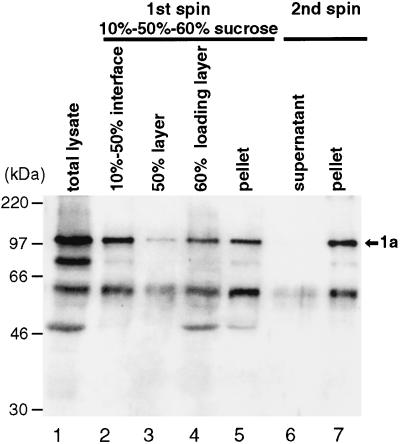
BMV 1a protein expressed in the yeast S. cerevisiae is functional, since it together with other viral and cellular components can mediate complete BMV RNA replication and subgenomic mRNA synthesis in vivo (22). Therefore, yeast should provide a good source of recombinant 1a protein for enzymatic studies. To prepare a 1a-enriched fraction for testing, we chose to take advantage of the fact that 1a expressed in yeast is membrane associated (32, 34). Yeast cells expressing 1a in the absence of other BMV components were spheroplasted, lysed, and total membranes were isolated by flotation of cell extracts in a discontinuous sucrose gradient with 10, 50, and 60% sucrose layers (Fig. 2). Some proteolytic degradation of 1a was always observed, as evidenced by faster-migrating bands reacting with 1a antibodies. Approximately 45% of full-length 1a was found in the fraction floating at the 10% sucrose/50% sucrose interface (lane 2). This interface layer was collected, and membranes were repelleted after dilution with buffer lacking sucrose (lanes 6 and 7). The final concentrated membrane preparation (lane 7) contained about 6% of total cellular protein and 40% of input 1a. Similar membrane preparations were used in all subsequent work.
FIG. 2.
Isolation of 1a-containing membranes. The total lysate from yeast cells expressing BMV 1a (lane 1) was subjected to flotation in a discontinuous sucrose gradient, consisting of 1 ml of 10% sucrose, 5 ml of 50% sucrose, and 6 ml of 60% sucrose (originally containing the lysate). After centrifugation, a floated membrane fraction (top 3 ml; lane 2), an intermediate fraction (next 3 ml; lane 3), the sample loading layer (bottom 6 ml; lane 4), and a resuspended pellet fraction (lane 5) were collected. The floated fraction was diluted with buffer and subjected to a second centrifugation to concentrate the membranes. This yielded supernatant (lane 6) and pellet (lane 7) fractions. All fractions were analyzed by SDS-PAGE and Western blotting and probed with antiserum against the N-terminal amino acids 1 to 502 of 1a (33). Equal percentages of each fraction were loaded on the gel, so that the recovery of 1a protein can be estimated from direct comparisons of the various lanes. Positions of molecular weight markers are shown at the left, and the position of full-length 1a is shown at the right.
Covalent binding of nucleotides by 1a.
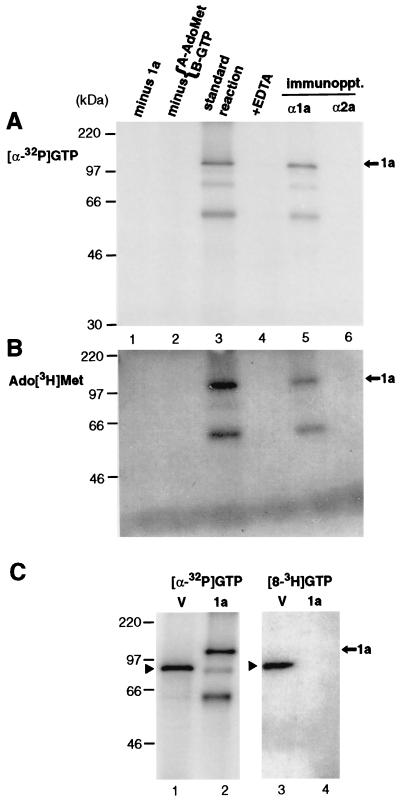
As a first test for enzymatic activity associated with 1a, 1a-enriched membrane fractions were prepared as described above, incubated with [α-32P]GTP, fractionated by SDS-PAGE, and analyzed by autoradiography. Under these conditions, only covalent complexes formed between proteins and the labeled nucleotide can survive and be visualized. A 32P-labeled band corresponding in size to full-length 1a protein (109 kDa) was readily observed, but only when the methyl donor AdoMet was present in the reaction mixture (Fig. 3A, lanes 2 and 3). This reaction also required divalent cations, as it did not take place in the presence of EDTA (lane 4). No labeled product was found in a control reaction using the analogous membrane fraction from yeast cells lacking 1a (lane 1). Furthermore, the labeled protein could be specifically immunoprecipitated with anti-1a antiserum (lane 5) but not with anti 2a-antiserum (lane 6). Two smaller products of approximately 60 and 80 kDa could also be labeled and immunoprecipitated (lanes 3 and 5). They correspond in size to 1a degradation products that were also detected by Western blotting with antiserum reactive against the N-terminal half of 1a (Fig. 2) and therefore contain at least portions of the N-terminal half of 1a.
FIG. 3.
Analysis of covalent complex formation between 1a and 7-methylated guanylate by SDS-PAGE and autoradiography. Portions of 1a-enriched membrane fractions prepared as in Fig. 2, lane 7, were incubated under standard conditions (see Materials and Methods) with [α-32P]GTP and unlabeled AdoMet (A) or Ado[methyl-3H]Met and unlabeled GTP (B) (lane 3). In control experiments, unlabeled AdoMet (A) or GTP (B) was omitted (lane 2), 5 mM EDTA was added to the standard reaction (lane 4), or membranes from cells not expressing 1a were incubated under standard conditions (lane 1). In each panel, the standard reaction mixture (lane 3) was also subjected to immunoprecipitation with anti-1a or anti-2a antiserum (lanes 5 and 6). (C) Vaccinia virus capping enzyme (lanes marked V) or 1a was incubated with [α-32P]GTP or [8-3H]GTP, as indicated, under the same conditions, in the presence of unlabeled AdoMet. Arrows mark the position of full-length 1a, arrowheads indicate the vaccinia virus capping enzyme, and positions of coelectrophoresed molecular weight markers are shown at the left.
1a also could be covalently labeled with AdoMet tritiated at the methyl group (Fig. 3B), suggesting that the covalent guanylate complex also contained a methyl group derived from AdoMet. Covalent 1a labeling with Ado[3H]Met took place under the same conditions as covalent 1a labeling with [α-32P]GTP (lane 3), required the presence of GTP (lane 2), and similarly was sensitive to EDTA (lane 4). The resulting complex was immunoprecipitable by anti-1a antibodies (lane 5).
The covalent nucleotide binding properties of 1a were compared with those of the well-characterized vaccinia virus capping enzyme, which is homologous to and functionally similar to cellular mRNA capping enzymes (40). Both 1a and the vaccinia virus enzyme could be labeled with 32P derived from [α-32P]GTP (Fig. 3C, lanes 1 and 2). However, unlike 1a (Fig. 3A), labeling of the vaccinia virus capping enzyme did not require AdoMet (data not shown), consistent with the prior finding that the nucleotide covalently bound to the vaccinia virus enzyme is unmethylated GMP (41). To further explore the basis of the 1a-specific AdoMet requirement and the structure of the covalently bound nucleotide, 1a and the vaccinia virus capping enzyme were each tested for covalent labeling after incubation with [8-3H]GTP (Fig. 3C, lanes 3 and 4). Based on the GTP-dependent labeling of 1a by Ado[3H]Met (Fig. 3B), this experiment tested for formation of m7G, since the hydrogen (or tritium) atom at position 8 of the guanosine ring is stable in GMP but becomes rapidly exchangeable with water if the guanosine is methylated at position 7 (18). As expected for binding unmethylated GMP, the vaccinia virus enzyme readily was covalently 3H labeled upon incubation with [8-3H]GTP (Fig. 3C, lane 3). However, although incubation with [α-32P]GTP under the same conditions covalently 32P labeled 1a and the vaccinia virus enzyme to similar levels (Fig. 3C, lanes 1 and 2), no 3H labeling of 1a could be visualized after incubation with [8-3H]GTP (Fig. 3C, lane 4). Therefore, this result indicates that before or simultaneously with covalent binding by 1a, all 1a-bound nucleotides were modified by methylation at position 7 of the guanosine ring.
Substrate requirements for covalent nucleotide binding.
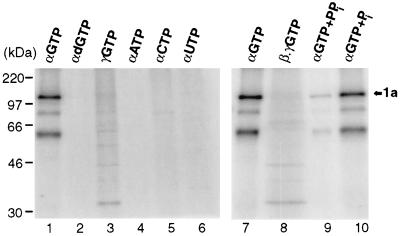
The substrate requirements for covalent complex formation with 1a were characterized by replacing [α-32P]GTP by various other 32P-labeled nucleotides (Fig. 4). The reaction was specific for GTP (lanes 1 and 7), since neither α-32P-labeled ATP, CTP, or UTP (lanes 4 to 6) or even [α-32P]dGTP (lane 2) could label 1a. Transfer of 32P label to 1a required its presence at the α position, as incubation with [γ-32P]GTP or [β,γ-32P]GTP did not lead to 1a labeling (lanes 3 and 8). Instead, other weak bands were observed, most prominently at 33 kDa. These appear likely to represent proteins phosphorylated by various kinases present in the reaction mixture. Thus, only the α-phosphate of GTP is present in the covalent complex with 1a. Furthermore, pyrophosphate but not phosphate inhibited covalent complex formation between 1a and [α-32P]GTP (lanes 9 and 10), suggesting that formation of the covalent complex involves the release of pyrophosphate. Taken together with the previous results, these findings imply that 1a forms a covalent complex with m7GMP.
FIG. 4.
Substrate specificity of covalent complex formation. 1a was incubated under standard conditions (see Materials and Methods) with the indicated, 32P-labeled nucleotide substrates in the presence of AdoMet. Greek letters indicate the labeled phosphate position in each case; 1 mM pyrophosphate (PPi; lane 9) or phosphate (Pi; lane 10) was included as indicated. All samples were analyzed by SDS-PAGE and autoradiography. The arrow marks the position of 1a, and positions of molecular weight markers are shown at the left.
1a methyltransferase activity.
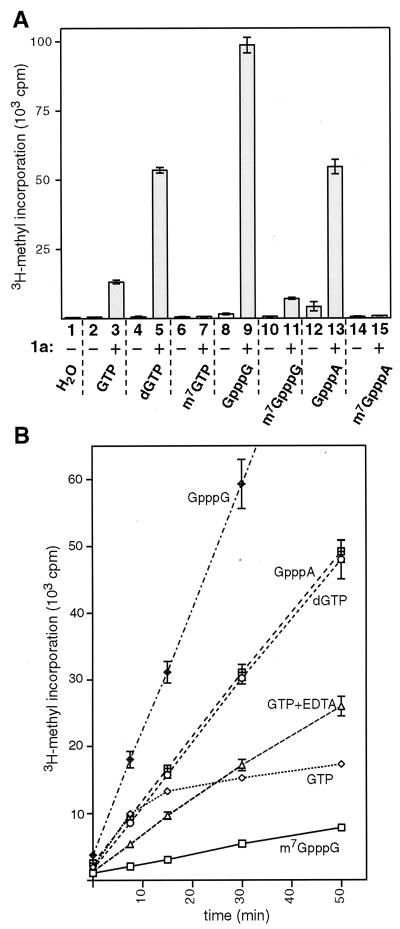
The requirement for AdoMet in the covalent guanylate binding by 1a and the presence of an AdoMet-derived methyl group in the resulting covalent complex suggested that guanosine methyltransferase activity might be associated with 1a protein. Therefore, we tested for the ability of 1a to methylate various nucleotide and dinucleotide acceptors, using AdoMet, 3H labeled at the methyl group, as the methyl donor (Fig. 5A). Membrane fractions from control cells lacking 1a were also assayed in each case. The methylated nucleotide reaction products were isolated from the reaction mixture by ion-exchange chromatography, and the amount of 3H-methyl label incorporated was measured by liquid scintillation. In initial reactions using GTP as the methyl acceptor, membrane fractions from yeast lacking 1a (Fig. 5A, bar 2) showed no methyltransferase activity above the 600-cpm background seen in parallel reactions with bovine serum albumin or water substituting for yeast extract (bar 1). In contrast, 1a-containing membranes showed robust activity, directing 3H-methyl transfer from AdoMet to GTP at a level 22 times above background (bar 3). Interestingly, dGTP appeared to be better than GTP as a methyl acceptor substrate for 1a in this reaction (bar 5). m7GTP, however, could not be further methylated by 1a (bar 7). As with the [8-3H]GTP results described above (Fig. 3), this indicates that 1a methylation was specific for position 7 of guanosine. In keeping with this inference, 1a showed no methylation activity on ATP, CTP, or UTP (results not shown).
FIG. 5.
Methyltransferase activity of 1a. (A) The substrates indicated below the bars were assayed as methyl group acceptors in standard reactions with membrane fractions isolated from yeast lacking 1a (−) and yeast expressing 1a (+). Two independent membrane preparations of each type were assayed, each in duplicate, with each of the substrates shown. Radioactivity incorporated to the substrate was measured by scintillation counting, and the average counts per minute for each condition is displayed by the histogram bars. Standard deviations are indicated by error bars. (B) Kinetics of the methyltransferase reaction catalyzed by 1a were studied by withdrawing aliquots from larger-scale reactions at 0, 7.5, 15, 30, and 50 min. The incorporated radioactivity was measured as described above, and the average counts per minute for each time point is displayed. The error bars indicate standard deviation and are included for all points, but in some cases they are obscured by the symbols used to plot average values. Two independent 1a-containing membrane preparations were assayed, each in duplicate, with each of the substrates shown. Standard reaction conditions including 2 mM MgCl2 were used except for the curve labeled GTP+EDTA, for which case the divalent cations were replaced with 5 mM EDTA. The curve for GpppG linearly continues to the 50-min time point (not shown).
The dinucleotide cap analogs GpppG and GpppA were good methyl acceptor substrates for 1a (Fig. 5A, bars 9 and 13). 1a also exhibited some activity toward m7GpppG, which has one nonmethylated guanosine (bar 11). However, this was less than 10% of the activity toward GpppG, suggesting that the 7-methylated guanosine already present in m7GpppG has an inhibitory effect on 1a. Similar to m7GTP, m7GpppA did not accept methyl groups in 1a-catalyzed reactions (bar 15). Membrane fractions from yeast not expressing 1a exhibited low levels of activity toward two of the substrates tested, the cap analogs GpppA (average, 4,240 cpm; bar 12) and GpppG (1,590 cpm; bar 8). This activity may be due to contamination of the membrane fraction with the yeast cap methyltransferase, since the eukaryotic enzyme has been shown to have activity toward these substrates (13).
To test whether the incorporation of methyl groups was constant over time (i.e., that the values obtained above in 50-min reactions accurately reflected the initial rates of reaction), time course experiments were performed with the substrates methylated by 1a (Fig. 5B). For all methyl-accepting substrates except GTP, the methyltransferase reaction was linear for the 50-min period. However, GTP showed a high initial rate of reaction, which rapidly declined to a much lower, constant rate after 15 min. Since GTP is the only nucleotide capable of forming a covalent complex with 1a (Fig. 4), we reasoned that covalent complex formation might inhibit the methyltransferase reaction. To test this, the methyltransferase reaction with GTP was performed under conditions where the covalent complex formation does not take place, i.e., in the absence of divalent cations (Fig. 5B, GTP+EDTA). In this case the initial rate of reaction was lower, but the reaction continued linearly for the 50-min period tested, yielding a final level of 3H-methyl incorporation above that for GTP under the standard reaction conditions.
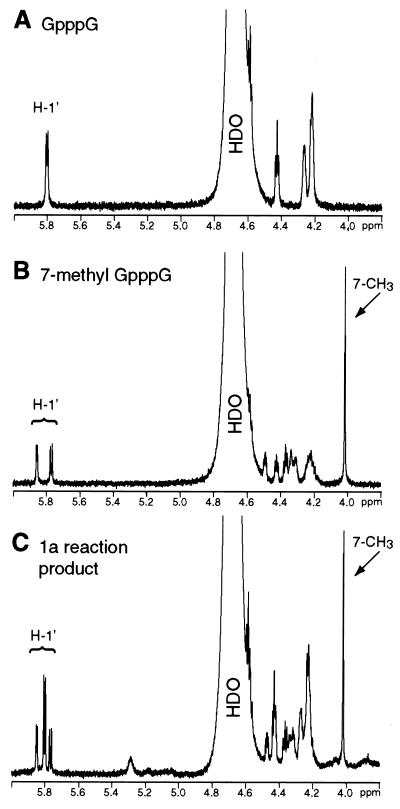
The product of the methyltransferase reaction was further studied after large-scale methylation of the best substrate, GpppG, by subjecting a partially purified substrate-product mixture to 1H NMR spectroscopy analysis. For comparison, commercially available GpppG (Fig. 6A) and 7-methyl-GpppG (the presumed product) (Fig. 6B) were also studied. As expected, the spectrum of the 1a-catalyzed reaction product (Fig. 6C) contained a prominent peak just above 4 ppm, exactly corresponding to the peak in the spectrum of 7-methyl-GpppG (Fig. 6B) due to the hydrogen atoms of the 7-methyl group (10). Other peaks in the spectrum of the product-substrate mixture also unambiguously supported the presence of 7-methyl-GpppG, such as the signals around 5.8 ppm derived from the hydrogen atoms at the 1′ positions of these dinucleotides (compare Fig. 6A to C) (10). Integration of the signals indicated that the partially purified reaction product is an approximately 1:1 mixture of GpppG and 7-methyl-GpppG. Thus, as close inspection reveals, the spectrum in Fig. 6C corresponds to the sum of spectra in Fig. 6A and B.
FIG. 6.
Identification of the 1a methyltransferase reaction product. Expansions (6.0 to 3.8 ppm) of 500-MHz 1H NMR spectra of 0.5 mM solutions of GpppG (A) and 7-methyl-GpppG (B) in D2O are shown. For panel C, a 1a-containing membrane fraction was incubated with GpppG and AdoMet, and the partially purified mixture of unreacted GpppG and the methylated reaction product was isolated as described in Materials and Methods. Similar expansion of the NMR spectrum of a D2O solution of this 1a reaction product-substrate mixture is shown. Peaks due to H-1′ and 7-CH3 (10) and the large peak due to residual 1H-containing water (HDO) are labeled.
Effects of 1a point mutations on guanylate methylation and covalent binding.
To further test the direct role of 1a in GTP methylation and to gain insight into the amino acids in 1a involved in GTP methylation and covalent m7GMP binding, we next constructed targeted point mutations in 1a. These were designed to individually change some of the most conserved amino acids in 1a to alanine (Fig. 1). We chose to alter three residues in the capping domain of BMV 1a, H80, D106, and R136. The corresponding alphavirus nsP1 residues have been previously targeted in point mutation studies and shown to be important for both activities (4, 47). It was also proposed that the conserved histidine could be the covalent guanylate binding residue, and it was shown that the conserved aspartate was involved in binding AdoMet (4). Thus, it was of interest to test the effects of mutating BMV 1a at these positions and to compare the results with those for alphavirus nsP1 data (see Discussion). An additional alanine substitution mutation was made in the helicase-like domain of 1a at K691, which corresponds to a universally conserved lysine residue essential for nucleotide binding by various helicases and nucleoside triphosphatases (16).
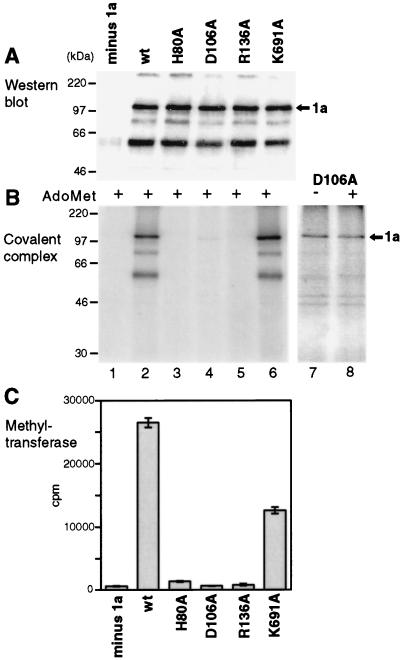
Western blotting showed that each of the mutant 1a proteins was expressed in yeast at levels similar to those for the wild-type (wt) 1a protein and that each was also found associated with the membrane fraction at levels similar to those for wt 1a (Fig. 7A). These wt and mutant 1a preparations were then assayed in parallel for covalent m7GMP binding (Fig. 7B) and guanine-7-methyltransferase activity (Fig. 7C). In the methyltransferase reaction, 1a mutants D106A and R136A were indistinguishable from the minus-1a control, whereas 1a mutant H80A reproducibly displayed approximately 3% of wt activity (averaging 1,400 cpm, compared to 26,500 cpm for wt 1a and 600 cpm (background) for the minus-1a control). Interestingly, mutation K691A in the helicase domain of 1a reproducibly reduced the methyltransferase activity by approximately 50%, implying some form of interaction between these two 1a domains, as previously suggested from genetic data (25). In the covalent m7GMP binding reaction (Fig. 7B), 1a mutants H80A and R136A were inactive and mutant K691A was fully active. 1a mutant D106A displayed 1% of covalent binding activity, as evidenced by a faint but specific band at the 1a position (Fig. 7B, lane 4), despite the fact that it had no detectable methyltransferase activity. To investigate this further, D106A was incubated with [α-32P]GTP in the presence and absence of AdoMet, and long exposures were taken (Fig. 7B, lanes 7 and 8). The low level of binding activity exhibited by this mutant appeared AdoMet-independent, in contrast to the wt 1a protein, which had no detectable activity in the absence of added AdoMet even when exposed to similar levels.
FIG. 7.
Effects of point mutations on enzymatic activities of 1a. Membrane fractions from yeast lacking 1a (lane 1), expressing wt 1a (lane 2), or expressing the indicated point mutant derivatives of 1a (lanes 3 to 6) were analyzed by Western blotting with 1a antiserum (A). The same fractions were assayed for covalent m7GMP complex formation in the presence of AdoMet (B) and for guanine-7-methyltransferase activity (C). The histogram bars of panel C show averages and standard deviations from assays of two complete sets of such fractions, each in duplicate. A longer exposure of the covalent guanylate binding activity of mutant D106A is shown in panel B, lanes 7 and 8, assayed in the absence or presence of AdoMet, as indicated. Arrows mark the position of 1a, and positions of molecular weight markers are shown at the left.
Covalent binding of nucleotides by 1a derived from BMV-infected plant cells.
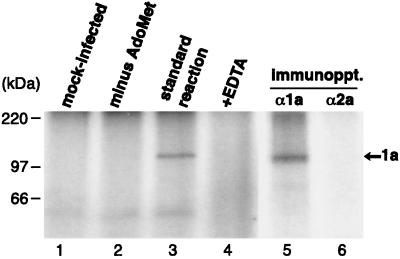
BMV 1a was also produced by infecting barley protoplasts with BMV virions, after which protoplast membranes were isolated and incubated with [α-32P]GTP. As in yeast cells, formation of a covalent guanylate complex with 1a required AdoMet (Fig. 8, lanes 2 and 3). No reaction was observed with extracts from mock-infected protoplasts (lane 1) or in the absence of divalent cations (lane 4). The covalent, labeled complex was immunoprecipitated specifically with 1a antibodies (lanes 5 and 6). Thus, BMV 1a produced in its natural host cells in the context of full viral infection is similarly active as 1a produced on its own in yeast cells, further confirming the AdoMet requirement and covalent complex formation only with methylated guanylate.
FIG. 8.
Covalent guanylate complex formation by 1a derived from BMV-infected plant cells. Portions of BMV-infected barley protoplast membrane fractions were incubated under standard conditions (see Materials and Methods) with [α-32P]GTP and unlabeled AdoMet (lane 3). In control experiments, unlabeled AdoMet was omitted (lane 2), 5 mM EDTA was added to the standard reaction (lane 4), or membranes from mock-infected barley protoplasts were incubated under standard conditions (lane 1). Four volumes of the standard reaction mixture (lane 3) was also subjected to immunoprecipitation with anti-1a or anti-2a antiserum (lanes 5 and 6). All samples were analyzed by SDS-PAGE and autoradiography.
DISCUSSION
Although the members of the large alphavirus-like superfamily of positive-strand RNA viruses are quite variable in genome organization, virion structure, and host specificity, they encode similar replicase proteins. This finding suggests that these viruses should use common mechanisms of RNA replication. However, the extent of such conservation is unclear, since knowledge of the replication process is still fragmentary and demonstrated instances of conserved functions are rare. The sequence relationships of RNA replication proteins within the superfamily are in some cases very distant; of the three conserved domains delineated in the introduction, the putative capping enzyme domain is the least conserved. However, divergence at the sequence level might be due to the rapid evolution of RNA viruses, so that only the essential, positively selected structural and functional properties of these proteins would remain well conserved despite sequence variation. Here we have provided evidence for the conservation of reactions involved in capping of the viral RNAs.
We have demonstrated two enzymatic activities of BMV replicase protein 1a, expressed in the yeast S. cerevisiae. 1a was able to methylate GTP, dGTP, GpppA, GpppG, and m7GpppG but not m7GTP or m7GpppA (Fig. 5A). This specificity suggested that methylation takes places at the 7 position of the guanosine ring, since compounds already fully methylated at this position could accept no further methyl groups. To prove this, the structure of a reaction product yielded by 1a-catalyzed methylation of GpppG was directly verified as 7-methyl-GpppG by NMR spectroscopy (Fig. 6). Evidence that 1a was responsible for methylation activity is twofold. First, membrane fractions containing 1a showed robust methyltransferase activity toward GTP, which was absent from parallel fractions from yeast lacking 1a (Fig. 5A). Second, point mutations in 1a abolished the activity (Fig. 7C). BMV 1a also covalently bound a nucleotide derived from GTP in an immunoprecipitable complex. Complex formation required the presence of the methyl donor AdoMet (Fig. 3A and 8), and labeling experiments showed that an AdoMet-derived methyl group, present at the 7 position of the guanosine ring (Fig. 3B and C), was part of the complex. 32P from [α-32P]GTP but not from [γ-32P]- or [β,γ-32P]GTP was transferred to the complex, and pyrophosphate was inhibitory to complex formation (Fig. 4). Collectively, the evidence indicates that the nucleotide bound was m7GMP.
Previously, within the superfamily, similar activities have been described for two alphavirus-encoded proteins, Semliki Forest virus (SFV) and SIN nsP1 (3, 26, 38). The SFV and SIN proteins are relatively closely related with each other, with 64% amino acid sequence identity (calculated for the entire protein), whereas BMV 1a is by comparison a distant homolog, sharing only 17% sequence identity with SFV nsP1 (conserved domain only [Fig. 1]). Therefore it may not be surprising that some differences were found in the substrate specificities of these enzymes. In the methyltransferase reaction, 1a was highly active toward cap analogs, or 5′-5′ triphosphate-linked dinucleotides (Fig. 5A), whereas these are poor substrates for SFV nsP1 (26). By contrast, in the covalent binding reaction, BMV 1a was highly specific for GTP, leading to binding of m7GMP, whereas SFV nsP1 but not BMV 1a can additionally bind m7dGMP, albeit at reduced efficiency (Fig. 4) (3).
Time course studies of the methyltransferase reaction (Fig. 5B) indicated that covalent complex formation by BMV 1a inhibits further methylation reactions. Like BMV 1a (Fig. 5B), SFV nsP1 appears to exhibit a decreased rate of methylation over time when GTP is used as a substrate in the presence of divalent cations (26). For BMV 1a, the initial rate of methylation of GTP is at least as high as that of dGTP and GpppA; GpppG may be an even better methyl acceptor due to its symmetrical nature (Fig. 5B).
The essential features of the reactions catalyzed by BMV 1a and alphavirus nsP1 are conserved. In both cases, the viral enzyme is able to methylate guanosine-containing mononucleotides, whereas the eukaryotic cellular mRNA guanine-7-methyltransferase involved in RNA capping is not active toward these substrates (13). Second, after or concomitantly with methylation, 1a and nsP1 covalently bind the methylated nucleotide m7GMP. This reaction strictly requires the presence of the methyl donor AdoMet, indicating that unmethylated nucleotides cannot be covalently bound. The conservation of AdoMet dependence between alphavirus nsP1s and bromovirus 1a is particularly notable in the light of their considerable sequence divergence, implying that AdoMet dependence is a central feature of the reaction path catalyzed by these enzymes. In contrast, cellular guanylyltransferases form a covalent complex only with unmethylated GMP (21, 30, 39, 46). This difference suggests that while methylation has been shown to be the last step of cellular cap formation, it precedes or coincides with covalent binding in the capping reactions catalyzed by alphavirus-like superfamily replicase proteins. It has been suggested that another superfamily member, tobacco mosaic virus replicase protein p126, exhibits covalent guanylate binding in the absence of AdoMet (12). However, it is possible that the concentrated cell lysates used by these authors provided the AdoMet. We also observed that BMV 1a, if assayed in total yeast cell extracts, was not dependent on externally added AdoMet; this dependence became apparent only when purified, 1a-containing membrane fractions were used.
Point mutations in conserved residues had similar effects on 1a and nsP1. Mutation of R136 or D106 (or the corresponding alphavirus residues) to alanine reduced both activities to very low or undetectable levels (Fig. 7) (4, 47). Cross-linking experiments have previously implicated SFV nsP1 D64 (equivalent to 1a D106 [Fig. 1B]) in binding the methyl donor AdoMet (4), whereas the specific function of the conserved arginine (R136 in 1a) is unclear. Interestingly, 1a mutant D106A exhibited low-level, AdoMet-independent, covalent binding of unmethylated GMP (Fig. 7B). This phenotype was not found among the SFV nsP1 mutants studied (4), but it is consistent with the proposed AdoMet-binding role of this residue. Mutation of the conserved histidine (H80 in 1a; Fig. 1B) reduced or abolished the methyltransferase activity for SIN nsP1 (47) and BMV 1a (Fig. 7C), whereas it increased this activity for SFV nsP1 (4). In all cases the histidine mutation destroyed the covalent binding activity completely, consistent with the suggestion that this histidine may be the covalent binding site for the nucleotide (4).
In conclusion, the enzymatic properties displayed by BMV 1a, SFV nsP1, and SIN nsP1 are similar in their essential features. This parallels other fundamental similarities in their RNA replication factors and RNA replication mechanisms, such as homology of the helicase- and polymerase-like domains; membrane association of their replication complexes (7, 14, 33); differential regulation of negative-strand, positive-strand, and subgenomic RNA synthesis; and early shutoff of negative-strand synthesis (25, 37). These and other findings imply that additional mechanistic similarities between RNA replication by the distantly related bromovirus and alphavirus groups are likely to emerge as their studies advance.
ACKNOWLEDGMENTS
We thank René Quadt for generously sharing preliminary observations on covalent 1a-guanylate complex formation made during his stay in this laboratory, and we thank Anja Lampio for help in acquiring reagents.
This research was supported by the National Institutes of Health through grant GM35072. This study made use of the National Magnetic Resonance Facility at Madison, Wis., which is supported by NIH grant RR02301. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P, Strauss E G, Rice C M, Strauss J H, Haseloff J, Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985;53:536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 7.Barton D J, Sawicki S G, Sawicki D L. Solubilization and immunoprecipitation of alphavirus replication complexes. J Virol. 1991;65:1496–1506. doi: 10.1128/jvi.65.3.1496-1506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Darzynkiewicz E, Stepinski J, Tahara S M, Stolarski R, Ekiel I, Haber D, Neuvonen K, Lehikoinen P, Labadi I, Lönnberg H. Synthesis, conformation and hydrolytic stability of p1,p3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogs. Nucleosides Nucleotides. 1990;9:599–618. [Google Scholar]
- 11.Dasgupta R, Harada F, Kaesberg P. Blocked 5′ termini in brome mosaic virus RNA. J Virol. 1976;18:260–267. doi: 10.1128/jvi.18.1.260-267.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunigan D D, Zaitlin M. Capping of tobacco mosaic virus RNA: analysis of a viral-coded guanylyltransferase-like activity. J Biol Chem. 1990;265:7779–7786. [PubMed] [Google Scholar]
- 13.Ensinger M J, Moss B. Modification of the 5′ terminus of mRNA by an RNA (guanine-7)-methyltransferase from HeLa cells. J Biol Chem. 1976;251:5283–5291. [PubMed] [Google Scholar]
- 14.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987;4:197–202. [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Cell Biol. 1993;3:419–429. [Google Scholar]
- 17.Haseloff J, Goelet P, Zimmern D, Ahlquist P, Dasgupta R, Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci USA. 1984;81:4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendler S, Fürer E, Srinivasan P R. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerases. Biochemistry. 1970;9:4141–4153. doi: 10.1021/bi00823a017. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa M, Díez J, Restrepo-Hartwig M, Ahlquist P. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh N, Mizumoto K, Kaziro Y. Messenger RNA guanlyltransferase from Saccharomyces cerevisiae. II. Catalytic properties. J Biol Chem. 1984;259:13930–13936. [PubMed] [Google Scholar]
- 22.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 24.Kroner P, Richards D, Traynor P, Ahlquist P. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroner P A, Young B M, Ahlquist P. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J Virol. 1990;64:6110–6120. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laakkonen P, Hyvönen M, Peränen J, Kääriäinen L. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J Virol. 1994;68:7418–7425. doi: 10.1128/jvi.68.11.7418-7425.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi S, Durbin R, Huang H V, Rice C M, Stollar V. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology. 1989;170:385–391. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 29.Mise K, Ahlquist P. Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 30.Mizumoto K, Kaziro Y, Lipmann F. Reaction mechanism of mRNA guanylyltransferase from rat liver: isolation and characterization of a guanylyl-enzyme intermediate. Proc Natl Acad Sci USA. 1982;79:1693–1697. doi: 10.1073/pnas.79.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Reilly E K, Tang N, Ahlquist P, Kao C C. Biochemical and genetic analyses of the interaction between the helicase-like and polymerase-like proteins of the brome mosaic virus. Virology. 1995;214:59–71. doi: 10.1006/viro.1995.9954. [DOI] [PubMed] [Google Scholar]
- 32.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 36.Russell P J, Hambidge S J, Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4949–4953. doi: 10.1093/nar/19.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicki D L, Sawicki S G, Keränen S, Kääriäinen L. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J Virol. 1981;39:348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheidel L M, Durbin R K, Stollar V. SVLM21, a Sindbis virus mutant resistant to methionine deprivation, encodes an altered methyltransferase. Virology. 1989;173:408–414. doi: 10.1016/0042-6822(89)90553-9. [DOI] [PubMed] [Google Scholar]
- 39.Shuman S. RNA capping by HeLa cell RNA guanylyltransferase. Characterization of a covalent protein-guanylate intermediate. J Biol Chem. 1982;257:7237–7245. [PubMed] [Google Scholar]
- 40.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 41.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc Natl Acad Sci USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan M, Ahlquist P. Cis-acting signals in bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin Virol. 1997;8:221–230. [Google Scholar]
- 44.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traynor P, Young B M, Ahlquist P. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkatesan S, Moss B. Eukaryotic mRNA capping enzyme-guanylate covalent intermediate. Proc Natl Acad Sci USA. 1982;79:340–344. doi: 10.1073/pnas.79.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H L, O’Rear J, Stollar V. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology. 1996;217:527–531. doi: 10.1006/viro.1996.0147. [DOI] [PubMed] [Google Scholar]