Abstract
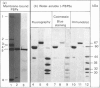
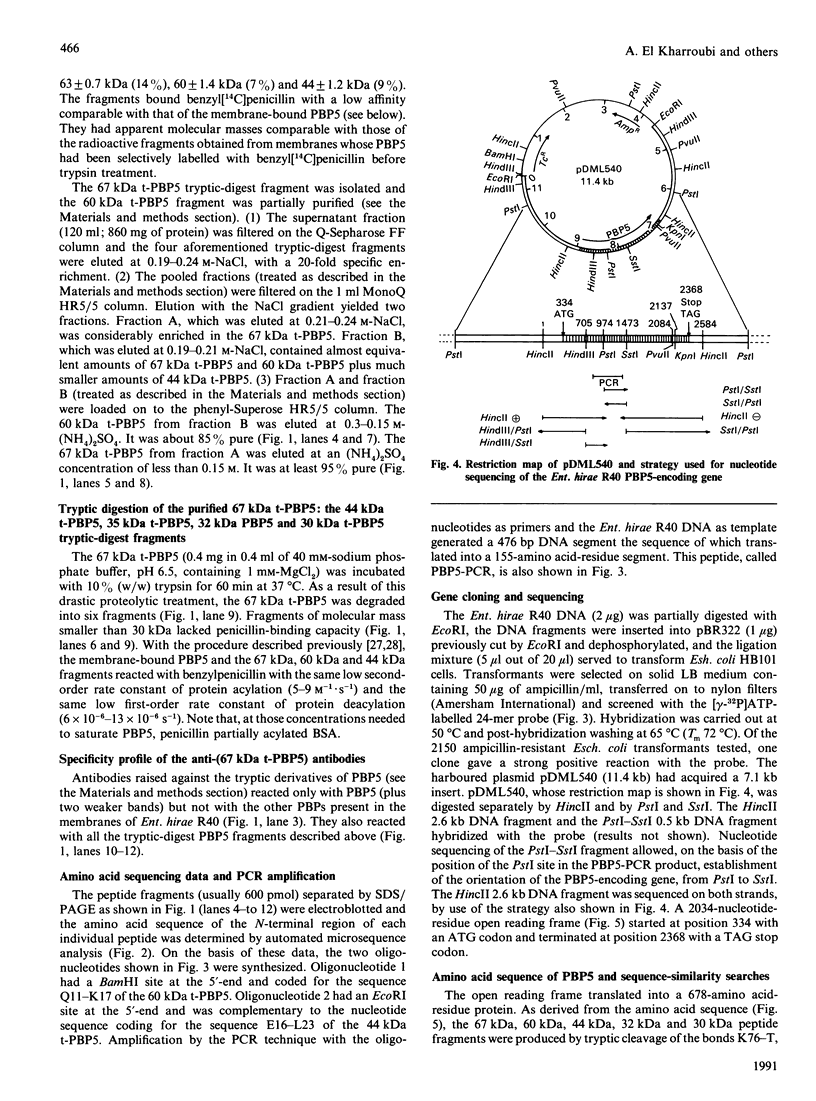
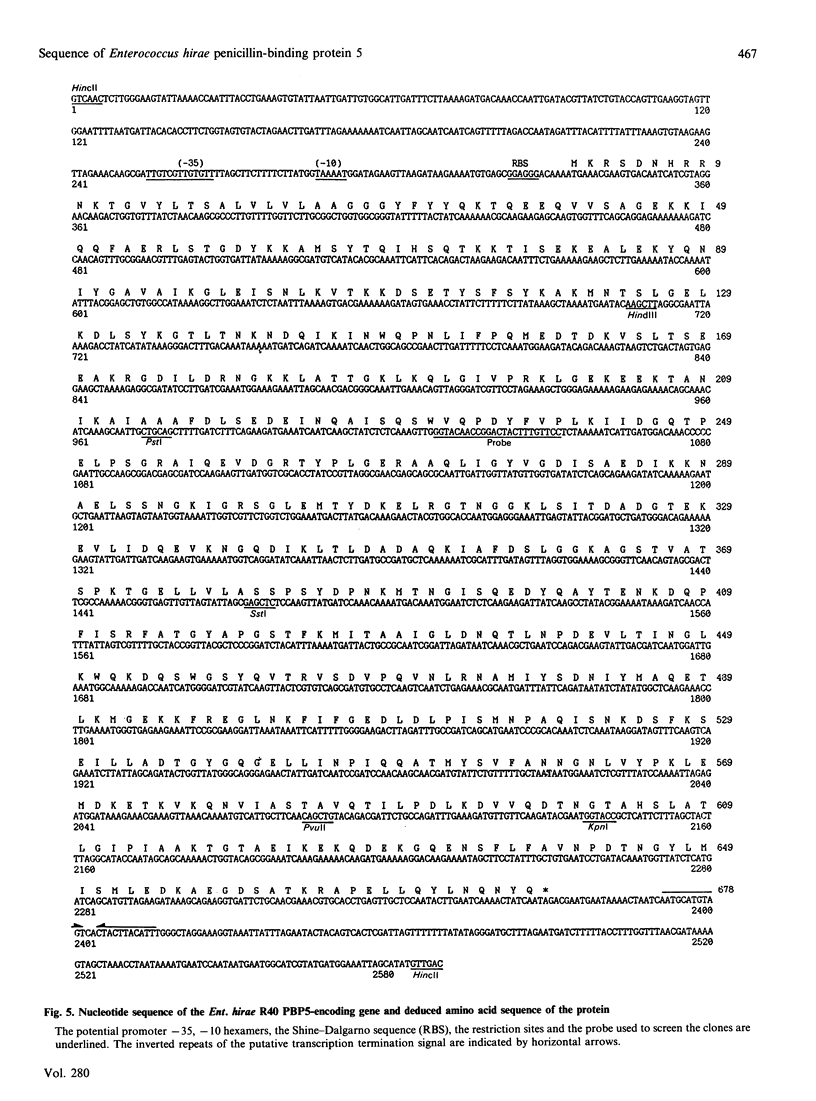
The penicillin-resistant Enterococcus hirae R40 has a typical profile of membrane-bound penicillin-binding proteins (PBPs) except that the 71 kDa PBP5 of low penicillin affinity represents about 50% of all the PBPs present. Water-soluble tryptic-digest peptides were selectively produced from PBP5, their N-terminal regions were sequenced and synthetic oligonucleotides were used as primers to generate a 476 bp DNA fragment by polymerase chain reaction. On the basis of these data, the PBP5-encoding gene was cloned in Escherichia coli by using pBR322 as vector. The gene, included in a 7.1 kb insert, had the information for a 678-amino acid-residue protein. PBP5 shows similarity, in the primary structure, with the high-molecular-mass PBPs of class B. In particular, amino acid alignment of the enterococcal PBP5 and the methicillin-resistant staphylococcal PBP2' generates scores that are 30, for the N-terminal domains, and 53, for the C-terminal domains, standard deviations above that expected for a run of 20 randomized pairs of proteins having the same amino acid compositions as the two proteins under consideration.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asoh S., Matsuzawa H., Ishino F., Strominger J. L., Matsuhashi M., Ohta T. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur J Biochem. 1986 Oct 15;160(2):231–238. doi: 10.1111/j.1432-1033.1986.tb09961.x. [DOI] [PubMed] [Google Scholar]
- Begg K. J., Spratt B. G., Donachie W. D. Interaction between membrane proteins PBP3 and rodA is required for normal cell shape and division in Escherichia coli. J Bacteriol. 1986 Sep;167(3):1004–1008. doi: 10.1128/jb.167.3.1004-1008.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyette J., Ghuysen J. M., Fontana R. Solubilization and isolation of the membrane-bound DD-carboxypeptidase of Streptococcus faecalis ATCC9790. Properties of the purified enzyme. Eur J Biochem. 1978 Jul 17;88(1):297–305. doi: 10.1111/j.1432-1033.1978.tb12450.x. [DOI] [PubMed] [Google Scholar]
- Coyette J., Ghuysen J. M., Fontana R. The penicillin-binding proteins in Streptococcus faecalis ATCC 9790. Eur J Biochem. 1980 Sep;110(2):445–456. doi: 10.1111/j.1432-1033.1980.tb04886.x. [DOI] [PubMed] [Google Scholar]
- Coyette J., Perkins H. R., Polacheck I., Shockman G. D., Ghuysen J. M. Membrane-bound DD-carboxypeptidase and LD-transpeptidase of Streptococcus faecalis ATCC 9790. Eur J Biochem. 1974 May 15;44(2):459–468. doi: 10.1111/j.1432-1033.1974.tb03504.x. [DOI] [PubMed] [Google Scholar]
- Dowson C. G., Hutchison A., Spratt B. G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989 Jan;3(1):95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- Dowson C. G., Hutchison A., Spratt B. G. Nucleotide sequence of the penicillin-binding protein 2B gene of Streptococcus pneumoniae strain R6. Nucleic Acids Res. 1989 Sep 25;17(18):7518–7518. doi: 10.1093/nar/17.18.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson C. G., Hutchison A., Woodford N., Johnson A. P., George R. C., Spratt B. G. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson C. G., Jephcott A. E., Gough K. R., Spratt B. G. Penicillin-binding protein 2 genes of non-beta-lactamase-producing, penicillin-resistant strains of Neisseria gonorrhoeae. Mol Microbiol. 1989 Jan;3(1):35–41. doi: 10.1111/j.1365-2958.1989.tb00101.x. [DOI] [PubMed] [Google Scholar]
- Fontana R., Cerini R., Longoni P., Grossato A., Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983 Sep;155(3):1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Grossato A., Rossi L., Cheng Y. R., Satta G. Transition from resistance to hypersusceptibility to beta-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob Agents Chemother. 1985 Nov;28(5):678–683. doi: 10.1128/aac.28.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J. M., Frère J. M., Leyh-Bouille M., Nguyen-Distèche M., Coyette J. Active-site-serine D-alanyl-D-alanine-cleaving-peptidase-catalysed acyl-transfer reactions. Procedures for studying the penicillin-binding proteins of bacterial plasma membranes. Biochem J. 1986 Apr 1;235(1):159–165. doi: 10.1042/bj2350159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J. M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge P. J., Spratt B. G. Resistance to beta-lactam antibiotics by re-modelling the active site of an E. coli penicillin-binding protein. Nature. 1985 Dec 5;318(6045):478–480. doi: 10.1038/318478a0. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Sato T., Wachi M., Jung H. K., Ishino F., Kobayashi Y., Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989 Nov;171(11):6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F., Park W., Tomioka S., Tamaki S., Takase I., Kunugita K., Matsuzawa H., Asoh S., Ohta T., Spratt B. G. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and rodA protein. J Biol Chem. 1986 May 25;261(15):7024–7031. [PubMed] [Google Scholar]
- Joris B., Dive G., Henriques A., Piggot P. J., Ghuysen J. M. The life-cycle proteins RodA of Escherichia coli and SpoVE of Bacillus subtilis have very similar primary structures. Mol Microbiol. 1990 Mar;4(3):513–517. doi: 10.1111/j.1365-2958.1990.tb00618.x. [DOI] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Kobayashi T., Lampen J. O., Ghuysen J. M. Expression in Escherichia coli of the carboxy terminal domain of the BLAR sensory-transducer protein of Bacillus licheniformis as a water-soluble Mr 26,000 penicillin-binding protein. FEMS Microbiol Lett. 1990 Jun 15;58(1):107–113. doi: 10.1016/0378-1097(90)90111-3. [DOI] [PubMed] [Google Scholar]
- Laible G., Hakenbeck R., Sicard M. A., Joris B., Ghuysen J. M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989 Oct;3(10):1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- Leyh-Bouille M., Nguyen-Distèche M., Pirlot S., Veithen A., Bourguignon C., Ghuysen J. M. Streptomyces K15 DD-peptidase-catalysed reactions with suicide beta-lactam carbonyl donors. Biochem J. 1986 Apr 1;235(1):177–182. doi: 10.1042/bj2350177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Morrison D. A., Jaurin B. Streptococcus pneumoniae possesses canonical Escherichia coli (sigma 70) promoters. Mol Microbiol. 1990 Jul;4(7):1143–1152. doi: 10.1111/j.1365-2958.1990.tb00689.x. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Piras G., el Kharroubi A., van Beeumen J., Coeme E., Coyette J., Ghuysen J. M. Characterization of an Enterococcus hirae penicillin-binding protein 3 with low penicillin affinity. J Bacteriol. 1990 Dec;172(12):6856–6862. doi: 10.1128/jb.172.12.6856-6862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E., Brown D. F. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett. 1985 Nov 11;192(1):28–32. doi: 10.1016/0014-5793(85)80036-3. [DOI] [PubMed] [Google Scholar]
- Reynolds P. E. Resistance of the antibiotic target site. Br Med Bull. 1984 Jan;40(1):3–10. doi: 10.1093/oxfordjournals.bmb.a071944. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. D., Wachi M., Doi M., Ishino F., Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987 Aug 31;221(1):167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Cromie K. D. Penicillin-binding proteins of gram-negative bacteria. Rev Infect Dis. 1988 Jul-Aug;10(4):699–711. doi: 10.1093/clinids/10.4.699. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature. 1988 Mar 10;332(6160):173–176. doi: 10.1038/332173a0. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Zhang Q. Y., Jones D. M., Hutchison A., Brannigan J. A., Dowson C. G. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8988–8992. doi: 10.1073/pnas.86.22.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner P. E., Huls P. G., Pas E., Woldringh C. L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988 Apr;170(4):1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Y., Jones D. M., Sáez Nieto J. A., Pérez Trallero E., Spratt B. G. Genetic diversity of penicillin-binding protein 2 genes of penicillin-resistant strains of Neisseria meningitidis revealed by fingerprinting of amplified DNA. Antimicrob Agents Chemother. 1990 Aug;34(8):1523–1528. doi: 10.1128/aac.34.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. F., Curran I. H., Joris B., Ghuysen J. M., Lampen J. O. Identification of BlaR, the signal transducer for beta-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 beta-lactamase (class D) of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):1137–1141. doi: 10.1128/jb.172.2.1137-1141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Kharroubi A., Piras G., Jacques P., Szabo I., Van Beeumen J., Coyette J., Ghuysen J. M. Active-site and membrane topology of the DD-peptidase/penicillin-binding protein no. 6 of Enterococcus hirae (Streptococcus faecium) A.T.C.C. 9790. Biochem J. 1989 Sep 1;262(2):457–462. doi: 10.1042/bj2620457. [DOI] [PMC free article] [PubMed] [Google Scholar]