Abstract
Sheep pulmonary adenomatosis (SPA), also known as jaagsiekte or ovine pulmonary carcinoma, is a contagious lung cancer of sheep, originating from type II pneumocytes and Clara cells. Previous studies have implicated a type D retrovirus (jaagsiekte sheep retrovirus [JSRV]) as the causative agent of SPA. We recently isolated a proviral clone of JSRV from an animal with a spontaneous case of SPA (JSRV21) and showed that it harbors an infectious and oncogenic virus. This demonstrated that JSRV is necessary and sufficient to induce SPA. A major impediment in research on JSRV has been the lack of an in vitro tissue culture system for the virus. The experiments reported here show the first successful in vitro infection with this virus, using the JSRV21 clone. JSRV21 virus was obtained by transiently transfecting human 293T cells with a plasmid containing the JSRV21 provirus driven by the human cytomegalovirus immediate-early promoter. Virus produced in this manner exhibited reverse transcriptase (RT) activity that banded at 1.15 g/ml in sucrose density gradients. Infection of concentrated JSRV21 into ovine choroid plexus (CP), testes (OAT-T3), turbinate (FLT), and intestinal carcinoma (ST6) cell lines resulted in establishment of infection as measured by PCR amplification. Evidence that this reflected genuine infection included the fact that heat inactivation of the virus eliminated it, the levels of viral DNA increased with passage of the infected cells, and the infected cells released active RT as measured by the sensitive product enhancement RT assay. The RT activity released from the infected cells banded at 1.15 g/ml, and JSRV21 provirus was transmitted from infected cells to uninfected ones by cocultivation. However, the amount of virus released from infected cells was low. These results suggest that the JSRV receptor is present on many ovine cell types and that the observed restriction of JSRV expression in vivo to tumor cells might be controlled by factors other than the viral receptor. Finally we tagged the U3 of pJSRV21 with the bacterial supF gene, an amber suppressor tRNA gene. The resulting clone, termed pJSRVsupF, is infectious in vitro. It may be a useful tool for future studies on viral DNA integration, since the normal sheep genome contains 15 to 20 copies of highly JSRV-related endogenous sequences that cross-react with many JSRV hybridization probes.
Cancers of the lungs and bronchus are the main cause of mortality among cancer patients in the United States (13, 22), and animal models to study the mechanisms of pulmonary carcinogenesis are greatly needed. Retroviruses are valuable tools to dissect the multistep events leading to cancer (28); however, the majority of oncogenic retroviruses are associated with tumors of the hematopoietic system, while the bulk of naturally occurring tumors originate from epithelial tissues (13). A type D retrovirus, jaagsiekte sheep retrovirus (JSRV), is unique among retroviruses because it is able to induce a naturally occurring contagious lung cancer in sheep (21) known as sheep pulmonary adenomatosis (SPA), jaagsiekte, or ovine pulmonary carcinoma (1, 6, 17, 19). SPA strongly resembles human bronchioloalveolar carcinoma. Both neoplasms have the same clinical, macroscopic, histopathological, and ultrastructural features (11, 23). Thus, SPA is a valuable experimental model for bronchioloalveolar carcinoma that could offer novel insights into oncogenic mechanisms in epithelial cells.
Studies on JSRV have been hampered by the lack of an infectious molecular JSRV clone and the lack of an in vitro infection system. The only available source for infectious JSRV has been lung fluid from SPA-affected sheep (25, 27). Recently we isolated an infectious and pathogenic JSRV molecular clone (JSRV21) (21). In vivo transfection and infections in newborn lambs proved conclusively that this virus is necessary and sufficient to induce neoplasia. In addition, we established a convenient method to prepare infectious JSRV in vitro. This involved transfection of human 293T cells with a plasmid DNA containing the JSRV21 provirus that been modified by replacement of the U3 sequences in the upstream long terminal repeat (LTR) with the human cytomegalovirus immediate-early promoter (pCMV2JS21) (21). Virus produced by this method was used to demonstrate the in vivo infectivity and oncogenicity of JSRV21.
In this study we used JSRV21 virus produced from transfected 293T cells to establish an infection system in several ovine cell lines.
MATERIALS AND METHODS
In vitro production of JSRV21 virions.
JSRV21 particles were obtained by transfection of the plasmid pCMV2JS21 into 293T cells as already described (21). Briefly, cells were transfected with pCMV2JS21 DNA (45 μg/10-cm dish) by using the CalPhos mammalian transfection kit (Clonetech). Medium was changed at 12 to 16 h and harvested at 24, 48, and 72 h after the first medium change. The medium was filtered through a 0.45-μm-pore-size filter, and virus was harvested by ultracentrifugation at 100,000 × g through a double layer of glycerol (25 and 50%, vol/vol) for 1 h at 4°C. The resulting viral pellet was resuspended in TNE buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA) at a 300-fold concentration relative to the original medium and stored in aliquots at −140°C.
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE and Western blotting analysis with a rabbit anti-serum to the JSRV major capsid protein (18) was used essentially as described previously (18, 26) except that an enhanced chemiluminescence detection system (Supersignal; Pierce) was used as recommended by the manufacturers. Concentrated lung fluid collected from an animal with a natural case of SPA was prepared as already described (18, 26).
Analysis of JSRV21 buoyant density.
Approximately 700 μl of JSRV21 concentrated particles was analyzed by isopycnic centrifugation on a linear 25 to 60% (wt/wt) continuous sucrose gradient in an SW41 rotor (Beckman) at 25,000 rpm for 16 h at 4°C. Fractions of approximately 450 μl were collected, and their density was determined by refractometry. Consecutive fractions were pooled two at a time and diluted with 10 ml of TNE buffer, and virus was recovered by centrifugation in an SW41 rotor at 35,000 rpm for 1 h at 4°C. Viral pellets were resuspended in 20 μl of TNE buffer and used in an conventional exogenous reverse transcriptase (RT) assay with poly(rA)-oligo(dT) essentially as previously described (31).
Biosynthetic labeling.
Dishes (diameter, 10 cm) of 293T cells were transfected with pCMV2JS21 as described above. At 24 h posttransfection, growth medium was replaced with 5 ml of labeling medium, consisting of 1 part of complete Dulbecco’s modified Eagle’s medium (DMEM) and 9 parts of methionine- and cysteine-deficient DMEM (Gibco-BRL) supplemented with 10% dialyzed fetal calf serum (Gibco-BRL) and 109 μCi of [35S]methionine per ml. After 24 h of labeling, virus was harvested as described above. Pellets harvested from five dishes of transfected 293T cells or from five dishes of mock-transfected cells were resuspended in 500 μl of TNE buffer and banded in a 20 to 55% continuous sucrose gradient. Gradient fractions corresponding to a buoyant density of 1.145 to 1.168 g/ml were pooled, and virus was recovered as above. Labeled viral pellets were resuspended in 35 μl of TNE buffer and resolved by SDS-PAGE (8.7 or 14% polyacrylamide gels) followed by autoradiography with an intensifying screen.
Infections of sheep cell lines with JSRV21.
The sheep choroid plexus (CP) cell line, the fetal lamb turbinates (FLT) cell line, and the ST-6 (16) cell line were obtained from the tissue culture service of the Moredun Research Institute (Edinburgh, United Kingdom) and were grown at 37°C and 5% CO2 in Eagle’s basal medium (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS). The OAT-T3 cell line (derived from sheep testis) was obtained from the American Type Culture Collection (CRL-6546-FL) and was grown in DMEM–10% FBS at 37°C and 5%CO2.
For infection, cells (1 × 105 to 5 × 105 cells) were plated in 5-cm tissue culture dishes and infected 4 to 16 h after plating. Infections were performed with 250 μl of concentrated JSRV21 diluted in 1 ml of either Eagle’s basal medium plus 10% FBS or DMEM plus 10% FBS, both supplemented with 8 μg of Polybrene per ml. A 2-ml volume of fresh medium with 8 μg of Polybrene per ml was added after 2 h, and the cells were incubated further for 16 h at 37°C. Medium was then replaced, and the cells were maintained and subsequently passaged in medium containing 2 μg of Polybrene per ml. As a control, cells were infected in parallel with the same amount of concentrated JSRV21 virus that had been heat inactivated either at 95°C for 5 min or at 65°C for 15 min.
As a control for possible plasmid DNA contamination in the concentrated JSRV21 stocks, 450 μg of pCMV2JS21 plasmid DNA was mixed with 150 ml of DMEM and then filtered and processed exactly as described for concentration of JSRV21 particles.
Cocultivation of JSRV21-infected CP cells with uninfected CP cells.
A total of 106 choroid plexus cells infected with JSRV21 (passage 10 postinfection) were γ-irradiated (100 Gy) and were mixed with 2 × 105 unirradiated choroid plexus target cells and passaged every 3 to 5 days. In parallel, 106 producer cells were X-irradiated (100 Gy) and cultured alone until 100% mortality occurred. Infection was assayed by PCR detection (see below) at 9 weeks postexposure.
PCR analysis.
The JSRV21 provirus in the infected cells was detected by the JSRV U3-PCR assay with primers PI (5′-TGGGAGCTCTTTGGCAAAAGCC-3′) and PIII (5′-CACCGGATTTTTACACAATCACCGG-3′) as already described (20). This assay is specific for exogenous JSRV.
PERT assay.
JSRV21 RT activity in supernatants from infected cells was detected by using the product enrichment RT (PERT) assay, in which the RNA of bacteriophage MS2 serves as template for RT-mediated cDNA synthesis, by using the method already described (24) with minor modifications. Briefly, 10-ml volumes of culture supernatants were clarified by centrifugation at 12,000 × g for 10 min and filtered through 0.22-μm-pore-size microfilters. Virus from the filtered supernatants was harvested by ultracentrifugation over a double cushion of glycerol (25 to 50%) as described above. The pellets were suspended in 30 μl of buffer A (50 mM KCl, 25 mM Tris-HCl [pH 7.5], 5 mM dithiothreitol, 0.25 mM EDTA, 0.025% Triton X-100, 50% glycerol). A 3-μl volume of resuspended sample was used in the reverse transcription of MS2 RNA and subsequent PCR amplification and Southern blot hybridization as already described (24).
The PERT assay was also used to investigate the buoyant density of the JSRV21 particles released by the CP cell line. A 100-ml volume of supernatant collected from CP cells at passage 21 postinfection was clarified and filtered, and virus was harvested by ultracentrifugation and banded in a continuous 20 to 55% sucrose gradient as above. Fractions of 500 μl were collected, and their density was determined. Material from each fraction was harvested as above, pellets were resuspended in 39 μl of buffer A, and 3 μl of each fraction was used in the PERT assay. A 10-μl volume of the final PCR products was blotted onto a nylon membrane (Hybond-N Plus; Amersham) by using a slot blot apparatus (Bio-Rad). The membrane was hybridized with a radioactively labeled MS2 DNA probe as described for the PERT procedure, and the hybridized radioactivity was quantified by phosphorimaging.
Tagging of pCMV2JS21.
Plasmid pCMV2JS21 was tagged with the supF gene, a bacterial suppressor tRNA gene. The supF gene was amplified from piAN7 (a gift of C. Holland and N. DiFronzo [15]) by PCR with primers containing BlnI adapters and then ligated into the unique BlnI site at −208 in the U3 region of pCMV2JS21. The resulting plasmid was called pJSRVsupF. The supF gene was chosen because it was successfully inserted into the U3 of the leukemogenic murine mink cell focus-inducing virus MCF 247 without changing the biological properties of the virus (3, 15). JSRVsupF viral particles were produced as described above in 293T cells. Choroid plexus cells were infected as described above, and infection was detected by JSRV U3-PCR.
RESULTS
Analysis of JSRV21 viral particles produced by transfection.
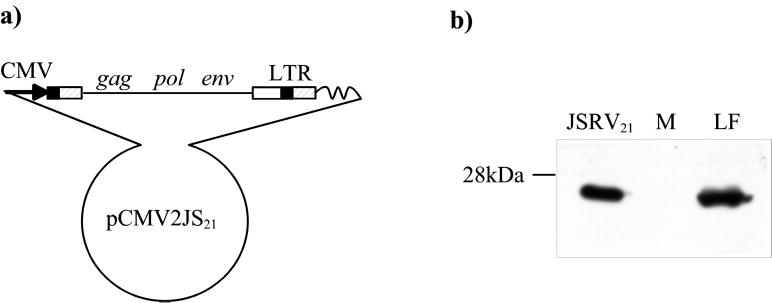
As described in Introduction, we previously showed that a molecular clone of JSRV, JSRV21, is infectious and pathogenic in sheep (21). Infectious virus was obtained by transfecting plasmid pCMV2JS21 into human 293T cells and harvesting culture supernatants. pCMV2JS21 contains an integrated JSRV21 provirus that has been modified by replacement of the upstream U3 region with the immediate-early promoter from human cytomegalovirus (Fig. 1a). The cytomegalovirus promoter is highly active in 293T cells, and it was inserted in a position such that the primary transcript would closely resemble native JSRV RNA. As a result, this procedure will yield replication-competent JSRV with infectivity equivalent to that of wild-type virus. The transfected 293T cells release substantial amounts of viral protein into the supernatant, as evident from Western blotting of concentrated supernatant for viral CA protein (Fig. 1b). Moreover, the CA protein in the supernatant was of the mature cleaved (26-kDa) size, which suggested that the bulk of viral protein is packaged into virus particles that can activate the viral protease necessary for polyprotein cleavage. Supernatants from the transfected 293T cells seemed to be appropriate material for in vitro infection experiments as well. Prior to these experiments, we further characterized the virus particles produced from the transfected 293T cells.
FIG. 1.
Organization of the pCMV2JS21 construct and in vitro synthesis of JSRV21 particles. (a) pCMV2JS21 is a plasmid containing a modified version of the integrated JSRV21 provirus in which the U3 region of the upstream LTR was replaced by the human cytomegalovirus immediate-early promoter. (b) SDS-PAGE and Western blotting of 300-fold-concentrated supernatant from 293T cells transiently transfected with pCMV2JS21. The filters were probed with a rabbit polyclonal antiserum toward the major capsid protein (CA) of JSRV. Lung fluid collected from an SPA-affected animal and concentrated in the same way as the 293T supernatant was used as a positive control (LF). Concentrated supernatant from mock-transfected 293T cells was used as a negative control (M). The 26-kDa JSRV-CA protein is indicated.
Supernatants from pCMV2JS21-transfected 293T cells were analyzed by isopycnic centrifugation in sucrose density gradients as already described (21). Supernatants from transfected cells contained RT activity that could be measured by an exogenous RT assay with poly(rA)-oligo(dT) as the template primer. RT assays across the sucrose gradient indicated a peak of RT activity with a buoyant density of approximately 1.15 g/ml, which was in accord with previous results in in vitro-synthesized pJSRV21 particles (21) although it was lower than the buoyant density of JSRV (1.16 to 1.18 g/ml) when the virus was isolated directly from the lung secretions of SPA-affected animals (7, 18, 26, 30). Supernatants from mock-transfected 293T cells showed no RT activity.
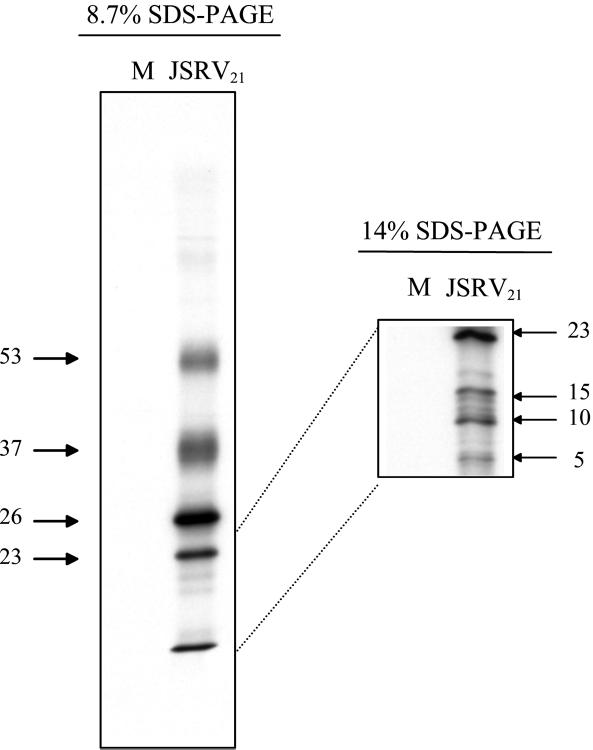
Proteins in JSRV21 virions were labeled by incubation of transfected 293T cells for 24 h with [35S]methionine. Labeled supernatant from transfected 293T cells was banded in an isopycnic sucrose gradient, and fractions corresponding to 1.14 to 1.16 g/ml were pooled and subjected to SDS-PAGE and autoradiography as shown in Fig. 2. Several prominent labeled proteins were detected, while supernatant from mock-transfected and labeled 293T cells yielded no detectable radioactivity in material banding at 1.14 to 1.16 g/ml. By analogy to the sizes of other type D and type B retroviral proteins, it seems possible that the 53- and 37-kDa proteins are envelope proteins (4, 14); their diffuse migration in SDS-PAGE would be consistent with glycosylation. Similarly, some of the lower-molecular-mass proteins (26, 23, 17, and 14 kDa) might correspond to mature Gag proteins or proteins encoded by other viral genes (2, 8, 9). Indeed, the 26-kDa protein had the same mobility as the 26-kDa CA protein present in lung fluid from SPA-affected sheep that is detectable by Western blots (Fig. 1b). More definitive characterization of the virion proteins is in progress.
FIG. 2.
Proteins in JSRV21 virions. Labeled JSRV21 was prepared from transfected 293T cells that had been labeled with [35S]methionine 24 h prior to supernatant harvest. Viral particles were purified by isopynic centrifugation, and the fractions corresponding to 1.14 to 1.16 g/ml were pooled and analyzed by SDS-PAGE on 8.7 and 15% polyacrylamide gels followed by autoradiography (JSRV21). 293T cells that were mock-transfected were labeled and processed in parallel (M). No radioactivity banded at 1.14 to 1.16 g/ml from tissue culture supernatant from the mock-transfected culture. JSRV21 virions contained major bands of 53, 37, 26, and 23 kDa visible in the 8.7% polyacrylamide gel (left). An additional band migrated at the bottom of the 8.7% polyacrylamide gel that resolved into three bands of 15, 10, and 5 kDa in the 14% polyacrylamide gel. The sizes of the radioactive proteins were calculated from the mobilities of protein size markers.
These results indicated that the bulk of viral protein released from pCMV2JS21-transfected 293T cells appeared to be in native virions, since the proteins appeared to be processed. Coupled with the fact that supernatants from transfected 293T cells are infectious and pathogenic in sheep, this material was used as a source of virus for establishing an in vitro infection system.
In vitro infection with JSRV21.
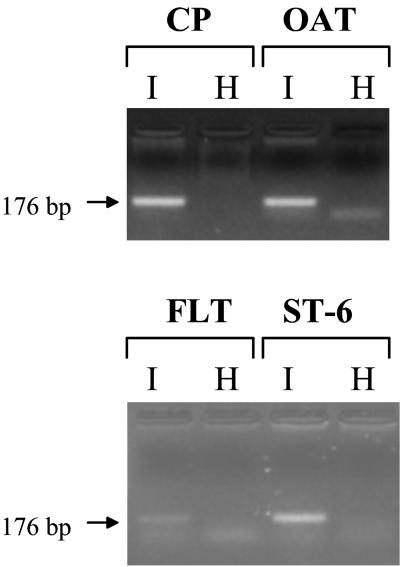
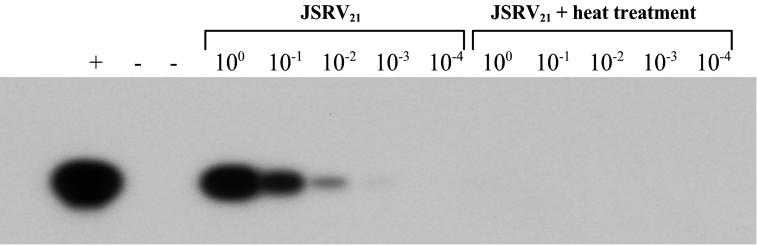
JSRV21 produced from transfected 293T cells (henceforth termed JSRV21 virus) was tested for infection of several ovine cell lines. These included the CP cell line, the OAT-T3 cell line, the FLT cell line and the ST-6 cell line (derived from an ovine intestinal carcinoma). The cells were incubated with virus in the presence of 8 μg of Polybrene per ml and serially passaged in the presence of 2 μg of Polybrene per ml. DNA was extracted from the cultures at different passages and tested for the presence of JSRV21 provirus by PCR. As shown in Fig. 3, JSRV21 DNA was detected in all four infected cell lines at passages ranging from 1 to 7. In contrast, JSRV21 DNA was not detected in the mock-infected controls or in cells infected with heat-inactivated JSRV21 (100°C for 5 min or 65°C for 15 min). Moreover, the PCR products were negative at passage 2 postinfection in FLT cells but became positive in the following passages (data not shown, but see below). These results suggested that JSRV21 was able to infect all four cell lines and to propagate in them.
FIG. 3.
In vitro infection of ovine cell lines. Four different ovine cell lines were infected with JSRV21 (I) or in parallel with heat-inactivated virus (H), and the cultures were serially passaged. PCR for exogenous JSRV with primers from the U3 region of the LTR was carried out on 500 ng of infected-cell DNA. The diagnostic 176-bp product is indicated by an arrow. PCR assays were carried out on CP cells at passage 7 after infection and on the other three cell lines at passage 5.
A major concern with the experiments in Fig. 3 was that PCR was used as the detection method. Given the sensitivity of this technique, it was important to rule out potential artifacts besides simple contamination during the PCR amplification. In particular, there was concern that pCMV2JS21 plasmid DNA used in the 293T cell transfections might have carried over into the JSRV21 stocks and led to the positive PCR signals. However, infection with heat-inactivated JSRV21 did not result in PCR-positive cells (Fig. 3), even when the virus was inactivated under conditions where contaminating plasmid DNA would not be denatured (65°C for 15 min). As additional test, CP cells were “infected” with material concentrated from growth medium into which pCMV2JS21 plasmid DNA was mixed before being processed for virus concentration as described in Materials and Methods. The amount of pCMV2JS21 DNA added corresponded to the total quantity of DNA used in the 293T transfection to generate 500 μl of concentrated JSRV21 virus—the amount of virus used in infection of each cell line. Both the “infected” cells and the cells infected with heat-treated material showed positive PCR signals only in the first one or two passages postinfection and then were consistently PCR negative until passage 7, when the experiment was terminated. Thus, even when large amounts of pCMV2JS21 DNA were initially present in the 293T supernatants, this did not lead to permanent propagation of the DNA signal under the infection conditions. This further supported the idea that in vitro JSRV21 infection had been achieved.
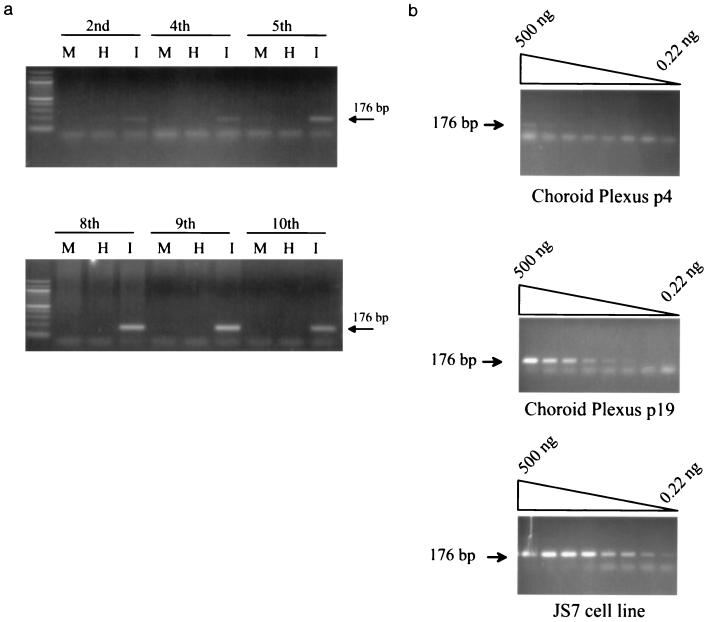
A more systematic characterization of JSRV21 infection in CP cells is shown in Fig. 4. As shown in Fig. 4a, JSRV sequences (the diagnostic 176-bp PCR product) were barely detectable at passages 2 and 4 but could be detected at passage 5 and in all subsequent passages (through passage 21). The intensity of the PCR signal also increased for the later passages, consistent with the spread of infection within the cultures. PCR on threefold dilutions of infected CP cells DNA at passages 4 and 19 postinfection indicated a ca. 34-fold (ca. 80-fold) increase in JSRV DNA content at passage 19 compared to passage 4. These results further indicated that JSRV21 was able to infect CP cells and propagate infection within them. In Fig. 4b, PCR for JSRV was also carried out on dilutions of DNA from the JS7 tumor cell line (12). JS7 cells are tumor cells derived from an animal with a natural case of SPA, and they contain one copy of JSRV provirus (2a). The level of the JSRV21 DNA signal in the passage 19-infected CP cells was approximately 10-fold lower than for the JS7 cells. Thus, if the infected CP cells also contained one copy of proviral DNA per cell, approximately 10% of them would be infected.
FIG. 4.
In vitro infection of CP cells. (a) PCR for JSRV DNA was carried out on 500 ng of DNA from infected CP cells at different passages. Analysis of cells infected with heat-treated JSRV21 (I) or mock-infected cells (M) of equivalent passage numbers was also carried out. The passages postinfection are indicated at the top of each gel. The intensity of the amplified 176-bp product intensified in the later passages (passages 8 through 10). (b) PCR for JSRV DNA on threefold dilutions of infected CP cell DNA at passage 4 postinfection are compared with the same cells at passage 19 postinfection. The specific amplimer of 176 bp is indicated by an arrow. The faster-migrating bands are primer-dimers. The passage 19 cells had approximately 34 times as much JSRV21 DNA as the passage 4 cells did. PCR on serial dilutions of JS7 cell line (9) DNA is also shown. JS7 cells contain one copy of JSRV provirus per cell (2a). The results indicate that passage 19-infected CP cells have approximately 32 less DNA per cell than do JS7 cells.
Virus production from in vitro-infected cell lines.
We next investigated if the in vitro-infected cell lines were expressing JSRV proteins or producing virus particles. Because our stocks of ST-6 and FLT cells grew very slowly, we concentrated our subsequent studies on the OAT-T3 and CP cell lines. Initial tests of infected cell lines for expression of viral CA protein by Western blotting were negative, and standard RT assays of infected cell supernatant were also negative (not shown). Therefore, if the infected cells produced virus, they were inefficient.
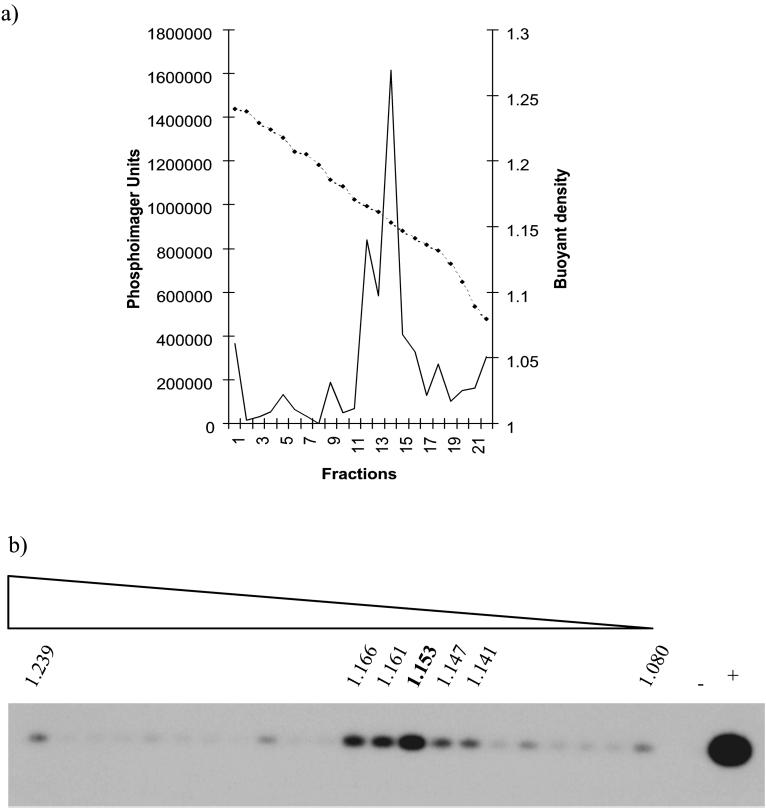
We next tested supernatants from infected CP and OAT-T3 cells for the presence of JSRV21 particles by using the highly sensitive PERT assay for RT (24). RT activity was detected in 33-fold-concentrated supernatant of both CP (passage 16 postinfection [Fig. 5]) and OAT-T3 (passage 7 postinfection [data not shown]) cells but not in matched passages of cells infected with heat-inactivated JSRV21 (Fig. 5). This indicated low-level production of virus from the infected CP and OAT-T3 cells. To further test if the released RT activity was present in virus particles, concentrated supernatant from the infected CP cells was subjected to isopycnic centrifugation and gradient fractions were assayed for RT activity by the PERT assay. As shown in Fig. 6, RT activity released from the infected CP cells had a peak buoyant density of 1.15 g/ml, the same as for the original JSRV21 used in the infections. RT-PCR analysis also confirmed that JSRV RNA was present in the 1.15-g/ml peak fractions (data not shown). Thus, the infected CP cells appear to be producing native JSRV21 at low level.
FIG. 5.
RT activity released from infected CP cells. RT activity was tested by PERT assay on 10-fold dilutions of 33-fold-concentrated supernatant from infected CP cells (passage 12) or on equivalently concentrated supernatant from CP cells infected with heat-treated JSRV21 at the same passage. Southern blot hybridization with an MS2-specific probe was used for PCR product detection. Note the presence of the specific MS2 band only in the JSRV21-infected cells. Controls: +, a PERT reaction with purified Moloney murine leukemia virus RT (10−4 U); −, a PERT reaction with buffer A alone.
FIG. 6.
Buoyant density of virus produced by infected CP cells. Supernatant from JSRV21-infected CP cells (passage 19) was concentrated and fractionated by isopycnic centrifugation in a 20 to 55% continuous sucrose gradient. Gradient fractions were assayed for RT activity by the PERT assay. (a) RT activity (solid line) expressed in PhosphorImager units taken from samples visualized in panel b; the broken line shows density (grams per milliliter). The peak of RT activity is in the fraction with a density of 1.15 g/ml. (b) PERT reactions from each gradient fraction visualized by Southern blotting for MS-2 DNA; +, Moloney murine leukemia virus RT (10−4 U); −, buffer A control.
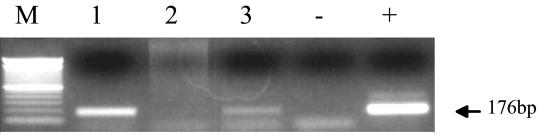
Finally, we tested whether the JSRV-infected CP cells could transfer infection in vitro to uninfected cells. Infected CP cells at passage 12 postinfection were γ-irradiated and then cocultured with uninfected CP cells at an initial ratio of 5:1 (infected/uninfected) in the presence of 2 μg of Polybrene per ml. After 8 weeks (and six passages), the coculture showed evidence of viral infection by the JSRV-U3 PCR assay (Fig. 7). By this time, a parallel culture of an equal number of γ-irradiated infected CP cells showed no survivors. This indicated that the cells in the coculture after 8 weeks of passage were progeny of initially uninfected CP cells in the coculture and that infection had been transferred to them from the infected CP cells. Thus, in vitro-infected CP cells were producing infectious JSRV.
FIG. 7.
Transfer of JSRV infection from infected CP cells. U3 PCR for exogenous JSRV DNA was carried out on 500 ng of genomic DNA. The parental infected and uninfected CP cells before cocultivation are shown in lanes 1 and 2, respectively. In lane 3 are shown CP cells 8 weeks after cocultivation. +, JSRV21 plasmid DNA; −, distilled water. M, 100-bp ladder (Gibco).
Tagged JSRV21 virus (JSRVsupF) is infectious.
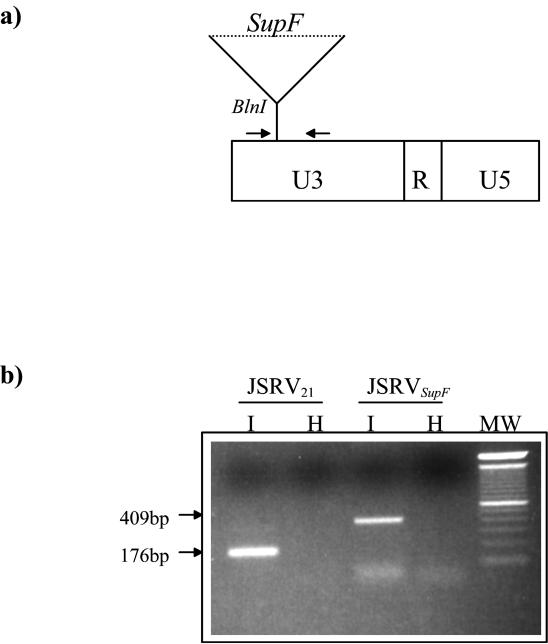
Finally, we tagged the JSRV21 virus with the supF gene to develop a tool that would allow us to readily detect exogenous JSRV virus and circumvent the background represented by endogenous JSRV-related retroviruses in the sheep genome. The supF gene was inserted in the U3 of pCMV2JS21 (Fig. 8a). Transfection into 293T cells yielded infectious JSRVsupF, as demonstrated by the successful detection of provirus in CP cells infected with JSRVsupF at passage 2 (Fig. 8b). The size of the PCR product in infected CP cells was that predicted for JSRVsupF, indicating retention of the supF sequences during in vitro infection. In an independent experiment, the JSRVsupF PCR product could be detected in infected CP cells for at least four passages postinfection and not in control cells infected with heat-inactivated virus.
FIG. 8.
JSRVsupF is infectious in vitro. (a) Schematic diagram of the JSRVsupF LTR. The supF bacterial tRNA suppressor gene was inserted into the U3 region of pCMV2JS21. The primers used in this study for the PCR detection of JSRV provirus (JSRV U3-PCR) are downstream and upstream of the supF gene (bold arrows); the resulting PCR product obtained from the amplification of JSRVsupF will be 409 bp, versus the 176 bp for the wild-type JSRV LTR. (b) CP cells were infected with JSRVsupF or wild-type JSRV21 obtained from transfected 293T cells and assayed for infection by PCR analysis. The results, from passage 2 after infection, show successful infection by JSRVsupF. Cells were infected in parallel with untreated virus (I) or virions heat inactivated at 65°C for 15 min (H). Note the different sizes of the PCR products obtained from choroid plexus cells infected with JSRV21 versus JSRVsupF.
DISCUSSION
In this paper, we describe the first successful in vitro infection of ovine cell lines with JSRV. Previously, research in the field was hampered by the lack of an in vitro infection system, and the results reported here provide a first solution. Several criteria indicated that infection and propagation of JSRV in vitro was achieved: (i) presence of JSRV21 DNA in the infected cultures and increase in viral DNA content with passage; (ii) lack of JSRV21 DNA in cultures infected with heat-inactivated virus; (iii) release from the infected cells of RT activity that banded at 1.15 g/ml in sucrose density gradients; and (iv) transmission of JSRV21 provirus to uninfected target cells by cocultivation of γ-irradiated infected cells.
These results have several implications. The first implication stems from the fact that multiple ovine cell lines of different lineages were infectable by JSRV21, including choroid plexus (CP), testes (OAT-T3), turbinates (FLT), and intestinal carcinoma (ST6) cell lines. This indicates that the cellular receptor for JSRV is expressed in a broad range of ovine cell types. When these experiments were initiated, it seemed possible that the JSRV receptor was relatively restricted, given the fact that the virus is associated only with tumors of type II pneumocytes and Clara cells. On the other hand, we have recently found low levels of JSRV infection (JSRV proviral DNA) in lymphoid tissues of SPA-affected sheep (20) and in several lineages of hematopoietic cells in experimentally infected lambs before and after tumor development (10). This would also be consistent with a broad distribution of JSRV receptors within the animal. In additional experiments, infection of JSRV21 into the MLE15 mouse epithelial lung cell line (29) was not successful (data not shown). This might suggest that functional JSRV receptors are not present on mouse cells.
Another implication is that in vitro infection in ovine cell lines may be useful to test other cloned JSRV proviruses for infectivity. Previously, the only way to demonstrate infectivity was intratracheal injection of cloned DNA or virus into newborn lambs (21). This method was extremely laborious, time-consuming, and expensive. The key to successful in vitro infection lay in the production of substantial amounts of JSRV21 virus by transfection of 293T cells with pCMV2JS21. The infectivity of other JSRV proviral clones could be tested by making analogous cytomegalovirus-driven expression plasmids, transfecting 293T cells, and using the supernatants to infect ovine cell lines, as performed here.
In this study, we also engineered a tagged virus with the bacterial tRNA suppressor supF gene inserted into the U3 of JSRV21. The resulting virus, termed JSRVsupF, is infectious in vitro, and we are currently testing its pathogenicity in vivo. If it is oncogenic, JSRVsupF will be a powerful tool to investigate the clonality of the JSRV-induced tumors and to analyze the sites of viral integration. Past studies have failed to detect novel integration sites into SPA tumors for the lack of sensitive and exogenous provirus-specific hybridizing probes that do not cross-react with the 15 to 20 copies of highly JSRV-related endogenous retroviruses present in the sheep genome (5).
The in vitro-infected ovine cell lines produced very low levels of virus (e.g., RT detection in cell supernatants required the highly sensitive PERT assay), even though there appeared to be significant levels of viral DNA in late-passage cells (Fig. 4). One possible explanation is that the JSRV LTR is preferentially active in differentiated type II pneumocytes and Clara cells. Indeed, immunohistochemistry of SPA-affected lungs reveals viral CA protein in the tumor cells in the lungs but not in surrounding cells of different differentiation types (e.g., type I pneumocytes and macrophages) (18). Moreover, Jassim (12) established two cell lines, JS7 and JS8, derived from SPA tumors. During the establishment of the cell lines, these cells simultaneously lost markers for type II pneumocyte differentiation and the ability to produce virus. However, they retained functional proviruses, since injection of JS7 or JS8 into the lungs of newborn lambs resulted in release of virus and induction of SPA (12). It will be interesting to test the transcriptional activity of the JSRV21 LTR in type II pneumocytes in comparison to other cell types and to identify cellular factors and LTR sequences responsible for high-level expression. Indeed, preliminary experiments indicate that the JSRV LTR is preferentially active in murine type II pneumocytes and Clara cell-derived cell lines (18a). Treatments or drugs that induce JSRV21 LTR activity in ovine cell types other than type II pneumocytes would also be of interest. This might lead to improved in vitro infection systems where higher levels of JSRV are produced.
ACKNOWLEDGMENTS
We are grateful to C. Holland and N. DiFronzo for providing plasmid piAN7 and to G. Whitsett for providing the MLE15 cell line.
M.P. is a recipient of a “Wellcome Prize Travelling Research Fellowship” from the Wellcome Trust. Additional funding was provided by the SOAEFD. Support from the UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center is acknowledged.
REFERENCES
- 1.Bai J, Zhu R Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradac J, Hunter E. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology. 1984;138:260–275. doi: 10.1016/0042-6822(84)90350-7. [DOI] [PubMed] [Google Scholar]
- 2a.DeMartini, J. C., and J. M. Sharp. Unpublished data.
- 3.DiFronzo N L, Holland C A. A direct demonstration of recombination between an injected virus and endogenous viral sequences, resulting in the generation of mink cell focus-inducing viruses in AKR mice. J Virol. 1993;67:3763–3770. doi: 10.1128/jvi.67.7.3763-3770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dion A S, Williams C J, Pomenti A A. The major structural proteins of murine mammary tumor virus: techniques for isolation. Anal Biochem. 1977;82:18–28. doi: 10.1016/0003-2697(77)90129-4. [DOI] [PubMed] [Google Scholar]
- 5.Hecht S J, Carlson J O, DeMartini J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and uneffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 6.Hecht S J, Sharp J M, Demartini J C. Retroviral aetiopathogenesis of ovine pulmonary carcinoma: a critical appraisal. Br Vet J. 1996;152:395–409. doi: 10.1016/s0007-1935(96)80034-0. [DOI] [PubMed] [Google Scholar]
- 7.Herring A J, Sharp J M, Scott F M, Angus K W. Further evidence for a retrovirus as the aetiological agent of sheep pulmonary adenomatosis (jaagsiekte) Vet Microbiol. 1983;8:237–249. doi: 10.1016/0378-1135(83)90076-7. [DOI] [PubMed] [Google Scholar]
- 8.Hizi A, Henderson L E, Copeland T D, Sowder R C, Hixson C V, Oroszlan S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc Natl Acad Sci USA. 1987;84:7041–7045. doi: 10.1073/pnas.84.20.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hizi A, Henderson L E, Copeland T D, Sowder R C, Krutzsch H C, Oroszlan S. Analysis of gag proteins from mouse mammary tumor virus. J Virol. 1989;63:2543–2549. doi: 10.1128/jvi.63.6.2543-2549.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland M J, Palmarini M, Garcia-Goti M, Gonzalez L, Mckendrick I, De las Heras M, Sharp J M. Jaagsiekte retrovirus is widely distributed in both T and B lymphocytes and mononuclear phagocytes of sheep with naturally and experimentally acquired pulmonary adenomatosis. J Virol. 1999;73:4004–4008. doi: 10.1128/jvi.73.5.4004-4008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ives J C, Buffler P A, Greenberg S D. Environmental associations and histopathologic patterns of carcinoma of the lung: the challenge and dilemma in epidemiologic studies. Am Rev Respir Dis. 1983;128:195–209. doi: 10.1164/arrd.1983.128.1.195. [DOI] [PubMed] [Google Scholar]
- 12.Jassim F A. Ph.D. thesis. University of Edinburgh; 1988. [Google Scholar]
- 13.Landis S H, Murray T, Bolden S, Wingo P A. Cancer statistics, 1998. CA Cancer J Clin. 1998;48:6–29. doi: 10.3322/canjclin.48.1.6. [DOI] [PubMed] [Google Scholar]
- 14.Li J K. Chromatographic purification and immunological analysis of viral polypeptides of mouse mammary tumor virus. J Virol Methods. 1987;17:299–310. doi: 10.1016/0166-0934(87)90140-6. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Holland C A, Hartley J W, Hopkins N. Viral integration near c-myc in 10–20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci USA. 1984;81:6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norval M, Head K W, Else R W, Hart H, Neill W A. Growth in culture of adenocarcinoma cells from the small intestine of sheep. Br J Exp Pathol. 1981;62:270–282. [PMC free article] [PubMed] [Google Scholar]
- 17.Palmarini M, Cousens C, Dalziel R G, Bai J, Stedman K, DeMartini J C, Sharp J M. The exogenous form of jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmarini M, Dewar P, De las Heras M, Inglis N F, Dalziel R G, Sharp J M. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 18a.Palmarini, M., and H. Fan. Unpublished results.
- 19.Palmarini M, Fan H, Sharp J M. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 20.Palmarini M, Holland M J, Cousens C, Dalziel R G, Sharp J M. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J Gen Virol. 1996;77:2991–2998. doi: 10.1099/0022-1317-77-12-2991. [DOI] [PubMed] [Google Scholar]
- 21.Palmarini M, Sharp J M, De las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkin D M. The global burden of cancer. Semin Cancer Biol. 1997;8:219–235. doi: 10.1006/scbi.1998.0080. [DOI] [PubMed] [Google Scholar]
- 23.Perk K, Hod I. Sheep lung carcinoma: an endemic analogue of a sporadic human neoplasm. J Natl Cancer Inst. 1982;69:747–749. [PubMed] [Google Scholar]
- 24.Pyra H, Boni J, Schupbach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc Natl Acad Sci USA. 1994;91:1544–1548. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp J M, Angus K W, Gray E W, Scott F M. Rapid transmission of sheep pulmonary adenomatosis (jaagsiekte) in young lambs. Brief report. Arch Virol. 1983;78:89–95. doi: 10.1007/BF01310861. [DOI] [PubMed] [Google Scholar]
- 26.Sharp J M, Herring A J. Sheep pulmonary adenomatosis: demonstration of a protein which cross-reacts with the major core proteins of Mason-Pfizer monkey virus and mouse mammary tumour virus. J Gen Virol. 1983;64:2323–2327. doi: 10.1099/0022-1317-64-10-2323. [DOI] [PubMed] [Google Scholar]
- 27.Verwoerd D W, Williamson A L, De Villiers E M. Aetiology of jaagsiekte: transmission by means of subcellular fractions and evidence for the involvement of a retrovirus. Onderstepoort J Vet Res. 1980;47:275–280. [PubMed] [Google Scholar]
- 28.Vogt P K. Historical introduction to the general properties of retroviruses. In: Coffin J M, editor. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1–25. [PubMed] [Google Scholar]
- 29.Wikenheiser K A, Vorbroker D K, Rice W R, Clark J C, Bachurski C J, Oie H K, Whitsett J A. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90:11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.York D F, Vigne R, Verwoerd D W, Querat G. Isolation, identification, and partial cDNA cloning of genomic RNA of jaagsiekte retrovirus, the etiological agent of sheep pulmonary adenomatosis. J Virol. 1991;65:5061–5067. doi: 10.1128/jvi.65.9.5061-5067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.York D F, Williamson A, Barnard B J, Verwoerd D W. Some characteristics of a retrovirus isolated from transformed bovine cells. Virology. 1989;171:394–400. doi: 10.1016/0042-6822(89)90607-7. [DOI] [PubMed] [Google Scholar]