Abstract
Polyomavirus induces a broad array of tumors when introduced into newborn mice of certain standard inbred strains, notably those bearing the H-2k haplotype. Susceptibility in these mice is conferred by an endogenous mouse mammary tumor virus superantigen (Mtv-7 sag) that acts to delete T cells required for polyomavirus-induced tumor immunosurveillance. In the present study we show that mice of two wild-derived inbred strains, PERA/Ei (PE) and CZECH II/Ei (CZ), are highly susceptible to polyomavirus but carry no detectable Mtv sag-related sequences and show no evidence of Vβ deletion. C57BR/cdJ (BR) mice, which are H-2k but lack the endogenous Mtv-7, are highly resistant based on an effective anti-polyomavirus tumor immune response. When crossed with BR, both PE and CZ mice transmit their susceptibility in a dominant fashion, indicating a mechanism(s) that overrides the immune response of BR. Susceptibility in PE and CZ mice is not based on interference with antigen processing or presentation since cytotoxic T cells from BR can efficiently kill F1-derived tumor cells in vitro. The expected precursors of polyomavirus-specific cytotoxic T cells are present in both the wild inbred animals and their F1 progeny. These findings indicate a novel basis of susceptibility that operates independently of endogenous superantigen and prevents the development of tumor immunity.
The murine polyomavirus can be a powerful oncogenic agent in its natural host, as evidenced by the rapid development of multiple solid tumors after inoculation of newborn animals (11, 19). Genetic backgrounds of both virus and host play important roles in determining the tumor response. By using a highly susceptible host, the effects of various determinants in the viral T (tumor) antigens involved in cell transformation (6, 14, 16, 38), as well as ones in the viral structural proteins with effects on receptor binding, cell penetration, and spread (2, 14, 36), have been investigated.
The role of the host genetic background is complex and less well understood. Earlier studies of susceptible and resistant strains established an important role of the major histocompatibility complex (MHC) type and the ability to generate antitumor cellular immune responses (4, 17, 25, 27). Most resistant mouse strains show a radiation-sensitive form of resistance and become susceptible after radiation or neonatal thymectomy (1, 9, 26). A radiation-resistant or nonimmunological form of host resistance that acts by curtailing virus spread has also been described (9).
In crosses between susceptible and resistant mice of the same MHC type (H-2k), susceptibility is inherited as a single dominant Mendelian trait (17, 30). This gene segregates with the endogenous mouse mammary tumor provirus Mtv-7 carried by the susceptible strain (28). C57BR/cdJ (BR) mice, used as the resistant parent, generate virus-specific cytotoxic T lymphocytes (CTLs) with a Vβ specificity (Vβ6) that would be deleted by the Mtv-7 superantigen (sag) present in all highly susceptible H-2k strains (28, 29).
In the present study we first isolated and characterized a polyomavirus-specific CTL line from a virus-infected BR mouse and used it to investigate possible mechanisms of immune evasion by rare tumors that arise in this resistant strain. We describe a genetic and immunological basis of susceptibility to polyomavirus tumors manifested by two wild-derived inbred mouse strains. These highly susceptible wild inbreds are shown to be free of endogenous Mtvs and to express no sag(s) and yet, in crosses with BR mice, they transmit their susceptibility in a dominant fashion. Tumor cells derived from F1 animals are killed by CTLs from strain BR mice, demonstrating that these mice are able to process and present the appropriate viral epitope and that the tumor cells themselves are not intrinsically resistant to CTL killing. The wild-derived inbreds and their F1s have normal levels of CD8+ Vβ6+ T cells that are the expected precursors of polyomavirus-specific CTLs in this system. These results suggest a novel basis of tumor susceptibility involving a mechanism that interferes with the development of tumor immunity.
MATERIALS AND METHODS
Tumor studies.
C57BR/cdJ, PERA/Ei (PE), CZECH II/Ei (CZ), and C57BL/6J (B6) mice were purchased from the Jackson Laboratory (Bar Harbor, Maine). C3H/BiDa mice were obtained from Clarence Reeder at the National Cancer Institute, Frederick, Md. All mice were bred and maintained in our virus antibody-free (VAF) barrier facility prior to virus inoculation. The A2 and A2+ high-tumor strains of polyomavirus were used (15). Newborn animals (<18 h old) were inoculated intraperitoneally with ∼50 μl of virus containing 2 × 106 to 10 × 106 PFU and were monitored for up to 6 months for tumor development. Animals were sacrificed when moribund and then necropsied; gross tumors, as well as apparently normal tissues, were examined histologically as described earlier (10). Tumor-derived cell lines were as follows: A-6215 is immunogenic, derived from a salivary gland tumor that arose in an irradiated virus-infected BR mouse; A-6241 and A-6689 are nonimmunogenic, derived from mammary tumors that arose in normal (unirradiated) virus-infected BR mice (28). Cultures of tumor cells from PE, CZ, and F1 animals were prepared by collagenase treatment of primary tumors and plating cells in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum.
Detection of antigens by PCR.
Genomic DNAs were prepared from tail tissue as described previously (24). PCR was conducted in a PTC-100 thermocycler (MJ Research, Inc., Watertown, Mass.). Briefly, 100 μl of a reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 100 μg of gelatin per ml, 200 μM concentrations of each of the deoxynucleoside triphosphates, and 0.5 μM concentrations of each primer was mixed with 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Foster City, Calif.) and 200 ng of genomic DNA. The PCR mixture was subjected to predenaturation at 94°C (5 min), followed by 35 cycles of 94°C (45 s), 58°C (45 s), and 72°C (1.5 min) and a final postcycling extension at 72°C for 5 min. PCR amplification of Mtv-7 sag-specific sequences was carried out with forward primer Mtv-7-2 (5′-TTACATCTACAGACCAACAGATGCCCCGT-3′) and reverse primer Mtv-7-1 (5′-GAAGCCAACGCGACCCCC-3′) based on published sequences (5, 35). Primers for conserved mouse mammary tumor virus (MMTV) sag sequences were forward MTV-cons (5′-GGGAATTCTCGAGATGCCGCGCCTGCAG-3′) and reverse MTV-cons (5′-GGGGATCCTCTAGAGGGAACCGCAAGGTTGGG-3′) (21). For polyomavirus sequences in tumor cell DNAs, primers were used to give a 542-bp product covering overlapping sequences for the large and middle T antigens: forward, 5′-GTATTTGGACATCCTAC-3′; and reverse, 5′-AAATGGTGCTGCGGTTACAA-3′. The annealing temperature for polyomavirus PCR was 55°C; other parameters were the same as those given above. As a PCR control, mouse-specific DNA was amplified by using Map Pair D8 Mit 223 from Research Genetics, Inc. (Huntsville, Ala.).
Cytofluorometric analysis: screening for Vβ expression in T cells from spleen.
Leukocytes from the spleen were isolated by centrifugation of a single cell suspension over Ficoll-Hypaque gradients (Accurate Chemicals, Westbury, N.Y.). T cells were enriched by passage through CD3 T-cell enrichment columns (R&D, Minneapolis, Minn.). The enriched T cells were first incubated with Vβ T-cell receptor-specific monoclonal antibodies (MAbs) (kindly provided by Martin E. Dorf, Department of Pathology, Harvard Medical School, Boston, Mass.) and then with fluorochrome-labeled second antibodies (Jackson Immunoresearch, West Grove, Pa.). The stained cells were analyzed by using a Coulter Profile II flow cytometer (Coulter, Hialeah, Fla.).
Analysis of MHC haplotype.
Splenocytes were prepared by ammonium chloride lysis of erythrocytes, followed by gravity sedimentation to remove cell clumps and debris. One million cells were stained with 1 μg of fluorescein isothiocyanate (FITC)-conjugated anti-H-2Kk or anti-H-2Dk MAbs (Pharmingen, San Diego, Calif.). A monoclonal rat immunoglobulin (immunoglobulin G1 [IgG1]) was used as a control. Flow cytometry was performed on a Becton Dickinson (Mountain View, Calif.) FACScan by using LYSIS II software, and the data were analyzed with CellQuest software.
In vitro one-way mixed lymphocyte reaction.
Splenocytes were prepared by ammonium chloride lysis of erythrocytes. Responder cells were from BR (H-2k) mice. Stimulator cells from different mouse strains were treated with mitomycin C (50 μg/50 million cells). Responder (2 × 105 cells/well) and stimulator (4 × 105 cells/well) cells were added together in wells of a 96-well tissue culture plate in DMEM containing 10% fetal calf serum. The cells were incubated in a humidified chamber at 37°C with 7% CO2 for 4 days and subsequently pulsed with 1 μCi of [3H]thymidine per well for an additional 16 h. Cells were collected on a glass fiber filter and counted in a beta scintillation counter.
In vitro CTL assay: generation of polyomavirus-specific CTL line LN-13.1.
Cell line LN-13.1 was generated from a virus-immunized BR mouse. A newborn mouse was infected in the footpad with 50 μl of A2 virus containing 108 PFU/ml. Two weeks later, lymphocytes from the draining lymph nodes were harvested and cultured in a 24-well plate in complete Iscove modified Dulbecco medium (IMDM) with 10% fetal bovine serum. Lymphocytes were stimulated weekly with A2 virus-infected and gamma-irradiated (2,000 rads) BR splenocytes. After three cycles of restimulation, bulk cultures were further established by using mitomycin-treated immunogenic BR salivary gland tumor cell line A-6215 and naive BR splenocytes as stimulator and feeder cells, respectively. The CTL line LN-13.1 thus generated was used along with 51Cr-labeled target cells in all CTL assays.
Polyomavirus tumor cell targets.
Cells derived from virus-induced tumors were maintained in DMEM containing 10% fetal bovine serum. The target cells were radiolabeled by the addition of 200 μCi of Na251CrO4 (NEN, Boston, Mass.) per ml in DMEM with 5% fetal bovine serum. Cells were incubated at 37°C for 1 h and washed three times in the same medium to remove the free 51Cr.
Targets prepared from normal cells by viral infection or peptide pulsing.
Adherent cells from the spleen were harvested from different mouse strains and selected by incubating the cell suspensions on a polystyrene plastic surface. Polyomavirus middle T peptide R389 (RRLGRTLLL) or E328 (EEQVPQLI) was added to the adherent cells at a concentration of 1 μM in DMEM with 10% fetal bovine serum, incubated at 37°C for 1 h, and then washed in the same medium three times. Alternatively, adherent cells were infected by virus at a multiplicity of infection of 1 to 2 PFU/cell; cells were then 51Cr-labeled as described above and used as targets at 16 to 20 h postinfection.
The radiolabeled targets (5 × 103 cells/well) were distributed into wells of 96-well U-bottom tissue culture plates along with effector CTLs (25 × 103 cells/well) at a 5:1 effector/target ratio. The assay medium was complete IMDM containing 10% fetal calf serum, 6% rat T-stimulation medium (Collaborative Research, Bedford, Mass.), 4 mM glutamine, 2 mM sodium pyruvate, and 50 μM 2-mercaptoethanol. After incubation for 4 h at 37°C, the supernatant (100 μl) from each well was collected and counted on a gamma counter (Wallac, Inc., Gaithersburg, Md.). Spontaneous and total 51Cr release were counted from supernatants of targets incubated with medium alone and 1% Triton X-100, respectively. The spontaneous release in all assays was less than 20% of the total release. The percent specific lysis was calculated as follows: [(lysis with effector cells − spontaneous lysis)/(total lysis − spontaneous lysis)] × 100. Each assay contained triplicate wells, and each experiment was repeated at least twice, with identical results.
RESULTS
Immune surveillance and evasion in the BR mouse.
The resistance of BR mice to tumor induction by polyomavirus is based on effective immune surveillance and the elimination of virus-induced tumors. BR mice show the radiation-sensitive form of resistance (9, 28) and mount an effective antitumor immune response dominated by CD8+ Vβ6+ T cells (28). These CTLs have been shown to be H-2Dk-restricted and specific for an immunodominant epitope derived from the middle T protein, the product of the major transforming gene of the virus (29). Rare tumors that arise in polyomavirus-infected BR mice appear to be “immune escape” variants by virtue of being transplantable in polyomavirus-immunized syngeneic hosts (28). The mechanism(s) of escape is unknown.
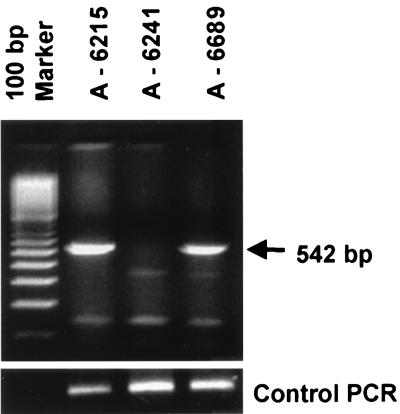
To investigate possible mechanisms of immune evasion, we first sought to recover viral DNA sequences by PCR. Primers were specific to a region of middle T containing the immunodominant epitope. Three tumors were tested: A-6215, a positive control, was derived from an irradiated virus-infected BR mouse and is known to express virus-specific transplantation antigen(s); and two variant tumors, A-6241 and A-6689, which arose in nonirradiated virus-infected BR mice and fail to express virus-specific transplantation antigen(s) (28). A-6215 and one of the variants were positive for viral DNA, while the other variant was negative (Fig. 1). The variant that failed to show viral DNA sequences (A-6241) is considered to be of spontaneous origin or possibly a virus-induced tumor that subsequently lost the viral DNA. Sequencing of the PCR product from the DNA-positive tumor A-6689 showed a wild-type middle T sequence, including an unaltered epitope. Immunoprecipitation of tumor cell extracts showed that A-6215 and A-6689 were both positive and A-6241 was negative for middle T protein expression (data not shown). These results demonstrate that A-6689 did not escape recognition due to a failure to express middle T or to antigenic variation.
FIG. 1.
Test for polyomavirus DNA sequences in tumors from BR mice. A-6215 is an immunogenic tumor from an irradiated virus-infected mouse, A-6241 is a nonimmunogenic tumor from an unirradiated virus-infected mouse, and A-6689 is another nonimmunogenic tumor from an unirradiated virus-infected mouse. On the left are molecular size markers. The arrow on the right indicates the position of the expected 542-bp DNA product. PCR was carried out for polyomavirus middle T-coding sequences. A PCR control is shown at the bottom of the figure.
Expression of MHC class I products by each of these tumor-derived cell lines was measured (Table 1). A-6215 and A-6241 showed moderate to high levels either with or without gamma interferon (IFN-γ) pretreatment. The viral DNA-positive variant A-6689, however, showed a nearly complete absence of expression without IFN-γ, although class I expression could be induced to high levels after the treatment. This finding raises the possibility that A-6689 escaped immune recognition by virtue of a failure to express class I adequately.
TABLE 1.
MHC class I expression on polyomavirus tumor cell surface
| Cell linea | IFN-γ | % Fluorescence-positive cellsb
|
|
|---|---|---|---|
| Anti-Kk | Anti-Dk | ||
| A-6215 | − | 80 | 14 |
| + | 98 | 81 | |
| A-6241 | − | 88 | 58 |
| + | 79 | 76 | |
| A-6689 | − | 2 | 1 |
| + | 97 | 59 | |
Polyomavirus tumor cell lines A-6215, A-6689, and A-6241 were described previously (28). Cells were treated with either medium alone or IFN-γ for 36 h.
Fluorescent staining (FITC-labeled anti-H-2Kk or anti-H-2Dk MAb) and FACS analysis were carried out. Values represent one of three experiments with similar results. The percent positive cells with mouse IgG1 isotype control MAb was less than 3%.
As a tool for further studies, we isolated and characterized a virus-specific CTL line from polyomavirus-immunized BR mice (see Materials and Methods). LN-13.1 cells were greater than 98% CD8+ Vβ6+. The results in Table 2 show that LN-13.1 killed A-6215 tumor cells and that killing could be prevented by pretreatment of the cells with anti-H-2Dk but not anti-H-2Kk MAb. In addition, pulsing of normal adherent spleen cells from BR mice with the H-2Dk immunodominant middle T peptide R389 sensitized the cells to killing by LN-13.1, while the control middle T peptide E328 (with a sequence bearing an H-2Kk epitope) had no effect (Table 2). LN-13.1 cells thus resemble the virus-specific CTLs previously described for this system (28, 29).
TABLE 2.
Specificities of LN-13.1 CTL cell line
| Type of specificity and antibody or peptide used | % Specific lysis |
|---|---|
| MHC restriction specificitya | |
| mIgG | 35 |
| Anti-H-2Kk | 25 |
| Anti-H-2Dk | 0 |
| Peptide specificityb | |
| mT-E328 | 11 |
| mT-R389 | 92 |
Tumor cells (A-6215) derived from an irradiated, virus-infected BR mouse were used as targets. These cells were pretreated with either anti-H-2Kk or anti-H-2Dk MAbs (5 μg/well). A monoclonal mouse Ig6 (mIgG; specific for TNP) was used as a control.
Plastic-adherent cells from naïve BR spleens were pulsed with middle-T peptides, and the CTL assay was carried out at a 5:1 effector-to-target ratio.
When tested for their susceptibility to killing by LN-13.1, only the immunogenic control tumor A-6215 was killed in the absence of added IFN (Table 3). Neither the spontaneous viral DNA-negative tumor A-6241 nor the escape variant A-6689 expressing wild-type middle T was killed under these conditions. However, when pretreated with IFN-γ, A-6689 became susceptible, a result consistent with evasion based on low constitutive expression of class I. As expected, exposure of A-6241 to IFN-γ had no effect on its resistance, although these cells could be sensitized to killing by exposure to the R389 peptide.
TABLE 3.
Susceptibility to CTL killing of “immune escape” tumor cell variants
| Tumor cellsa | Treatment | % Specific lysisb
|
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| A-6215 | Medium | 36 | 37 |
| IFN-γ | 49 | 50 | |
| A-6689 | Medium | 7 | 0 |
| IFN-γ | 44 | 42 | |
| A-6241 | Medium | 3 | 9 |
| IFN-γ | 2 | 0 | |
| A-6241 | mT-R389 | 39 | 41 |
| A-6241 | mT-E328 | 7 | 9 |
Tumor cells were treated with either medium alone or IFN-γ (100 U/ml) for 36 h. A-6241 cells (without IFN-γ treatment) were pulsed for 1 h at 37°C with either R389 (RRLGRTLLL) or E328 (EEQVPQLI) peptide (1 μM) derived from middle-T (mT) antigen.
The CTL assay was carried out with the LN-13.1 CTL line at a 5:1 effector/target ratio. The percent specific lysis was calculated.
Dominant susceptibility to polyomavirus tumors in PE and CZ mice.
PE and CZ mice are highly susceptible to tumor induction by polyomavirus, with 97 to 100% of the animals developing multiple tumors by 3 to 4 months of age (Table 4). Neither of these strains develops spontaneous tumors in the relatively short time period of studies with the virus. With respect to the range and histological features of tumor types and the time required for tumor development, these wild-derived inbred mice resemble strain C3H/BiDa and other standard inbreds (CBA/J, AKR, RF/J, and C58), which possess an H-2k/Mtv-7 basis of susceptibility (10, 16, 28). Also similar to the standard inbreds, PE and CZ mice transmit their susceptibility in a dominant fashion when crossed with BR mice. Results with F1 mice were independent of the mother-father pairing of the parental strains, indicating an autosomal basis of susceptibility.
TABLE 4.
Inheritance of tumor susceptibility of wild inbred mouse strainsa
| Mouse strain | Fraction of mice with tumor(s) (%) |
|---|---|
| Parental | |
| PE | 41/42 (97) |
| CZ | 30/30 (100) |
| BR | 2/47 (4)b |
| F1 | |
| (PE × BR)F1 | 36/40 (90) |
| (CZ × BR)F1 | 28/30 (93) |
| Backcross | |
| (PE × BR) × BR | 64/121 (53) |
| (CZ × BR) × BR | 26/70 (37) |
Newborn mice were inoculated intraperitoneally with either the A2 or A2+ strain of polyomavirus. These two viral strains differ with respect to a 40-bp duplication in the noncoding region which promotes gross thymic tumor development (15). No differences were noted among susceptible mice in terms of their responses to the A2 or A2+ strain, apart from those expected for thymic tumor development. Mice were monitored for development of all tumor types, and the data in each group for the two viruses were pooled.
Data from Lukacher et al. (28).
In backcross mice, 53% [(PE × BR) × BR] and 37% [(CZ × BR) × BR] developed at least single, and most often multiple, tumors (Table 4). These frequencies are consistent with a single dominant susceptibility gene in PE mice and one or two genes in CZ mice. Susceptibility is also reflected in the average number of tumors per affected animal, referred to as the tumor frequency index (TFI). In the PE cross, the TFIs for parental, F1, and backcross mice were 4.4, 2.6, and 1.3, respectively. For the CZ cross, the TFIs were 3.6, 2.3, and 1.7, respectively. The drop in TFI between parental and F1 mice indicates codominance with a clear dosage effect of the susceptibility gene(s). The further drop in TFI in the affected backcross animals suggests possible additive or interactive gene effects. The latencies of tumor development are reflected in the average ages at which tumor-bearing animals come to necropsy. In both crosses, latencies were found to increase from an average of 68 days (parental mice) to 95 days (F1s) to 112 days (affected backcross animals). Although “age at necropsy” is a crude estimate of tumor latency, the trend of increasing time for tumor development with dilution of the genetic contribution from the PE and CZ parental backgrounds is consistent with gene dosage and possible gene interaction. A similar pattern of inheritance with decreasing TFIs and increasing latencies in F1 and backcross animals was observed previously in crosses with mice carrying Mtv-7 as the dominant susceptibility factor (28).
Absence of endogenous superantigens in PE and CZ mice.
It was important in the present study to establish whether either PE or CZ mice carried endogenous MMTV sag sequences or showed any evidence of Vβ deletion that might account for their susceptibility. CZ mice have previously been screened for endogenous MMTV sequences by Southern hybridization and found to be negative, although MMTV-related sequences were detected under low-stringency conditions (7). A milk-borne MMTV was transmitted in CZ mice originally (18) but was no longer carried by CZ mice used in this study. PE mice have not been characterized for endogenous MMTVs to the best of our knowledge.
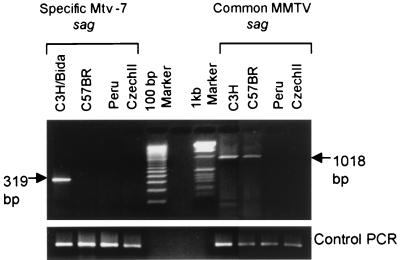
PCR was carried out on genomic DNAs by using primers specific for MMTV sag sequences (Fig. 2). Two primer pairs were used. The first was designed to amplify a 319-bp fragment specific to the carboxy terminus of Mtv-7 sag based on published sequences (5, 35). The other pair was designed to amplify a 1,018-bp fragment by using a forward primer complementary to a conserved or common region among MMTV sags and a reverse primer outside the coding region in the 3′ long terminal repeat (21). PE and CZ mice proved to be negative with both sets of probes. The standard susceptible C3H/BiDa strain used as a positive control gave products of the expected size with both probes. BR mice were positive for the conserved region probe and negative for the Mtv-7-specific probe, as expected. These results indicate that neither PE nor CZ mice carry detectable Mtv-7 sag or other MMTV sag sequences.
FIG. 2.
Tests for Mtv-7 Sag and other MMTV sag sequences in genomic DNAs of various mouse strains. (Left) PCR product for Mtv-7 sag-specific sequences. Lanes (left to right): C3H/BiDa, BR, PE, CZ, and markers. The arrow indicates the position of the predicted 319-bp product. (Right) PCR product for common MMTV sag sequences. Lanes (left to right): markers, C3H/BiDa, BR, PE, and CZ. The arrow indicates the position of the predicted 1,018-bp product. A PCR control is shown at the bottom of the figure. PCR was carried out as described in Materials and Methods.
Splenic T cells from parental and F1 mice were examined for Vβ gene expression by fluorescence-activated cell sorter (FACS) analysis by using a series of Vβ-specific antibodies (Table 5). BR mice express a limited repertoire of Vβ genes and are therefore useful as a “sag indicator” strain in genetic crosses with strains of uncertain status with respect to endogenous MMTVs. BR mice carry a large genomic deletion of Vβ genes, including Vβ genes 5, 8, 9, 11, 12, and 13 (3). They also carry several endogenous Mtvs which cause somatic deletion of Vβ genes 5, 11, 12, 16, and 17a (13, 22, 37). All Vβ genes expressed by BR mice were found to be expressed by PE and CZ mice, with BR mice expressing most types at higher percentages, a finding consistent with its more limited repertoire. The percentages in the F1s were intermediate and close to the average of the parental values, indicating the absence of deleting elements introduced by either the PE or CZ parents. This is particularly clear for Vβ6, which is expected to be of critical importance in this virus-host system. Direct measurement of CD8+ Vβ6+ cells showed these to be present at 4 to 5% in F1 mice, compared to 2% in PE and CZ mice and 7% in BR mice. The results shown in Table 5 confirm the absence of Vβ5 and Vβ8 in T cells from BR mice and their presence in PE and CZ mice. In the F1s with BR mice, Vβ5 is absent due to somatic deletion, while Vβ8 is present as expected. Of the remaining Vβ genes expressed by BR mice, only two (Vβ1 and -15) could not be examined due to the absence of available antibody reagents. These data show that neither PE nor CZ mice carry endogenous sags encoded by MMTVs or indeed by any other agent(s).
TABLE 5.
T-cell receptor Vβ representation in splenic T cellsa
| TCR antibody | % Fluorescence-positive cells (mean ± SE)
|
||||
|---|---|---|---|---|---|
| BR | PE | CZ | (PE × BR)F1 | (CZ × BR)F1 | |
| αβ | 73 ± 2 | 74 ± 3 | 72 ± 5 | 76 ± 4 | 73 ± 5 |
| Vβ2 | 19 ± 0 | 4 ± 0 | 4 ± 0 | 11 ± 0 | 12 ± 0 |
| Vβ3 | 7 ± 0 | 3 ± 0 | 1 ± 0 | 4 ± 1 | 3 ± 0 |
| Vβ4 | 10 ± 1 | 3 ± 0 | 6 ± 1 | 7 ± 0 | 10 ± 1 |
| Vβ5 | 0 | 8 ± 1 | 3 ± 0 | 0 | 0 |
| Vβ6 | 15 ± 1 | 5 ± 0 | 4 ± 0 | 11 ± 1 | 11 ± 2 |
| Vβ7 | 8 ± 0 | 3 ± 0 | 4 ± 0 | 4 ± 1 | 4 ± 1 |
| Vβ8 | 0 | 15 ± 1 | 17 ± 0 | 12 ± 1 | 13 ± 0 |
| Vβ10 | 0b | 3 ± 0 | 3 ± 0 | 3 ± 0 | 3 ± 0 |
| Vβ14 | 11 ± 1 | 4 ± 1 | 4 ± 0 | 7 ± 1 | 7 ± 0 |
Enriched splenic T cells were screened for the Vβ T-cell receptor (TCR) profile by using clonotypic specific MAbs. Positive cells were enumerated in spleens of different mouse strains as indicated. Fluorochrome-stained cells were analyzed on a Coulter Profile II flow cytometer. Each datum point represents the mean ± the standard error of four animals in each group. Zero corresponds to <0.2% positive cells.
Vβ10 shows considerable diversity among inbred strains and the antibody used here may react poorly with the Vβ10a present in BR mice (31).
MHC typing of PE and CZ mice.
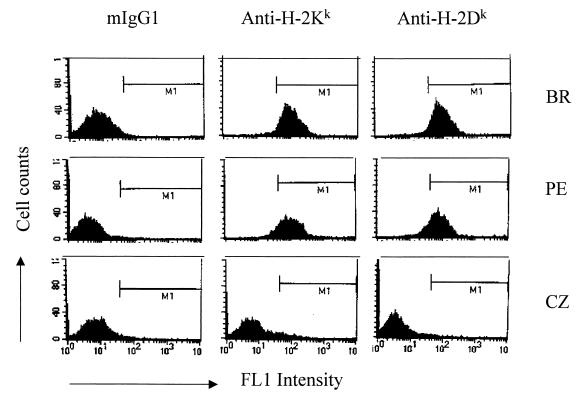
To determine whether either PE or CZ mice might share the H-2k haplotype common to highly susceptible standard inbreds, splenic leukocytes were analyzed by FACS with MAbs specific for H-2Dk and H-2Kk class I molecules (Fig. 3). Leukocytes from PE mice stained well with both antibodies, with results resembling the positive control from BR mice. Those from CZ mice were clearly negative. Similar results were found using antibody specific for class II (anti-IEk [data not shown]).
FIG. 3.
FACS analysis of H-2 haplotype in wild-derived mouse strains. Splenocytes from BR, PE, and CZ mice were stained with the FITC-labeled anti-MHC class I (H-2k) MAb. A monoclonal mouse IgG1 was used as a negative control. Spleen cells of BR mice served as a positive control. The analysis was performed by using a flow cytometer after gating on the viable cell population as determined by forward- and side-scatter analysis. Positive staining and fluorescence intensity (log scale) for 10,000 events are shown. The histograms represent one of three experiments. Similar results were obtained by using anti-class II (H-2k) MAb. The levels of surface positive cells were <1% with anti-H-2b class I MAb compared to >90% with anti-H-2k MAb (BR and PE mice).
Mixed lymphocyte reactions were also carried out as a functional test of histocompatibility (Table 6). In one-way tests, splenocytes from PE mice were unable to stimulate BR mice while those splenocytes from CZ and B6 mice, as a known MHC-incompatible strain (H-2b), were both positive. These data indicate that PE mice carry the H-2k haplotype and that CZ mice do not.
TABLE 6.
Mixed lymphocyte reactionsa
| Stimulator lymphocyte group | [3H]thymidine uptake (cpm)
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Medium | 2,228 | 2,191 |
| PE | 1,052 | 1,776 |
| CZ | 28,523 | 28,749 |
| BR | 950 | 931 |
| B6 | 37,304 | 37,882 |
Stimulator lymphocytes from spleens of different mouse strains were treated with mitomycin C. Responder cells were splenocytes from BR mice throughout. Reactions were performed in 96-well plates containing 4 × 105 stimulator lymphocytes plus 2 × 105 responder lymphocytes per well. The cells were incubated for 4 days and then for an additional 16 h in the presence of 1 μCi of [3H]thymidine per well. Thymidine incorporation was measured on a beta scintillation counter. Values are the means of triplicate cultures.
Antigen processing and presentation by PE and CZ mice.
Adherent cells from spleens of parental and F1 mice were pulsed with middle T-derived peptides and incubated with LN-13.1 virus-specific CTLs (Table 7). The R389 peptide sensitized normal cells from PE, PE × BR, and CZ × BR mice to killing by LN-13.1 but did not sensitize those from CZ mice. The E328 peptide had no effect. Similar results were obtained by pulsing peritoneal exudate cells with peptides (data not shown). These results are in line with those of MHC typing (Fig. 3 and Table 6) and demonstrate that killing is restricted to cells expressing H-2k. They also show that normal cells from tumor-susceptible F1 mice are able to present the immunodominant viral peptide on H-2Dk class I molecules.
TABLE 7.
Killing by CTLs of middle T peptide-pulsed targetsa
| Host strain | % Specific lysis of cells pulsed with peptide:
|
|
|---|---|---|
| mT-R389 | mT-E328 | |
| PE | 82 | 5 |
| CZ | 0 | 0 |
| (PE × BR)F1 | 74 | 2 |
| (CZ × BR)F1 | 93 | 0 |
Plastic-adherent cells from naive spleens were prepared from different mouse strains, pulsed with middle T (mT) peptides, and used as targets in the CTL assay at a 5:1 effector/target ratio.
To test directly for a possible defect in antigen processing, adherent spleen cells from parental and F1 mice were infected by polyomavirus in vitro and incubated with LN-13.1 cells (Table 8). Infected target cells from BR, PE, PE × BR, and CZ × BR mice were killed, while those from CZ mice were not, a finding consistent with the known pattern of MHC expression. Tumor-derived cells were also used as targets and found to follow the same pattern. Cells derived from a variety of different tumors (mammary gland, salivary gland, kidney, and fibrosarcoma) were tested; killing was MHC restricted but independent of the tissue origin or type of tumor. These results, achieved with both normal cells infected in vitro and cells derived from primary tumors as targets, establish that neither PE nor CZ mice (the latter deduced from results with CZ × BR mice) are defective in antigen processing and presentation with respect to the immunodominant viral peptide.
TABLE 8.
Killing by CTLs of infected targets and tumor cellsa
| Host strain | % Specific lysis
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Infected targets | ||
| BR | 33 | 47 |
| PE | 34 | 31 |
| CZ | 2 | 7 |
| (PE × BR)F1 | 21 | 29 |
| (CZ × BR)F1 | 22 | 25 |
| Tumor cell targets | ||
| BR (salivary) | 54 | 46 |
| PE (salivary) | 43 | 31 |
| CZ (fibrosarcoma) | 0 | 0 |
| (PE × BR)F1 (mammary) | 38 | 33 |
| (CZ × BR)F1 (mammary) | 35 | 39 |
The susceptibility of polyomavirus-infected and polyomavirus tumor cells to killing in vitro by LN-13.1 was checked in CTL assay at a 5:1 effector/target ratio. Splenic adherent cells were prepared and infected with A2 virus. Tumor cells were derived from polyomavirus-infected mice and maintained in DMEM containing 10% fetal calf serum. The 51Cr-labeled cells were incubated with the effector CTL line LN-13.1, and the specific lysis was calculated from triplicate wells. Specific lysis of tumor cells by concanavalin A-stimulated naive lymphocytes as a control was 2 to 4%. There was no detectable level of lysis when uninfected adherent cells were used as targets.
DISCUSSION
As a general rule, inheritance of immune responsiveness to a defined antigen is expected to show dominance over immune nonresponsiveness. With respect to polyomavirus-specific transplantation antigen(s), host resistance based on an effective immune response to virus-induced tumors should be dominant over susceptibility. An exception to this rule was first noted in standard inbred mouse strains where an endogenous superantigen gives rise to dominant nonresponsiveness by preventing the generation of polyomavirus tumor immunity (28, 30).
Here we describe a distinctly different genetic and immunological basis of susceptibility in two unrelated strains of wild-derived inbred mice. These mice are highly susceptible to tumor induction by polyomavirus but show no evidence of endogenous sags. In crosses with immunologically resistant BR mice, the wild-derived inbreds transmit their susceptibility in a dominant manner. We conclude that these mice have a sag-independent mechanism(s) operating to suppress the generation of polyomavirus tumor immunity.
No gross features indicative of immunological abnormality (splenomegaly, lymphadenopathy, or absence of thymic involution) in either PE or CZ mice have been observed by us, and none have been reported by others. A defect(s) in CTL effector functions (e.g., granzyme or perforase) could in principle explain the failure of PE and CZ mice to eliminate viral tumors. A simple absence of these functions, however, would not be transmitted as a dominant susceptible trait. Several additional observations rule against a defect at the level of CTL-tumor cell interaction in these mice. Tumor cells derived from PE, PE × BR, and CZ × BR F1 mice (but not from CZ mice which are non-H-2k) are susceptible to killing by virus-specific CTLs from BR mice. Normal cells from PE, PE × BR, and CZ × BR mice that are either infected in vitro by polyomavirus or incubated with specific viral peptide also become susceptible targets for these CTLs. Tumor cells derived from these mice thus express normal costimulatory functions and are not intrinsically resistant to T cell killing. Processing and presentation of viral antigen also appear to proceed normally. An absence of appropriate CTL precursors is ruled out since PE and CZ mice and their F1 progeny all show normal levels of CD8+ T cells bearing Vβ6.
A mechanism leading to CTL tolerance to viral antigen could account for the dominant susceptibility of PE and CZ mice. This could arise in principle by molecular mimicry in which a self-antigen effectively tolerizes the host against the virus, although the likelihood is remote that this would arise in two unrelated mouse strains. Alternatively, tolerance to viral antigen could be induced by cross-presentation (8, 23, 34), aided by virus-induced cytolysis and the high viral antigen load present in neonatally infected mice. Virus-specific CTLs without effector function can arise in chronically infected hosts under certain conditions (39). Other mechanisms not based on tolerance or nonresponsiveness are also possible. For example, expression of Fas ligand by tumor cells can lead to induction of apoptosis in tumor infiltrating lymphocytes (TIL) and thus to immune escape (20, 32, 33). Although not apparent in our assays of tumor cell killing in vitro, Fas ligand expression in vivo by tumors in PE and CZ mice, or other mechanisms leading to TIL dysfunction (12), may occur. Further experiments employing immunological as well as genetic approaches to map and identify the gene(s) should help to clarify the basis of susceptibility to virus-induced tumors in these mice.
ACKNOWLEDGMENTS
P.V. and I.Y. have contributed equally to this work.
We thank Martin Dorf for providing antibody reagents, for the use of a FACS, and for helpful discussions during preparation of the manuscript.
This work has been supported by a grant from the National Cancer Institute (R35 CA44343).
REFERENCES
- 1.Allison A C, Monga J N, Hammond V. Increased susceptibility to virus oncogenesis of congenitally thymus-deprived nude mice. Nature (London) 1974;252:746–747. doi: 10.1038/252746a0. [DOI] [PubMed] [Google Scholar]
- 2.Bauer P H, Cui C, Stehle T, Harrison S C, DeCaprio J A, Benjamin T L. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J Virol. 1999;73:5826–5832. doi: 10.1128/jvi.73.7.5826-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behlke M A, Chou H S, Huppi K, Loh D Y. Murine T-cell receptor mutants with deletions of β-chain variable region genes. Proc Natl Acad Sci USA. 1986;83:767–771. doi: 10.1073/pnas.83.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berke Z, Wen T, Klein G, Dalianis T. Polyoma tumor development in neonatally polyoma-virus-infected CD4−/− and CD8−/− single knockout and CD4−/−8−/− double knockout mice. Int J Cancer. 1996;67:405–408. doi: 10.1002/(SICI)1097-0215(19960729)67:3<405::AID-IJC15>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Beutner U, Frankel W N, Cote M S, Coffin J M, Huber B T. Mls-1 is encoded by the long terminal repeat open reading frame of the mouse mammary tumor provirus Mtv-7. Proc Natl Acad Sci USA. 1992;89:5432–5436. doi: 10.1073/pnas.89.12.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronson R, Dawe C, Carroll J, Benjamin T. Tumor induction by a transformation-defective polyoma virus mutant blocked in signaling through Shc. Proc Natl Acad Sci USA. 1997;94:7954–7958. doi: 10.1073/pnas.94.15.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callahan R, Drohan W, Gallahan D, D’Hoostelaere L, Potter M. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc Natl Acad Sci USA. 1982;79:4113–4117. doi: 10.1073/pnas.79.13.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone F R, Kurts C, Bennett S R M, Miller J F A P, Heath W R. Cross-presentation: a general mechanism for CTL immunity and tolerance. Immunol Today. 1998;19:368–373. doi: 10.1016/s0167-5699(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 9.Carroll J P, Fung J S, Bronson R T, Razvi E, Benjamin T L. Radiation-resistant and radiation-sensitive forms of host resistance to polyomavirus. J Virol. 1999;73:1213–1218. doi: 10.1128/jvi.73.2.1213-1218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawe C J, Freund R, Mandel G, Balmer-Hofer K, Talmage D A, Benjamin T L. Variations in polyoma virus genotype in relation to tumor induction in mice: characterization of wild type strains with widely differing tumor profiles. Am J Pathol. 1987;127:243–261. [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy B E. Polyoma virus. In: Gard S, Hallawer C, Meyer K F, editors. Virology monograph 7. New York, N.Y: Springer-Verlag; 1969. pp. 1–114. [Google Scholar]
- 12.Finke J, Ferrone S, Frey A, Mufson A, Ochoa A. Where have all the T cells gone? Mechanisms of immune evasion by tumors. Immunol Today. 1999;20:158–160. doi: 10.1016/s0167-5699(98)01435-2. [DOI] [PubMed] [Google Scholar]
- 13.Frankel W N, Rudy C, Coffin J M, Huber B T. Linkage of Mls genes to endogenous mammary tumour viruses of inbred mice. Nature (London) 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 14.Freund R, Calderone A, Dawe C J, Benjamin T L. Polyomavirus tumor induction in mice: effects of polymorphisms of VP1 and large T antigen. J Virol. 1991;65:335–341. doi: 10.1128/jvi.65.1.335-341.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund R, Dawe C J, Benjamin T L. Duplication of noncoding sequences in polyomavirus is required for the development of thymic tumors in mice. J Virol. 1988;62:3896–3899. doi: 10.1128/jvi.62.10.3896-3899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freund R, Dawe C J, Carroll J P, Benjamin T L. Changes in frequency, morphology and behavior of tumors induced in mice by a polyoma virus mutant with a specifically altered oncogene. Am J Pathol. 1992;141:1409–1425. [PMC free article] [PubMed] [Google Scholar]
- 17.Freund R, Dubensky T, Bronson R, Sotnikov A, Carroll J, Benjamin T. Polyoma tumorigenesis in mice: evidence for dominant resistance and dominant susceptibility genes of the host. Virology. 1992;191:724–731. doi: 10.1016/0042-6822(92)90248-n. [DOI] [PubMed] [Google Scholar]
- 18.Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross L G. Oncogenic viruses. 3rd ed. Vol. 2. Oxford, England: Pergamon Press, Inc.; 1983. pp. 737–828. [Google Scholar]
- 20.Hahne M, Rimoldi D, Schroter M, Romero P, Schreier M, French L E, Schneider P, Bornand T, Fontana A, Lienard D, Cerottini J-C, Tschopp J. Melanoma cell expression of Fas (Apo-1/CD95) ligand: implications for tumor immune escape. Science. 1996;274:1363–1366. doi: 10.1126/science.274.5291.1363. [DOI] [PubMed] [Google Scholar]
- 21.Ignatowicz L, Kappler J W, Marrack P, Scherer M T. Identification of two Vβ7-specific viral superantigens. J Immunol. 1994;152:65–71. [PubMed] [Google Scholar]
- 22.Kozak C, Peters G, Pauley R, Morris V, Michalides R, Dudley J, Green M, Davisson M, Prakash O, Vaidya A, Hilgers J, Verstraeten A, Hynes N, Diggelmann H, Peterson D, Cohen J C, Dickson C, Sarkar N, Nusse R, Varmus H, Callahan R. A standardized nomenclature for endogenous mouse mammary tumor viruses. J Virol. 1987;61:1651–1654. doi: 10.1128/jvi.61.5.1651-1654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurts C, Miller J F A P, Subramaniam R M, Carbone F R, Heath W R. Major histocompatibility complex class I-restricted cross-presentation is biased towards high dose antigens and those released during cellular destruction. J Exp Med. 1998;188:409–414. doi: 10.1084/jem.188.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laird P W, Zijderveld A, Linders K, Rudnicki M, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law L W. Immunologic responsiveness and the induction of experimental neoplasms. Cancer Res. 1966;26:1121–1132. [PubMed] [Google Scholar]
- 26.Law L W, Dawe C J. Influence of total body X-irradiation on tumor induction by parotid tumor agent in adult mice. Proc Soc Exp Biol Med. 1960;105:414–419. doi: 10.3181/00379727-105-26127. [DOI] [PubMed] [Google Scholar]
- 27.Law L W, Ting R C, Leckband E. Prevention of virus-induced neoplasms in mice through passive transfer of immunity by sensitized syngeneic lymphoid cells. Proc Natl Acad Sci USA. 1967;57:1068–1075. doi: 10.1073/pnas.57.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukacher A E, Ma Y, Carroll J P, Abromson-Leeman S R, Laning J C, Dorf M E, Benjamin T L. Susceptibility to tumors induced by polyoma virus is conferred by an endogenous mouse mammary tumor virus superantigen. J Exp Med. 1995;181:1683–1692. doi: 10.1084/jem.181.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukacher A E, Wilson C S. Resistance to polyoma virus-induced tumors correlates with CTL recognition of an immunodominant H-2Dk-restricted epitope in the middle T protein. J Immunol. 1998;160:1724–1734. [PubMed] [Google Scholar]
- 30.Lukacher A E, Freund R, Carroll J P, Bronson R T, Benjamin T L. Pyvs: a dominantly acting gene in C3H/BiDa mice conferring susceptibility to tumor induction by polyoma virus. Virology. 1993;196:241–248. doi: 10.1006/viro.1993.1472. [DOI] [PubMed] [Google Scholar]
- 31.Necker A, Rebai N, Matthes M, Jouvin-Marche E, Cazenave P A, Swarnworawong P, Palmer E, MacDonald H R, Malissen B. Monoclonal antibodies raised against engineered soluble mouse T cell receptors and specific for Vα8-, Vβ2- or Vβ10-bearing T cells. Eur J Immunol. 1991;21:3035–3040. doi: 10.1002/eji.1830211220. [DOI] [PubMed] [Google Scholar]
- 32.O’Connell J, Bennet M W, O’Sullivan G C, Collins J K, Shanahan F. The fas counterattack: cancer as a site of immune privilege. Immunol Today. 1999;20:46–52. doi: 10.1016/s0167-5699(98)01382-6. [DOI] [PubMed] [Google Scholar]
- 33.O’Connell J, O’Sullivan G C, Collins J K, Shanahan F. The fas counterattack: Fas-mediated T cell killing by colon cancer cells expressing fas ligand. J Exp Med. 1996;184:1075–1082. doi: 10.1084/jem.184.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolph M S, Matthaei K I, Carbone F R, Heath W R, Ramshaw I A. Loss of antiviral cytotoxic T-lymphocyte activity during high level antigen stimulation. Viral Immunol. 1998;11:183–195. doi: 10.1089/vim.1998.11.183. [DOI] [PubMed] [Google Scholar]
- 35.Rudy C K, Kraus E, Palmer E, Huber B T. Mls-1-like superantigen in the MA/MyJ mouse is encoded by a new mammary tumor provirus that is distinct from Mtv-7. J Exp Med. 1992;175:1613–1621. doi: 10.1084/jem.175.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahli R, Freund R, Dubensky T, Garcea R, Bronson R, Benjamin T. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology. 1993;192:142–143. doi: 10.1006/viro.1993.1016. [DOI] [PubMed] [Google Scholar]
- 37.Sherer M T, Ignatowicz L, Winslow G M, Kappler J W, Marrack P. Superantigens: bacterial and viral proteins that manipulate the immune system. Annu Rev Cell Biol. 1993;9:101–128. doi: 10.1146/annurev.cb.09.110193.000533. [DOI] [PubMed] [Google Scholar]
- 38.Talmage D A, Freund R, Young A T, Dahl J, Dawe C J, Benjamin T L. Phosphorylation of middle T by pp60c-src: a switch for binding of phosphatidylinositol 3-kinase and optimal tumorigenesis. Cell. 1989;59:55–65. doi: 10.1016/0092-8674(89)90869-6. [DOI] [PubMed] [Google Scholar]
- 39.Zajac A J, Blattman J N, Murali-Krishna K, Sourdive D J D, Suresh M, Altman J D, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]