Abstract
Purpose of review
To review novel experimental approaches for studying host:microbe interactions and their role in intestinal and systemic inflammation in people living with HIV (PLWH).
Recent findings
Inflammation in PLWH is impacted by interactions between the microbiome, the intestinal epithelium, and immune cells. This complex interplay is not fully understood and requires a variety of analytical techniques to study. Using a multiomic systems biology approach provides hypothesis generating data on host:microbe interactions that can be used to guide further investigation. The direct interactions between host cells and microbes can be elucidated using peripheral blood mononuclear cells (PBMCs), lamina propria mononuclear cells (LPMC's) or human intestinal organoids (HIO). Additionally, the broader relationship between the host and the microbiome can be explored using animal models such as nonhuman primates and germ-free and double humanized mice.
Summary
To explore complex host:microbe relationships, hypotheses are generated and investigations are guided by multiomic data, while causal components are identified using in-vitro and in-vivo assays.
Keywords: HIV, human intestinal organoids, humanized mice, microbiome dysbiosis, multiomic systems biology
INTRODUCTION
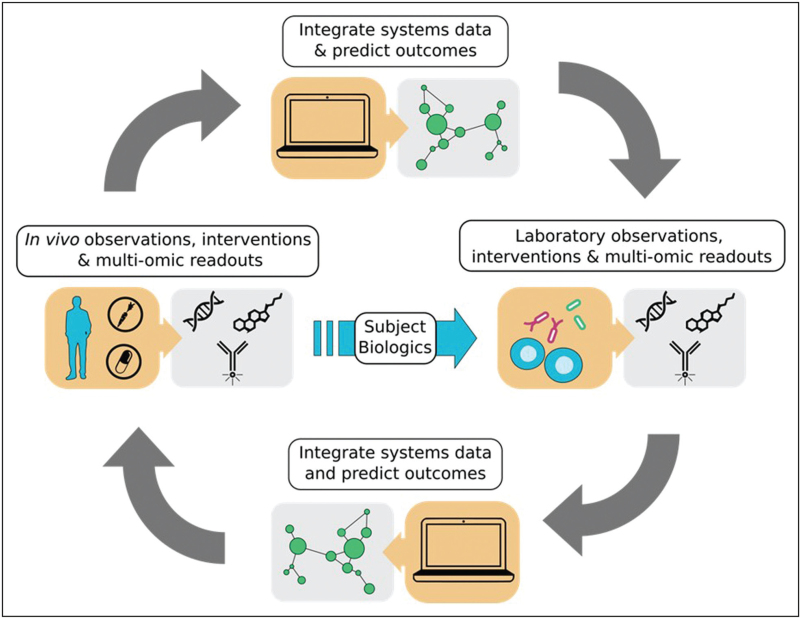
It is well understood that during initial HIV infection, CD4+ T cells in the gut-associated lymphoid tissue (GALT) are infected and depleted. The loss of CD4+ T cells and the ensuing inflammation associated with HIV infection leads to the breakdown of the intestinal barrier and translocation of bacterial products from the intestinal lumen into extraintestinal sites, resulting in systemic chronic immune activation and disease progression. While antiretroviral therapy (ART) improves CD4+ T cell numbers, decreases immune activation, and reduces viral reservoirs, changes in the intestinal microbiome, viral replication and incomplete immune reconstitution persists, including reduced frequencies of Th17 cells in the lamina propria which have pro-barrier and antimicrobial functions [1]. Furthermore, the majority of people living with HIV (PLWH) in the United States and Europe are MSM, who possess a unique inflammatory gut microbiome as well as more subtle microbiome differences associated with HIV infection itself. [2,3▪]. Thus, understanding host-microbiome interactions is vital to determine their contribution to intestinal inflammation, disease pathogenesis, and multiple comorbidities in PLWH. While there has been progress towards understanding the degree to which an altered host microbiome may directly drive intestinal and peripheral immune activation, the mechanisms by which bacteria and their metabolites mediate inflammation is still not fully understood, mainly due to the complexity of studying these interactions. Studying these interactions can effectively be accomplished using a combination of hypothesis generating multiomic tools and in-vitro and in-vivo methods to determine causality (Fig. 1).
FIGURE 1.
Workflow from systems data to exploratory experiments. Multiomics systems data can be used to generate hypotheses that can be investigated using in-vitro and in-vivo experimental approaches to elucidate the mechanisms of the host:microbiome interactions.
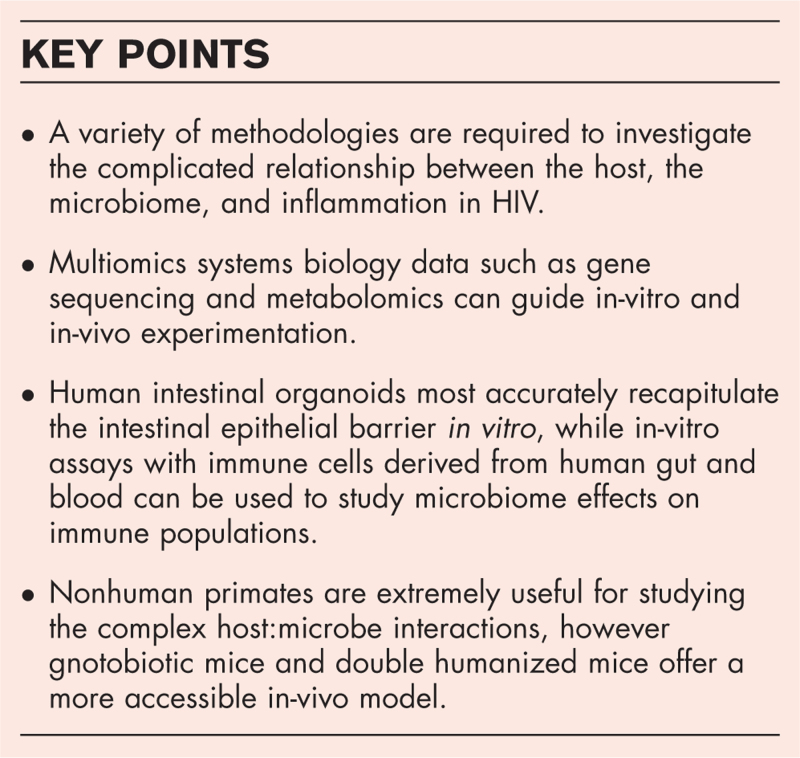
Box 1.
no caption available
MULTIOMIC SYSTEMS BIOLOGY APPROACHES
By using samples collected from PLWH, multiomic systems biology data can guide our understanding of the complex interplay between HIV infection, the gut microbiome, intestinal and peripheral inflammation, and downstream effects on disease pathogenesis and co-morbidities. In one recent study that analyzed fecal samples from MSM with and without HIV in Thailand using 16S ribosomal RNA (rRNA) gene sequencing, Sortino et al.[4] revealed that during untreated infection, pro-inflammatory bacteria were enriched as individuals transitioned from acute infection, and that Fusobacteria, which were enriched post-ART, positively correlated with levels of soluble CD14. Other 16S-based studies have revealed that chronic HIV infection, ART-treated and untreated, consistently confers specific microbial signatures, including reduced alpha diversity as well as reduced abundance of butyrate-producing bacteria [2,5–8]. This combined with the fact that Fusobacterium and Serratia abundances have been shown to be strong predictors of immune recovery in PLWH on ART, underscores the continued role the microbiome may play in immune reconstitution [9,10].
There has been recent interest in the contribution of other components of the microbiome, including fungi and viruses, to HIV pathophysiology. β-D-glucan in serum has been shown to be positively correlated with markers of systemic inflammation, indicating that fungal translocation may contribute to intestinal inflammation in HIV infection, even after administration of ART [11]. Shotgun metagenomics analysis of fecal samples has also shown HIV infection to be associated with a decrease in richness of bacteriophages, and increases in HIV-related viruses, including anelloviruses and adenovirus, which both exhibited strong relationships with HIV viral load expansion and CD4+ T cell count depletion [12▪,13].
In addition to characterization of gut microbiome composition, useful information regarding microbial function in the context of chronic HIV-related immune activation has been provided by transcriptomic, metagenomic, and metabolomic analyses. Vázquez-Castellanos et al.[14] combined fecal metagenomic and meta-transcriptomic analyses to reveal that HIV infection associates with microbial over-expression of genes associated with resistance to oxidative stress and under-expression of anti-inflammatory processes, including short chain fatty acid (SCFA) biosynthesis and indole production. Furthermore, untargeted metabolomics analysis of fecal samples in PLWH revealed that microbially-associated metabolites differ in individuals with HIV and correlate with microbial translocation and immune activation [6]. The gut microbiome exhibits increased expression of tryptophan metabolism genes and decreased expression of fatty acids metabolism genes during HIV infection, which both appear to be associated with disturbance of barrier integrity and bacterial translocation [15,16]. PLWH have also been shown to have reduced levels of certain microbially-produced metabolites in serum, including secondary bile acids and propionate, and that the latter preceded mortality [17▪▪,18▪].
Recent ex-vivo studies have highlighted potential mechanisms linking the gut microbiome to modulation of innate and adaptive host immune response in HIV infection. Production of SCFAs, which has been shown to be reduced in the gut of PLWH, is important for innate and adaptive immunity, as SCFAs promote gut homeostasis, gut barrier integrity, gut epithelial cell energy sourcing, and limited inflammation [17▪▪,19,20]. Characterization of gut immune cells has revealed elevated neutrophils in rectal biopsy tissue from PLWH [21▪]. This enhanced accumulation was associated with a reduced Lactobacillus:Prevotella ratio, and Lactobacillus was shown in vitro to decrease neutrophil survival and frequency [22]. Additionally, a study that examined levels of 28 fecal soluble immune factors (sIFs) in MSM with and without HIV and non-MSM without HIV found a significant relationship between these immune factors and fecal microbiome compositions [3▪]. These findings suggest there is a clear relationship between HIV related gut microbiome dysbiosis and alterations in innate immunity. Adaptive immune phenotypes are also associated with distinct features of the HIV-associated gut microbiome. SahBandar et al.[23] revealed that Fusobacterium abundance in PLWH is associated with dysfunction in CD8+ T cell responses, while Nowak et al.[24] found that reduced alpha diversity in PLWH correlated with reduced CD4+ T cell count and increased monocyte activation.
IN-VITRO METHODS
While exploring associations in multiomic data is useful for generating hypotheses regarding relationships between the host immune system and intestinal microbes, they do not typically provide information on causality. In contrast, in-vitro assays allow for causal mechanisms to be tested directly. In-vitro assays can be performed using immune cells isolated from the periphery, peripheral blood mononuclear cells (PBMC), or the intestine, lamina propria mononuclear cells (LPMC) [25,26]. These immune cells can be stimulated with killed or live bacteria; killed bacteria cannot overgrow cultures but are unable to produce biologically active metabolites while the opposite is true for live bacteria. For example, PBMC stimulated with fixed bacteria isolates induced MAIT cell activation and a subsequent loss of MAIT cells, which is observed during HIV infection [27]. Fecal bacteria communities (FBC), which are made by isolating the whole gut microbial community from stool, have been used to show that fecal microbiota from MSM with and without HIV induce more activation and pro-inflammatory cytokines from PBMCs than fecal microbiota from men who sex with women (MSW) and women without HIV [25]. To model gut immune response more accurately, LPMCs, which are typically derived from otherwise discarded tissue from gut resection surgeries, can be used. Using LPMC stimulated with single bacterial cultures, Castleman et al.[26] found that live Actinetobacter junii, Prevotella stercorea, and Salmonella typhimurium increased IL-22 production from ILC3 s and that the production of IL-22 was linked to the presence of myeloid dendritic cells. In another study, LPMCs stimulated with heat-killed FBCs from MSM with and without HIV and challenged with HIV were found to promote elevated levels of HIV infection than FBCs from MSW without HIV [28].
While studies using PBMC and LPMC are valuable for understanding immune responses, they ignore epithelial cells, which are important for fully understanding host:microbe interactions. Epithelial cells isolated directly ex vivo exhibit poor survival in culture. To address this, immortalized epithelial cell lines, such as Caco-2 or T84 in a monolayer, are used to investigate membrane integrity and barrier function in the settings of bacterial infection or bacterial products [3▪,29▪,30]. These cells can be used to monitor barrier function by measuring trans-epithelial electrical resistance (TEER) and quantifying cytokine and tight junction protein expression using RT-qPCR [29▪,30]. Culturing these cell lines with bacteria and their metabolites can have varying effects on membrane integrity. For example, Lacticaseibacillus paracasei improved the barrier integrity of Caco-2 s treated with LPS and reduced the pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 [29▪]. Soluble immune factors isolated from feces from PLWH are pro-inflammatory and induce barrier disruption in T84 when compared to those from seronegative individuals [3▪]. Butyrate also has a dose dependent effect on Caco-2 s with low doses increasing barrier function while high doses decrease integrity [30]. However, transformed cell lines have limitations in their usefulness because they do not express the full repertoire of epithelial cells contained in the intestinal barrier.
Human intestinal organoids (HIOs) derived from primary epithelial tissue contain all of the cell types found in the epithelium including enteroendocrine cells, Paneth cells, goblet cells, enterocytes, and intestinal stem cells, as well as rarer cell types such as tuft cells and m-cells [31,32]. This broad repertoire of epithelial cells better recapitulates the intestine. HIO can be plated into a monolayer to investigate barrier function, and a greater repertoire of cytokine production [33]. When used as intact vesicles, bacteria or metabolites are injected into the lumen exposing the apical side as it is in the gut [34]. Injecting live probiotic Escherichia coli strain Nissle protected the epithelial barrier from degradation by live pathogenic E. coli strains CFT073 and 0157:H7 as measured by a FITC-dextran assay and prevented reactive oxygen species generation and apoptosis [34]. However, microinjections are technically difficult, so apical out organoids can be generated by treating HIO with EDTA and suspending organoids in media for 3 days [35]. Apical out HIO that tolerate hypoxic environments have been developed, allowing for co-culture of anaerobic bacteria to model the host epithelial and microbiome interactions [36▪▪]. Using an apical out HIO, the commensal bacteria Lactobacillus casei and Bifidobacterium longum have been shown to colonize the apical side of the organoid and display probiotic benefits such as increased barrier integrity measured using FITC-dextran [36▪▪]. HIOs allow for the study of the host epithelial and microbiome interactions but are missing the influence of innate and adaptive immune cells that reside in the lamina propria and intestinal epithelium.
IN-VIVO METHODS
While in-vitro models provide valuable insights, they cannot fully replicate all the dynamic interactions between the microbiome and host. Thus, animal models offer a more comprehensive system to better understand how microbes interact with the host while still allowing for the manipulation of experimental variables. Germ-free or gnotobiotic mice have become the standard for assessing the impact of the microbiome on host parameters. Fecal microbial communities or cultured individual microbes can be introduced to gnotobiotic mice via oral gavage into the sterile intestine [37]. These mice are bred in isolators to maintain a germ-free environment and mice can be housed separately to prevent cross contamination [38]. Several gnotobiotic mouse studies have found correlations between microbiome composition and host factors such as genetics, diet, and environmental exposures, including pathogens [39–42]. Germ-free mice have been used to study how the HIV associated microbiome impacts the immune system. For example, germ-free mice transplanted with feces from MSM with and without HIV exhibited higher immune activation than mice transplanted with feces from MSW without HIV [28].
While valuable information on the microbiome can be gleaned with these models, most gnotobiotic mice are not suited for studying HIV infection directly as HIV does not typically infect nonhuman species. To overcome this, nonhuman primates (NHP), which are physiologically similar to humans, can be infected with a retrovirus akin to HIV, namely Simian immunodeficiency virus (SIV) or the chimeric virus Simian-HIV (SHIV). Using NHPs, a temporary spike in the abundance of Proteobacteria was observed in acutely SIV-infected macaques, providing support that retroviral infection impacts the microbiome; however, no significant change in gut bacteria was observed in animals with chronic infection [43–46]. More recently, intestinal epithelial barrier damage and microbial translocation were found in ART-treated SIV-infected rhesus macaques [47]. Interestingly, these effects were associated with a continued loss of the butyrate-producing bacterial species Faecalibacterium prausnitzii and Treponema succinifaciens, suggesting a potential metabolic contribution to gut inflammation even with effective control of viremia. In a recent study, vancomycin was used to induce dysbiosis before exposing macaques to low-dose intrarectal SIV challenge [48▪▪]. This antibiotic-induced dysbiosis disrupted antimicrobial immunity, including reduced frequencies of Th17 and Th22 cells and increased expression of host bacterial sensors and antibacterial peptides which increased susceptibility to SIV infection. Interestingly, in another study, fecal microbiota transplants from healthy macaque donors into antibiotic-treated SIV-infected macaques were associated with increases in Th17 and Th22 cells as well as a decrease in the frequency of activated CD4+ T cells when compared to pre-FMT within the same animal [49]. Diet modification has also been shown to affect SIV disease progression in NHP, with a high fat diet exacerbating disease through increased cell-associated SIV DNA and RNA [50].
A less expensive alternative to NHPs is humanized mice (Hu-mice), which are engineered to have a human immune system by engrafting immunodeficient mice with human immune cells and/or tissues. Unlike traditional mouse models and NHPs, Hu-mouse models can effectively sustain HIV infection while also recapitulating relatively accurate in-vivo immune responses. Gnotobiotic humanized or “double humanized” mice, are humanized immune system mice that have also been colonized with human microbiomes via gavage of human fecal material and are a promising model system for testing the effects of human microbes on HIV infection and disease progression. Recently, using Hu-mice, Whal et al. showed that the presence of resident microbiota markedly increased the susceptibility to HIV infection and enhanced viral replication [51▪]. Using a double Hu-mouse model, Lohani et al.[52▪] showed that a high fat diet increased systemic inflammation, immune activation, and rectal HIV transmission, a finding that is consistent with that of NHP models. These findings highlight the importance of the gut microbiome in the systemic immune activation, inflammation, and transmission in HIV infection and the viability of double Hu-mice as a model to investigate the immune, epithelial, and microbiome compartments simultaneously.
CONCLUSION
Investigating the link between inflammation and the microbiome in PLWH involves complex methods. Multiomic systems biology approaches can be used to identify novel associations and generate hypotheses to test. In-vitro studies offer insights into causal components of host:microbe relationships using both immune cells to study host immune responses and organoids which provide information on epithelial cell interactions with the microbiome. For even more comprehensive studies, in-vivo models provide a more complete picture of host:microbe interactions. Together, these approaches can be used to better understand the mechanisms by which the microbiome can modulate inflammation in PLWH.
Acknowledgements
None.
Financial support and sponsorship
This study was funded by grants R01-DK131581 (B.E.P, C.A.L.) and K01DK121864 (C.P.N.).
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Lazzaro A, Innocenti GP, Santinelli L, et al. Antiretroviral therapy dampens mucosal CD4(+) T lamina propria lymphocytes immune activation in long-term treated people living with HIV-1. Microorganisms 2021; 9:1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong AJS, Shaffer M, Nusbacher NM, et al. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2018; 6:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3▪.Littlefield KM, Schneider JM, Neff CP, et al. Elevated inflammatory fecal immune factors in men who have sex with men with HIV associate with microbiome composition and gut barrier function. Front Immunol 2022; 13:1072720. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigated the relationship between soluble immune factors from feces and serum cytokine levels, finding that pro-inflammatory soluble immune factors were higher in feces from MSM, regardless of HIV status, and increased transcellular gut epithelial permeability in vitro which is associated with increased bacterial translocation.
- 4.Sortino O, Phanuphak N, Schuetz A, et al. Impact of acute HIV infection and early antiretroviral therapy on the human gut microbiome. Open Forum Infect Dis 2020; 7:ofz367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocafort M, Noguera-Julian M, Rivera J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome 2019; 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Xie Z, Zhou J, et al. The altered metabolites contributed by dysbiosis of gut microbiota are associated with microbial translocation and immune activation during HIV infection. Front Immunol 2022; 13:1020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali Z, Shahzadi I, Majeed A, et al. Comparative analysis of the serum microbiome of HIV infected individuals. Genomics 2021; 113:4015–4021. [DOI] [PubMed] [Google Scholar]
- 8.Su F, Xia Y, Huang M, et al. Expression of NLPR3 in psoriasis is associated with enhancement of interleukin-1beta and Caspase-1. Med Sci Monit 2018; 24:7909–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SC, Chua LL, Yap SH, et al. Enrichment of gut-derived Fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Sci Rep 2018; 8:14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nganou-Makamdop K, Talla A, Sharma AA, et al. Translocated microbiome composition determines immunological outcome in treated HIV infection. Cell 2021; 184:3899–3914. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner LD, Retuerto M, Hager CL, et al. Fungal translocation is associated with immune activation and systemic inflammation in treated HIV. AIDS Res Hum Retroviruses 2019; 35:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪.Villoslada-Blanco P, Perez-Matute P, Iniguez M, et al. Impact of HIV infection and integrase strand transfer inhibitors-based treatment on the gut virome. Sci Rep 2022; 12:21658. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigated the gut virome of PLWH and found that HIV decreased phage richness, and that decreased richness could be partially remedied with integrase strand transfer inhibitors.
- 13.Liu K, Li Y, Xu R, et al. HIV-1 infection alters the viral composition of plasma in men who have sex with men. mSphere 2021; 6:e00081-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez-Castellanos JF, Serrano-Villar S, Jimenez-Hernandez N, et al. Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J 2018; 12:1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alzahrani J, Hussain T, Simar D, et al. Inflammatory and immunometabolic consequences of gut dysfunction in HIV: parallels with IBD and implications for reservoir persistence and non-AIDS comorbidities. EBioMedicine 2019; 46:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellano P, Prevedel L, Valdebenito S, Eugenin EA. HIV infection and latency induce a unique metabolic signature in human macrophages. Sci Rep 2019; 9:3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17▪▪.Sereti I, Verburgh ML, Gifford J, et al. Impaired gut microbiota-mediated short-chain fatty acid production precedes morbidity and mortality in people with HIV. Cell Rep 2023; 42:113336. [DOI] [PMC free article] [PubMed] [Google Scholar]; Studying comordities and mortality among PLWH, they found that a reduction of serum short chain fatty acids in PLWH as well as an altered microbiome preceded comorbity onset and mortality.
- 18▪.Fulcher JA, Li F, Tobin NH, et al. Gut dysbiosis and inflammatory blood markers precede HIV with limited changes after early seroconversion. EBioMedicine 2022; 84:104286. [DOI] [PMC free article] [PubMed] [Google Scholar]; This longitudinal study investigated the microbiome, serum metabolome and cytokines. They found that elevated pro-inflammatory cytokines, bioactive lipids and an altered microbiome may precede HIV infection.
- 19.Lu W, Feng Y, Jing F, et al. Association between gut microbiota and CD4 recovery in HIV-1 infected patients. Front Microbiol 2018; 9:1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Zhang P, Shen L, et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci 2020; 21: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Van Doren VE, Smith SA, Hu YJ, et al. HIV, asymptomatic STI, and the rectal mucosal immune environment among young men who have sex with men. PLoS Pathog 2023; 19:e1011219. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study examined blood and tissue samples and showed correlations between bacterial STI and HIV infection and inflammation and that HIV is associated with cellular composition changes in the rectum.
- 22.Hensley-McBain T, Wu MC, Manuzak JA, et al. Increased mucosal neutrophil survival is associated with altered microbiota in HIV infection. PLoS Pathog 2019; 15:e1007672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SahBandar IN, Chew GM, Corley MJ, et al. Changes in gastrointestinal microbial communities influence HIV-specific CD8+ T-cell responsiveness to immune checkpoint blockade. AIDS 2020; 34:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowak P, Troseid M, Avershina E, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 2015; 29:2409–2418. [DOI] [PubMed] [Google Scholar]
- 25.Neff CP, Krueger O, Xiong K, et al. Fecal microbiota composition drives immune activation in HIV-infected individuals. EBioMedicine 2018; 30:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castleman MJ, Dillon SM, Purba CM, et al. Commensal and pathogenic bacteria indirectly induce IL-22 but not IFNgamma production from human colonic ILC3 s via multiple mechanisms. Front Immunol 2019; 10:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberhard JM, Hartjen P, Kummer S, et al. CD161+ MAIT cells are severely reduced in peripheral blood and lymph nodes of HIV-infected individuals independently of disease progression. PLoS One 2014; 9:e111323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SX, Sen S, Schneider JM, et al. Gut microbiota from high-risk men who have sex with men drive immune activation in gnotobiotic mice and in vitro HIV infection. PLoS Pathog 2019; 15:e1007611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29▪.Xie Z, Zhang G, Liu R, et al. Heat-killed Lacticaseibacillus paracasei repairs lipopolysaccharide-induced intestinal epithelial barrier damage via MLCK/MLC pathway activation. Nutrients 2023; 15: [DOI] [PMC free article] [PubMed] [Google Scholar]; This study found that Caco-2 barrier function can be improved with culture of Lactocaseibacillus paracasei after treatment of LPS.
- 30.Peng L, He Z, Chen W, et al. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res 2007; 61:37–41. [DOI] [PubMed] [Google Scholar]
- 31.Yin X, Farin HF, van Es JH, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 2014; 11:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011; 141:1762–1772. [DOI] [PubMed] [Google Scholar]
- 33.Roodsant T, Navis M, Aknouch I, et al. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front Cell Infect Microbiol 2020; 10:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pradhan S, Weiss AA. Probiotic properties of Escherichia coli nissle in human intestinal organoids. mBio 2020; 11:e01470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Co JY, Margalef-Catala M, Monack DM, Amieva MR. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat Protoc 2021; 16:5171–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36▪▪.Kakni P, Jutten B, Teixeira Oliveira Carvalho D, et al. Hypoxia-tolerant apical-out intestinal organoids to model host-microbiome interactions. J Tissue Eng 2023; 14:20417314221149208. [DOI] [PMC free article] [PubMed] [Google Scholar]; This novel method of generating an apical out organoid that tolerates hypoxic environments allows for the culturing of epithelial cells with live anaerobic bacteria.
- 37.Bokoliya SC, Dorsett Y, Panier H, Zhou Y. Procedures for fecal microbiota transplantation in murine microbiome studies. Front Cell Infect Microbiol 2021; 11:711055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol 2018; 9:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dabrowska K, Witkiewicz W. Correlations of host genetics and gut microbiome composition. Front Microbiol 2016; 7:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009; 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu K, Warner G, Nowak RA, et al. The impact of environmental chemicals on the gut microbiome. Toxicol Sci 2020; 176:253–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darnaud M, De Vadder F, Bogeat P, et al. A standardized gnotobiotic mouse model harboring a minimal 15-member mouse gut microbiota recapitulates SOPF/SPF phenotypes. Nat Commun 2021; 12:6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glavan TW, Gaulke CA, Hirao LA, et al. SIV-infection-driven changes of pattern recognition receptor expression in mesenteric lymph nodes and gut microbiota dysbiosis. J Med Primatol 2015; 44:241–252. [DOI] [PubMed] [Google Scholar]
- 44.Klase Z, Ortiz A, Deleage C, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol 2015; 8:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handley SA, Thackray LB, Zhao G, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 2012; 151:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna P, Hoffmann C, Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog 2008; 4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanes C, Walker EM, Slisarenko N, et al. Gut microbiome changes associated with epithelial barrier damage and systemic inflammation during antiretroviral therapy of chronic SIV infection. Viruses 2021; 13:1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪▪.Ortiz AM, Baker PJ, Langner CA, et al. Experimental bacterial dysbiosis with consequent immune alterations increase intrarectal SIV acquisition susceptibility. Cell Rep 2023; 42:112020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Macaques were treated with vancomycin inducing dysbiosis and then challenged with SIV. Dysbiosis lowered TH17 and TH22 cell frequencies and the disrupted antimicrobial immunity and increased lentiviral infection.
- 49.Hensley-McBain T, Zevin AS, Manuzak J, et al. Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus-infected macaques. J Virol 2016; 90:4981–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He T, Xu C, Krampe N, et al. High-fat diet exacerbates SIV pathogenesis and accelerates disease progression. J Clin Invest 2019; 129:5474–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51▪.Wahl A, Yao W, Liao B, et al. A germ-free humanized mouse model shows the contribution of resident microbiota to human-specific pathogen infection. Nat Biotechnol 2023; [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study examined HIV infection in germ-free and conventional humanized mice and found that conventional humanized mice had increased HIV infection compared to germ-free mice, which was due to the microbiome.
- 52▪.Lohani SC, Ramer-Tait AE, Li Q. High-fat diet feeding exacerbates HIV-1 rectal transmission. mSystems 2024; 9:e0132223. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study using double humanized mice showed that a high fat diet elevates inflammation, immune activation, and rectal HIV infection.