Abstract
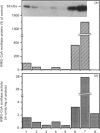
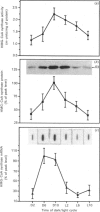
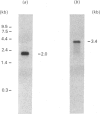
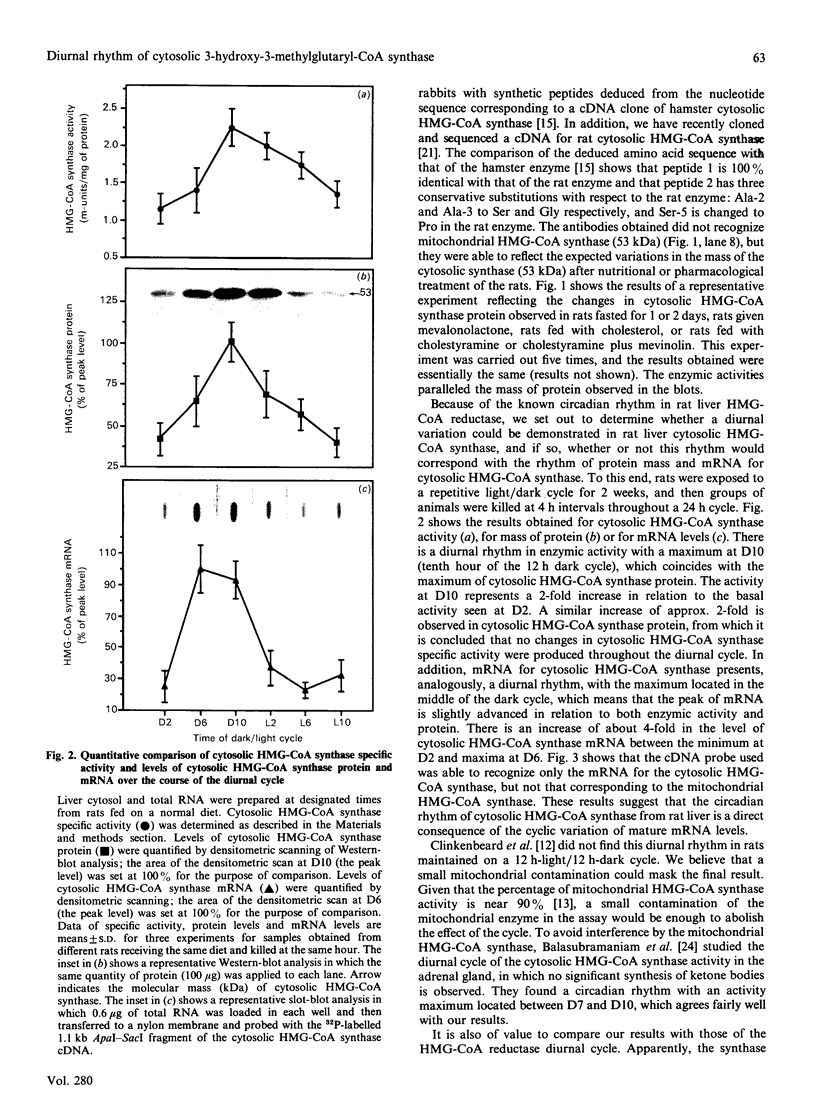
Rat liver cytosolic 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase exhibits a diurnal rhythm of enzyme activity which coincides with the diurnal rhythm of HMG-CoA synthase protein. The peaks of activity and protein (determined by SDS/PAGE and immunoblotting) both occur at D10 (the tenth hour of the daily 12 h dark cycle). The peak of mRNA levels (measured by slot-blot hybridization of liver RNA) is slightly advanced with respect to that of protein, by about 4 h, and shows a maximum at D6. Cytosolic HMG-CoA synthase activity and protein in rats fed on a normal diet were approx. 2-fold higher during the peak at D10 than in the nadir at D2. HMG-CoA synthase mRNA levels were approx. 4-fold higher during the peak at D6 than in the nadir at D2. These results point to a transcriptional and translational regulation of the cytosolic HMG-CoA synthase.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayté J., Gil-Gómez G., Haro D., Marrero P. F., Hegardt F. G. Rat mitochondrial and cytosolic 3-hydroxy-3-methylglutaryl-CoA synthases are encoded by two different genes. Proc Natl Acad Sci U S A. 1990 May;87(10):3874–3878. doi: 10.1073/pnas.87.10.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam S., Goldstein J. L., Brown M. S. Regulation of cholesterol synthesis in rat adrenal gland through coordinate control of 3-hydroxy-3-methylglutaryl coenzyme A synthase and reductase activities. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1421–1425. doi: 10.1073/pnas.74.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg Z. H., Allmann D. W., Gibson D. M. Modulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity with cAMP and wth protein fractions of rat liver cytosol. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1362–1369. doi: 10.1016/0006-291x(73)91137-6. [DOI] [PubMed] [Google Scholar]
- Beisiegel U., Schneider W. J., Brown M. S., Goldstein J. L. Immunoblot analysis of low density lipoprotein receptors in fibroblasts from subjects with familial hypercholesterolemia. J Biol Chem. 1982 Nov 10;257(21):13150–13156. [PubMed] [Google Scholar]
- Bové J., Hegardt F. G. Reversible modulation of rat liver 3-hydroxy 3-methyl glutaryl coenzyme A reductase. Evidence for an enzyme-catalyzed phosphorylation-dephosphorylation system. FEBS Lett. 1978 Jun 15;90(2):198–202. doi: 10.1016/0014-5793(78)80368-8. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Diurnal rhythm of rat liver mRNAs encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase. Correlation of functional and total mRNA levels with enzyme activity and protein. J Biol Chem. 1984 Aug 25;259(16):10439–10447. [PubMed] [Google Scholar]
- Clinkenbeard K. D., Reed W. D., Mooney R. A., Lane M. D. Intracellular localization of the 3-hydroxy-3-methylglutaryl coenzme A cycle enzymes in liver. Separate cytoplasmic and mitochondrial 3-hydroxy-3-methylglutaryl coenzyme A generating systems for cholesterogenesis and ketogenesis. J Biol Chem. 1975 Apr 25;250(8):3108–3116. [PubMed] [Google Scholar]
- Douvas A. S., Stumph W. E., Reyes P., Tan E. M. Isolation and characterization of nuclear ribonucleoprotein complexes using human anti-nuclear ribonucleoprotein antibodies. J Biol Chem. 1979 May 10;254(9):3608–3616. [PubMed] [Google Scholar]
- Gibbons G. F., Björnsson O. G., Pullinger C. R. Evidence that changes in hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase activity are required partly to maintain a constant rate of sterol synthesis. J Biol Chem. 1984 Dec 10;259(23):14399–14405. [PubMed] [Google Scholar]
- Gil G., Goldstein J. L., Slaughter C. A., Brown M. S. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. I. Isolation and sequencing of a full-length cDNA. J Biol Chem. 1986 Mar 15;261(8):3710–3716. [PubMed] [Google Scholar]
- Long B. H., Huang C. Y., Pogo A. O. Isolation and characterization of the nuclear matrix in Friend erythroleukemia cells: chromatin and hnRNA interactions with the nuclear matrix. Cell. 1979 Dec;18(4):1079–1090. doi: 10.1016/0092-8674(79)90221-6. [DOI] [PubMed] [Google Scholar]
- Lowe D. M., Tubbs P. K. 3-Hydroxy-3-methylglutaryl-coenzyme A synthase from ox liver. Purification, molecular and catalytic properties. Biochem J. 1985 Apr 15;227(2):591–599. doi: 10.1042/bj2270591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskey K. L., Chin D. J., MacDonald R. J., Liscum L., Goldstein J. L., Brown M. S. Identification of a cholesterol-regulated 53,000-dalton cytosolic protein in UT-1 cells and cloning of its cDNA. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6210–6214. doi: 10.1073/pnas.79.20.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marglin A., Merrifield R. B. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1970;39:841–866. doi: 10.1146/annurev.bi.39.070170.004205. [DOI] [PubMed] [Google Scholar]
- Mehrabian M., Callaway K. A., Clarke C. F., Tanaka R. D., Greenspan M., Lusis A. J., Sparkes R. S., Mohandas T., Edmond J., Fogelman A. M. Regulation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A synthase and the chromosomal localization of the human gene. J Biol Chem. 1986 Dec 5;261(34):16249–16255. [PubMed] [Google Scholar]
- Peffley D., Sinensky M. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase synthesis by a non-sterol mevalonate-derived product in Mev-1 cells. Apparent translational control. J Biol Chem. 1985 Aug 25;260(18):9949–9952. [PubMed] [Google Scholar]
- Reed W. D., Clinkenbeard D., Lane M. D. Molecular and catalytic properties of mitochondrial (ketogenic) 3-hydroxy-3-methylglutaryl coenzyme A synthase of liver. J Biol Chem. 1975 Apr 25;250(8):3117–3123. [PubMed] [Google Scholar]
- Rodwell V. W., Nordstrom J. L., Mitschelen J. J. Regulation of HMG-CoA reductase. Adv Lipid Res. 1976;14:1–74. doi: 10.1016/b978-0-12-024914-5.50008-5. [DOI] [PubMed] [Google Scholar]
- Schneider W. J., Slaughter C. J., Goldstein J. L., Anderson R. G., Capra J. D., Brown M. S. Use of antipeptide antibodies to demonstrate external orientation of the NH2-terminus of the low density lipoprotein receptor in the plasma membrane of fibroblasts. J Cell Biol. 1983 Nov;97(5 Pt 1):1635–1640. doi: 10.1083/jcb.97.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L. W., Rudney H. Regulation of 3-hydroxy-3-methylglutarate and mevalonate biosynthesis by rat liver homogenates. Effects of fasting, cholesterol feeding, and triton administration. Biochemistry. 1970 Jun 23;9(13):2725–2731. doi: 10.1021/bi00815a021. [DOI] [PubMed] [Google Scholar]