Abstract
The mechanisms underlying the selective regional vulnerability to neurodegeneration in Huntington’s disease (HD) have not been fully defined. To explore the role of astrocytes in this phenomenon, we used single-nucleus and bulk RNAseq, lipidomics, HTT gene CAG repeat-length measurements, and multiplexed immunofluorescence on HD and control post-mortem brains. We identified genes that correlated with CAG repeat length, which were enriched in astrocyte genes, and lipidomic signatures that implicated poly-unsaturated fatty acids in sensitizing neurons to cell death. Because astrocytes play essential roles in lipid metabolism, we explored the heterogeneity of astrocytic states in both protoplasmic and fibrous-like (CD44+) astrocytes. Significantly, one protoplasmic astrocyte state showed high levels of metallothioneins and was correlated with the selective vulnerability of distinct striatal neuronal populations. When modeled in vitro, this state improved the viability of HD-patient-derived spiny projection neurons. Our findings uncover key roles of astrocytic states in protecting against neurodegeneration in HD.
Subject terms: Genetics of the nervous system, Astrocyte, Huntington's disease
Huntington’s disease (HD) is a neurodegenerative disease that shows selective regional vulnerability. Here, the authors show that postmortem brain HD astrocytes are regionally diverse, with a striatal disease-associated state and a cortical compensatory state that mitigated neural death.
Introduction
Huntington’s disease (HD) is an incurable neurodegenerative disease characterized by accumulation of the mutant huntingtin protein and selective loss of specific neuronal populations leading to motor, cognitive and psychiatric impairment1–6. Although the disease-causing mutation is present in all brain cells, the severity of the disease varies across brain regions7,8. New evidence suggests that the CAG repeats in exon 1 of the Huntingtin gene (HTT) can expand during a lifetime, and in different cell types at different rates, which may contribute to disease progression9–11. Moreover, research implicates mutant HTT (mHTT) expression in cortical and striatal neurons as a necessary substrate for striatal neurodegeneration12,13. Furthermore, increased vulnerability of striatal neurons deprived of the cortical-derived, neuroprotective BDNF has shed light on striatal neuronal vulnerability in HD14. However, despite recent advances in our understanding of regional heterogeneity of neurodegeneration in HD, there is much to be learned about how astrocytes contribute to neuronal loss.
In addition to neurons, HD affects oligodendrocytes, microglia, and astrocytes. We previously described oligodendrocytic pathology using single-nucleus RNA-seq (snRNAseq) in HD15. Astrocytes, the subject of this study, have long been noted to be “reactive” in HD, as judged by their enlargement and the increase in markers such as GFAP, present particularly in severely affected areas like the striatum (ref. 16 for review), but astrocytes also contribute to the pathology in several ways. Studies in human brain and in mouse models suggest a loss of function phenotype, including, for example, a decrease in the major glutamate transporter EAAT2/GLT1 in mouse striatum17–20, human striatum20, and in the cortex21. In murine models of HD, mHTT expression in astrocytes can exacerbate HD pathology22,23, impair baseline astrocyte function19, and elicit inflammation24. In contrast, downregulating HTT in astrocytes can slow disease progression25. Human fetal-derived astrocytes with mHTT display several transcriptional abnormalities, including low levels of metallothionein-3 (MT3) and genes involved in fatty acid synthesis26. In addition, HD astrocyte pathology extends from function to morphology; in fact, HD astrocytes are structurally abnormal27,28, and astrocyte maturational defects have been described in HD iPSC models28. Taking all of this together, there is evidence for cell-autonomous astrocyte pathology that contributes to neuronal injury in HD.
The interaction between astrocytes and neurons is central to HD pathology. The response of astrocytes to injury in HD appears most robust when both neurons and astrocytes express the mutant protein in murine models29, suggesting that a large component of astrocyte pathology in HD is secondary to neuronal injury in the setting of compromised basic astrocyte functions29,30. However, there is evidence to support cell-autonomous or primary roles of astrocytes in HD. For example, human glial progenitor cell-derived astrocytes expressing mHTT can recapitulate motor aspects of the HD phenotype when implanted into mice, while control astrocytes can ameliorate HD pathology when implanted into HD mice31. Finally, genetically modified astrocytes can be leveraged to rescue HD symptoms32 and impart neuroprotection in HD33–35.
The goal of this study is to map astrocyte pathology across different brain regions in human HD. Building on our previous results showing that mHTT protein aggregates in astrocytes, and that cingulate cortex astrocytes exhibit phenotypic heterogeneity even within the same region21, we generated multimodal omics data and performed analyses to characterize astrocytic phenotypic heterogeneity in HD across multiple brain regions. We dissected post-mortem human brain tissue from the severely affected caudate nucleus, and relatively less severely affected cingulate cortex and nucleus accumbens7. Next, we generated bulk RNAseq from 76 samples from 10 controls and 20 patients across different HD grades, including 8 juvenile-onset HD patients. We also performed single-nucleus RNAseq (snRNAseq) on a subset of these samples. We combined these transcriptomics data with lipidomics from 27 cingulate cortex samples to extract disease severity signatures that correlate with CAG repeat length and HD grade. In particular, we analyzed astrocyte states and correlated them with neuronal vulnerability across different brain regions. We found that protoplasmic astrocytes differed from fibrous-like CD44-positive (CD44+) astrocytes in their association with neurodegeneration. Moreover, we discovered regional heterogeneity in protoplasmic astrocyte pathology across vulnerable and resilient brain regions, particularly related to metallothionein gene expression. We further analyzed existing GWAS data and identified single nucleotide polymorphisms (SNPs) in the metallothionein gene locus that were associated with delayed disease onset. We then performed validation and functional experiments, mapping metallothionein protein expression in vivo and confirming that metallothionein-3 in astrocytes protected neurons against death and degeneration in vitro. Finally, we discovered a previously undescribed function of astrocyte metallothionein-3 on microglial function and gene expression. Together, our results pave the way for astrocyte-centric therapeutic strategies to treat HD.
Results
Transcriptomic analysis of multiple anatomic regions of HD brains identifies disease severity-associated gene signatures
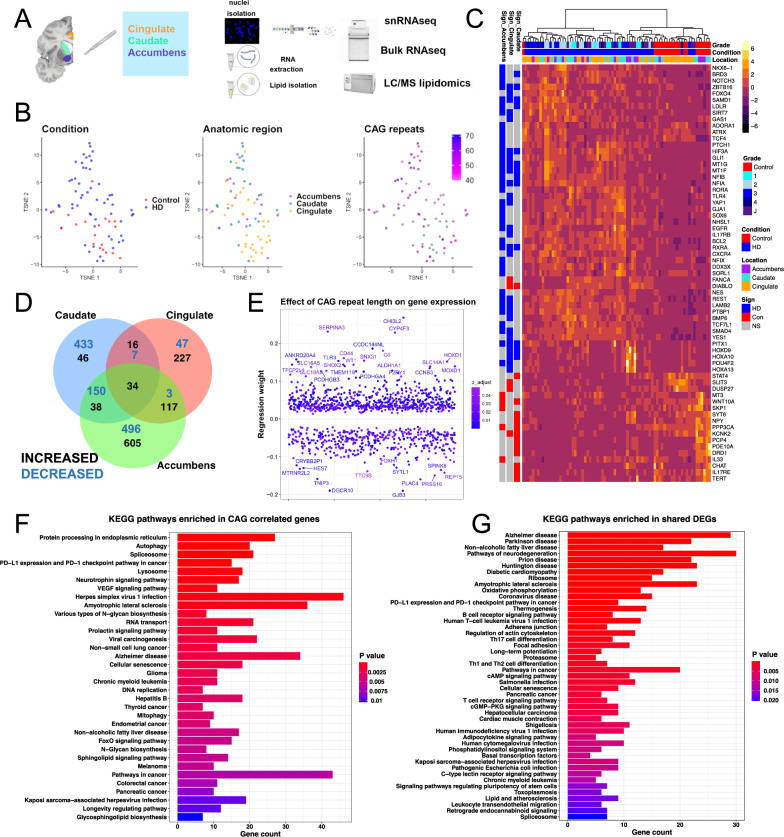
The pathology of HD has been studied most in the caudate nucleus, one of the earliest and most severely affected brain regions. Transcriptional pathology has been described in the caudate nucleus and other brain regions including the frontal cortex, motor cortex, and cerebellum using bulk RNAseq36,37, and in the caudate nucleus using snRNAseq38,39. To define the transcriptional signatures of human HD that are dependent on disease severity, we performed transcriptional and CAG repeat length analysis of different brain regions including the severely affected caudate nucleus, and less severely involved cingulate cortex and nucleus accumbens. The methodology is illustrated in Fig. 1A. We analyzed 76 brain samples from controls (n = 20 samples from 11 donors) and individuals with HD (n = 56 samples from 24 donors, including 16 samples with juvenile-onset HD – Supplementary Dataset-1) using bulk RNA sequencing. We further performed lipidomic studies on a subset of these donors to validate pathologies described below, and snRNAseq to define pathology in glial cells (see below – Fig. 1A). Furthermore, we measured the CAG repeat lengths in DNA extracted from the same tissue blocks we used for RNAseq across the three brain regions (Supplementary Dataset-1 and Fig. S1C). The CAG repeats in the brain samples ranged from 40 to 71, and in most cases showed either an increase of one repeat or no difference from blood/cerebellum-derived CAG lengths (Fig. S1C). A general inspection of the bulk RNA-seq dataset in the reduced dimensional space (t-SNE) shows separation of HD samples from controls in the tSNE space (Fig. 1B, Fig. S1A).
Fig. 1. Transcriptomic analysis of HD identifies cross-regional and CAG-correlated gene signatures.
A Schematic depicting experimental plan. B t-distributed stochastic neighbor (t-SNE) embedding of bulk RNAseq samples used in the study color-coded by condition (left), anatomic region (middle), and CAG repeat length (right). Control samples and ones with no available CAG repeat lengths are shown in grey. C Heatmap of normalized gene expression showing a select subset of differentially expressed genes (DEGs). The DEGs (rows) are color-coded on the right by the direction of differential expression in the specified region (left columns). DEGs Increased in Control: red, DEGs increased in HD: blue, non-significant (NS) genes – grey. The samples (Columns) are also color-coded by HD grade/Condition (Con: Control, HD1–4: 1–4, J: Juvenile onset HD) and anatomic region (top horizontal bars). D Venn diagram showing the overlap between DEGs with FDR-adjusted p value < 0.05 across the three anatomic regions. The numbers of increased – black, and decreased – blue, genes are indicated. E Scatter plot showing genes with significant regression weights for CAG repeat length (y-axis). The order of genes on the x-axis is arbitrary. Color indicates Benjamini–Hochberg adjusted p value. Genes with coefficients two standard deviations above the mean are indicated. F–G EnrichR barplots of KEGG pathways enriched in genes that positively or negatively correlate with CAG repeat length (F) or in DEGs that are shared across two or three anatomic regions (increased and decreased—G). Gene count is indicated on the y-axis. Color indicates Benjamini–Hochberg adjusted p value. A was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
There were many differentially expressed genes (DEGs) between HD and control bulk RNA-seq samples, a subset of which are shown in Fig. 1C. These include the upregulation of several genes involved in control of gliogenesis/stemness such as NES, EGFR, GLI1, PTCH1, YAP1, POU4F2, SMAD4, and REST, and downregulation of several genes involved in oxidative phosphorylation and mitochondrial function, and also several known HD spiny projection neuron (SPN) genes such as the dopamine receptor genes DRD1 and DRD2, as well as PCP4, which is known to be most highly expressed in the basal ganglia40 (Fig. 1C and Supplementary Dataset-2). We found that most of the DEGs were region-specific and were not shared with other brain regions (Fig. 1D and Supplementary Dataset-2). However, the overlap between DEGs among brain regions was most notable between the accumbens and caudate – both striatal regions – and accumbens and cingulate – both less severely involved in HD (Fig. 1D).
We next determined the effect of CAG repeat length on gene expression using a multi-variate regression analysis, taking into account sex, age, and anatomic region as co-variates. The results identified 1092 genes with either positive (672) or negative (420) regression weights (Fig. 1E and Supplementary Dataset-2, we refer to these genes as “CAG-correlated”). The genes with the most positive regression weights included astrocytic genes (AQP4, CD44, LIFR, P2RY1, OSMR, SERPINA341 - also called Alpha1-antichemotrypsin) and microglial genes (TLR3, VSIG4, IL13RA1, and complement genes). Using immunostaining, we confirmed that CD44, a membrane protein expressed in astrocytes42, was indeed increased in the HD caudate nucleus (Supplementary Fig. 4A, B). Next, we performed KEGG pathway enrichment analysis of the CAG-correlated genes and DEGs shared between two or three regions, both increased and decreased (Fig. 1F, G and Supplementary Dataset-2). As expected, CAG-correlated genes were enriched in pathways related to splicing, protein processing, autophagy, neurodegeneration, and lysosomal function (Fig. 1F). Interestingly, in both the CAG-correlated and multi-region disease-associated gene sets, KEGG pathways involved in inflammation, cancer-related pathways, and lipid metabolism were also significantly enriched (Fig. 1F, G). Moreover, weighted gene co-expression network analysis (WGCNA) identified gene modules that correlated with CAG repeats, modules enriched in genes involved in DNA damage response and T-cell mediated inflammation, and loss of connectivity of key astrocytic and immune gene modules (Fig. S2 – see Supplementary Results). Together, these results point to a significant contribution of glia, namely astrocytes and microglia, in the pathology of HD.
Integration of lipidomic analysis and transcriptomics implicates long-chain fatty acids and ceramides in HD neuropathology
Several studies have previously identified abnormalities in lipid species abundance in HD43–49. We therefore sought to associate gene expression and the lipidomic changes in HD that were correlated with disease progression, given that lipid fatty acid and lipid metabolism are key functions of astrocytes50. We focused on the cingulate cortex because it is less severely degenerated than the caudate, thus allowing for more robust correlation analyses between lipid species abundance and disease progression, the latter determined by different HD Vonsattel grades. This analysis is less feasible in the caudate nucleus because it shows pronounced neurodegeneration in most of our cases, which were Vonsattel grades 2–4, where more than 50% of neurons are lost. Thus, it is likely that lipidomics findings in the caudate may represent an “end-stage” phenotype.
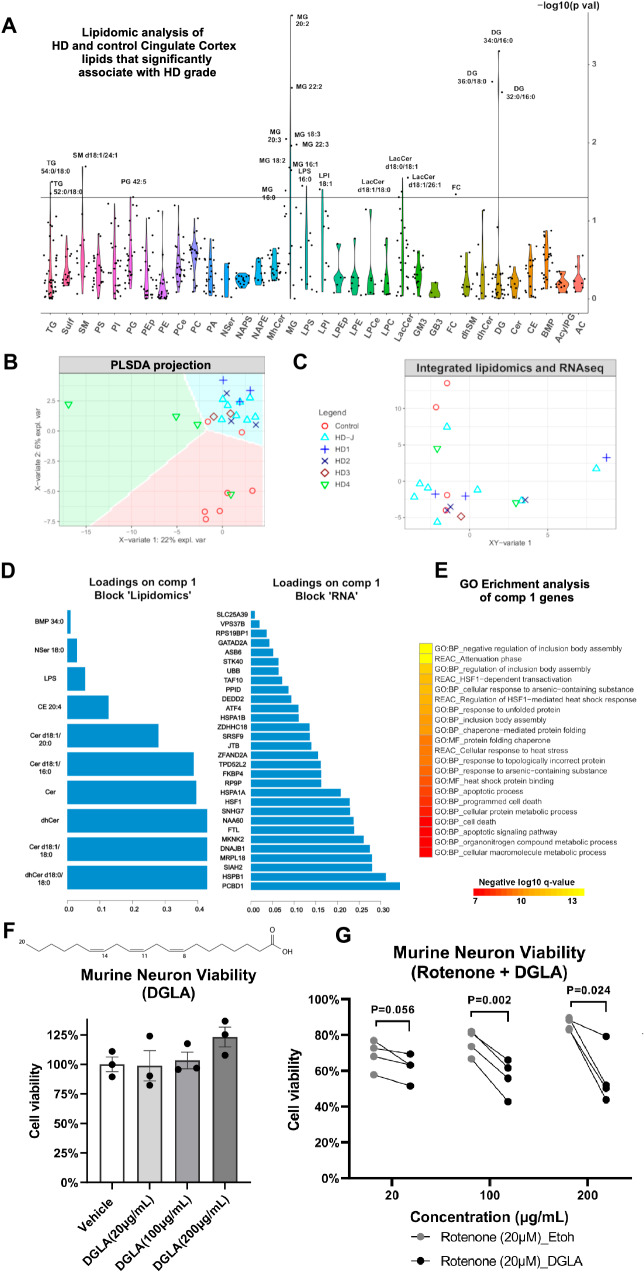
To this end, we performed lipidomic analysis of the cingulate cortex of 27 control and HD donors across different HD grades (Supplementary Dataset-1). Regression analysis (with grade as the explanatory variable and normalized lipid abundance as the response variable) identified several lipid species that significantly varied over grade, including several long-chain and very long-chain fatty acids, diacylglycerides (DG), monoacylglycerides (MG), cholesterol, and lactosyl-ceramides (Fig. 2A). Several of the very long chain monoglycerides were higher across different HD grades compared to control (Fig. S3A– Supplementary Dataset-4). Given the differences between HD and control in lipidomic signatures, we asked if we could predict HD grade from the lipidomics dataset. Using a sparse partial least square discriminant analysis (sPLSDA – Fig. 2B), we found that HD grade could be accurately predicted from the loadings in the first four sPLSDA components – (Fig. S3B). In addition, our data analysis did not show significant effects of sex on lipid species expression as visualized in the sPLSDA space (Fig. S3C).
Fig. 2. Lipidomic analysis of HD cingulate cortex.
A Violin plot of the −log10 of the ANOVA p values (y-axis) of lipid species (x-axis) that correlate with HD grade – see related Fig. S3A for direct comparisons between HD and controls, and Supplementary Dataset-4 for abbreviations. B Scatter plot showing the projection of brain samples analyzed by lipidomics in the first two latent variables of the sparse-partial least squares (sPLS) discriminant analysis model. The variance explained by each latent variable is indicated on the axes. The samples are color- and shape-coded by condition/grade. The condition can be predicted to a high degree of accuracy in the colored background regions - see related Fig. S3B. C Integration of lipidomics data and matched bulk RNAseq data generated from adjacent samples of the same brain region using sparse projection to latent space analysis. The samples are color- and shape-coded as per B and projected in the combined integrated space. D Barplots showing the loadings of the lipid species (left) and RNA transcripts (right) in the first sparse projection to latent space variable that predicts grade. E Gene ontology enrichment analysis of component 1 genes (from D). Negative log10 of the adjusted p values are indicated. P value adjustment was done in gprofiler using the g:SCS method. F Quantification of the percentage of viable murine neurons treated for 24 h with the indicated concentrations of di-homo gamma lenolenic acid (DGLA), a poly-unsaturated fatty acid increased in HD. Illustration of the structure of DGLA is indicated on the top. N = 3 experiments. Unpaired two-tailed test. P values = 0.936 for 20 µg/ml vs vehicle, p = 0.746 for 100 µg/ml vs vehicle and p = 0.091 for 200 µg/ml vs vehicle. Data are shown as mean ± SEM. G Quantification of the viability of murine neurons co-treated with 20 µM Rotenone and the indicated concentrations of DGLA (black dots) or Ethanol (grey dots). N = 4 experiments. Paired two-tailed t-test. P = 0.056 for 20 µg/ml, p = 0.002 for 100 µg/ml and p = 0.024 for 200 µg/ml.
To identify correlations between gene expression and altered lipid metabolism in HD, we analyzed RNAseq and lipidomics measurements from the same tissue samples. We integrated a subset of 21 matched samples from both datasets using sparse-projection to latent space (PLS) analysis, and projected the data by HD grade and sex in the integrated space (Figs. 2C and S3C, respectively). Unlike control samples, HD samples showed positive loadings in the integrated x-y variate-1 dimension (Fig. 2C). Several lipid species and RNA transcripts had positive loadings with the x-y variate-1, including ceramides, dihydroceramides, and cholesterol esters, as well as several genes such as PCBD1, HSPB1, DNAJB1, HSP1A1, HSF1, and MKNK2 (Fig. 2D). Gene ontology enrichment analysis revealed the genes correlated with the x-y variate-1 component were enriched in ontologies related to cell death, apoptosis, inclusion body assembly, and response to unfolded protein (Fig. 2E). Network analysis also highlighted correlations between RNA transcripts and lipids (Fig. S3D). Altogether, our data indicate that lipid metabolism is altered in the cingulate cortex and can be linked to transcriptomic signatures associated with HD pathology.
We next sought to determine whether the lipids correlated with HD grade were toxic or protective. Given that gene ontologies related to cell death were enriched in the integrated lipidomics-RNAseq signature, we asked if HD-associated lipids could induce neuronal death or increase the sensitivity of neurons to death. Accordingly, we performed functional in vitro studies and tested murine neuronal viability when treated with di-homo gamma lenolenic acid (DGLA), a poly-unsaturated fatty acid increased in HD (Fig. S3A). The results showed that, on its own, DGLA did not induce cell death (Fig. 2F). However, when combined with the mitochondrial toxin Rotenone, 100 µg/ml and 200 µg/ml of DGLA sensitized neurons to death (Fig. 2G). Together, these results support that the changes in lipid species including poly-unsaturated fatty acids in the HD cortex may drive or exacerbate neurodegeneration. Since astrocytes are the major cells that take up, synthesize51, and secrete poly-unsaturated fatty acids52, future studies will test whether astrocyte fatty acid synthesis functions can be a therapeutic target for HD.
snRNAseq of HD brain reveals regional astrocytic heterogeneity
One of the genes from our bulk RNA-seq analysis that was highly positively correlated with CAG-repeat length was CD44 (Fig. 1E). We previously showed that CD44 labels specific regionally heterogeneous subsets of astrocytes, including white matter and interlaminar astrocytes42. Thus, we initially confirmed that CD44 was increased in caudate nucleus astrocytes in HD at the protein level (Fig. S4A, B, F). We also found that CD44+ astrocytes were present in the pencil fibers in both HD and controls, as we reported before42, but only in HD were CD44+ astrocytes present in the caudate nucleus parenchyma, where they normally do not reside (Fig. S4A, B, F). These data lay the foundation for further examination of astrocyte heterogeneity in HD.
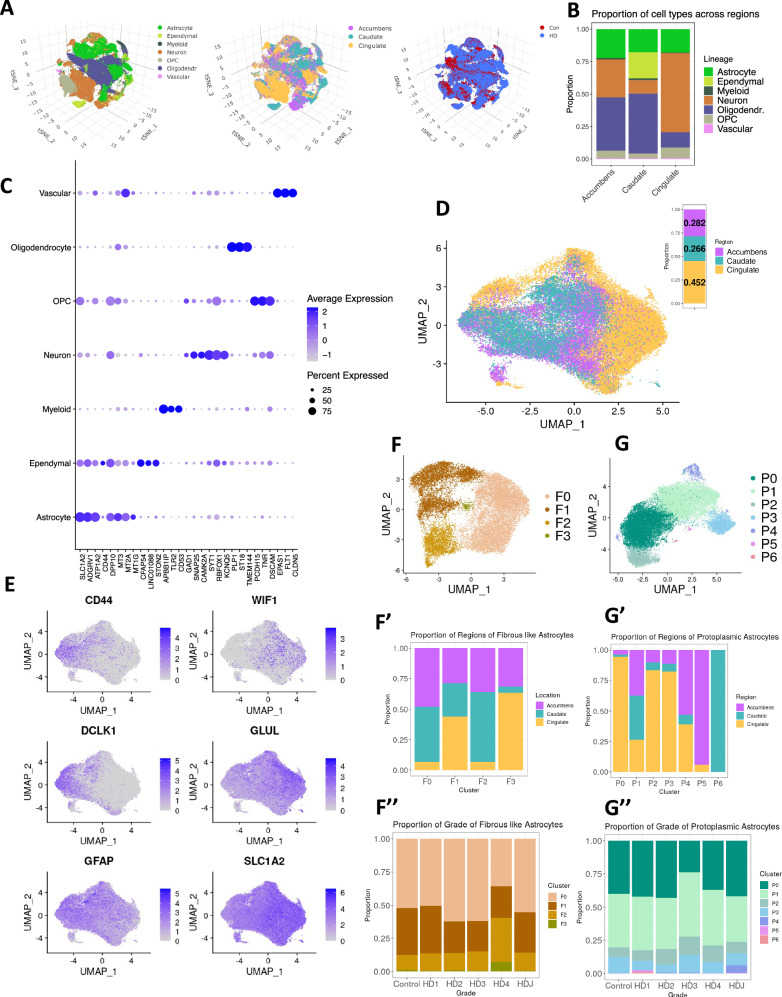
To characterize the heterogeneity of astrocytes along the axis of regional disease burden, we generated snRNAseq data from control and HD brain samples from the nucleus accumbens, cingulate cortex, and caudate nucleus. We previously reported the findings on oligodendrocytes and oligodendrocyte precursors in this dataset;15 here we present our findings involving astrocytes, microglia, and neurons. Filtering and initial QC led to analysis of a total of 281,099 nuclei (Supplementary Dataset-1). Unsupervised clustering of these nuclei identified the major cell lineages, which could be visualized in t-SNE plots color-coded by lineage, region, and disease condition (Fig. 3A), and by donor, batch, and HD grade (Fig. S5A). The proportions of each lineage per anatomic region and donor (Fig. 3B and Supplementary Dataset-1, respectively) show that neurons predominated in the cingulate cortex, and ependymal cells were discovered only in the caudate nucleus. A subset of the canonical gene markers of cell type/lineage is shown in Fig. 3C. Of all nuclei, 53,219 were astrocytes; after additional rounds of QC to exclude doublets, ambiguous nuclei, or low-quality cells, we recovered 45,101 high-quality astrocytes (Fig. 3D), of which 7556 were from control donors and 37,545 came from HD donors (Fig. S5B). The proportions of astrocytes were roughly evenly distributed between cortical and striatal regions, allowing a comprehensive analysis of astrocyte regional diversity (Fig. 3B, D).
Fig. 3. snRNAseq data analysis of HD and control astrocytes.
A t-SNE projection of snRNAseq samples across all lineages (left), brain regions (middle), and condition (right). B Stacked bar plots depicting the proportion (y-axis) of each cell lineage (color-coded) in different brain regions (x-axis). C Dot plot showing expression of select cell type/lineage marker genes. D UMAP plot of astrocytes projected in isolation of other cell types, and color-coded by region. The bar plot on the top right shows the distribution of astrocyte between the three brain regions. E Feature plots of normalized gene expression projected in the UMAP embeddings to highlight genes that differentiate fibrous-like (left) and protoplasmic astrocytes (right) – see also Fig. S5C. F UMAPs of sub-clusters of fibrous-like astrocytes (defined by highest expression of CD44 - cluster 0 see Fig. S5C). The barplots below show the proportion of different brain regions in each sub-cluster (F′) and the proportion of the sub-clusters in each HD grade/Condition (F″). G UMAPs of protoplasmic astrocytic sub-clusters (defined as all astrocytes except cluster 0 in Fig. S5C). The barplots below show the proportion of different brain regions in each sub-cluster (G′) and the proportion of the sub-clusters in each HD grade/Condition (G″).
In order to discover the underlying heterogeneity in the major astrocyte subtypes and characterize their response to injury in HD, we classified astrocytes as protoplasmic, the most commonly studied astrocyte type, and fibrous-like, which includes CD44+ astrocytes that reside in the white matter, subependymal zone, perivascular regions, and subpial regions42,53. In low-dimensional UMAP space, astrocytes varied along an axis with one end expressing higher levels of CD44, GFAP, and DCLK1, and the other end of the axis expressing high levels of Wnt-inhibitory factor 1 (WIF1), Glutamine synthetase (GLUL), and SLC1A2 (Fig. 3E). Sub-clustering the astrocytes identified five clusters (Fig. S5B), where subcluster 0 harbored the majority of the CD44 expressing nuclei (Fig. S5B, C). We designated subcluster 0 as fibrous-like (n = 18,700 nuclei), and remerged the remaining clusters into a protoplasmic group (n = 26,401 nuclei) before re-clustering (see below).
In parallel, we used pseudotime analysis on all astrocytes as a whole (Fig. S5D). Pseudotime analysis is a computational approach that orders cells along trajectories of gene expression, and can uncover modes of continuous gene expression variation, in contrast to discrete groupings afforded by clustering approaches. Along a given pseudotime trajectory, cells exhibit coordinated variation of the genes associated with that trajectory. We expected to find a trajectory of gene expression variation where protoplasmic and fibrous-like genes would fall on opposite ends. When projecting astrocytes in low-dimensional PHATE53 space, we found a spectrum of astrocytes distributed between cells with high SLC1A2 expression on one end, and high CD44 expression on the other end, which was captured using trajectory analysis (trajectory-1, Fig. S5D). Along this trajectory, HD astrocytes showed higher pseudotime values across multiple grades (Fig. S5D). The genes that varied along trajectory-1 were enriched in pathways related to glutamate receptor activity and depleted in pathways related to metal ions/metallothioneins (Fig. S5E, Supplementary Dataset-5). This suggests that as protoplasmic astrocytes transition to fibrous-like (CD44-high), metallothionein levels are reduced. In addition, genes associated with trajectory-2 were depleted in ontologies related to glutamate receptor activity and enriched for ontologies related to protein translation (Fig. S5E). The significance of this second trajectory is unclear.
Because of the anatomic differences in the localization of these astrocyte types, we analyzed protoplasmic and fibrous-like astrocytes independently. Re-clustering both the fibrous-like and protoplasmic groups separately identified four fibrous-like astrocyte clusters (F0–F3) and seven protoplasmic astrocyte clusters (P0-P6) (Fig. 3F, G), with distinct markers distinguishing these subgroups from each other (Fig. 4A, D, Supplementary Dataset-5).
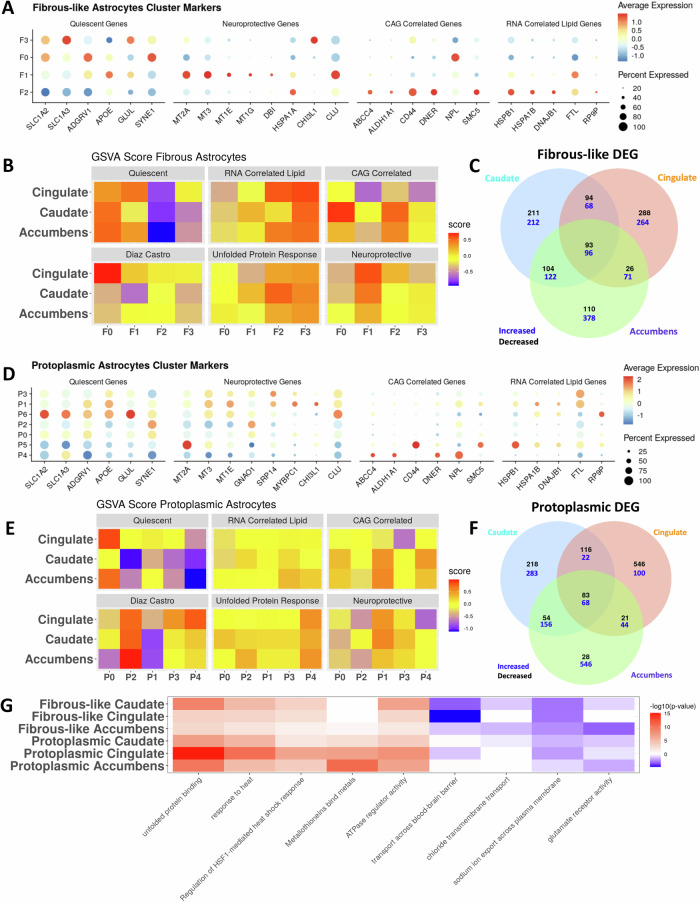
Fig. 4. Astrocytes are regionally heterogeneous in HD.
A Dot plot displaying the expression of select genes in fibrous-like astrocytic clusters. The genes were selected from four gene sets (Quiescent: baseline astrocyte genes, Neuroprotective: predicted from our previous work – see main text, CAG-Correlated: genes with significant positive regression weights - see Fig. 1E for more details, RNA-Correlated Lipid.: set of genes that correlated with lipid abundance from Fig. S3D. B Heatmap of the average enrichment scores determined by gene set variation analysis of select gene sets within each fibrous-like astrocytic sub-cluster per brain region. The gene sets of interest include the four in A, a core astrocytic signature described in Diaz-Castro et al. 20, and a GO term for response to unfolded protein (GO:0006986). C Venn diagram showing overlap between the differentially expressed genes (DEGs) in HD vs control in all fibrous-astrocytes across the three brain regions (increased: blue; decreased: black). D Similar to (A) but for protoplasmic astrocytic sub-clusters. E Similar to (B) but for protoplasmic astrocytic sub-clusters. F Similar to (C) but for protoplasmic astrocytes. G Heatmap displaying the negative log10(p-value) of enrichment of select GO terms (columns) in DEGs from fibrous-like and protoplasmic astrocytes DEGs per region (C, F) – rows. Red indicates terms significantly enriched in DEGs increased in HD, blue indicates GO terms enriched in DEGs significantly decreased in HD, and white indicates no significance. P value adjustment was done in gprofiler using the g:SCS method.
Fibrous-like astrocytic clusters
Fibrous-like astrocytes were distributed between clusters F0 (56%), F1 (29%), F2 (14%), and F3 (1%). Cells from the accumbens and caudate were significantly enriched in clusters F0 and F2 (ANCOMBC p-values 2.50 × 10−5 and 1.26 × 10−7, respectively), whereas clusters F1 and F3 were more enriched in the cingulate (p values: 4.83 × 10−7 and 1.92 × 10−2, respectively) (Fig. 3F′). Cluster F2 was significantly enriched in HD in the accumbens (Fig. S5F, G – Supplementary Dataset-5 and Supplementary Results). HD Vonsattel grades were represented in all clusters. Notably, grade 4 samples contained a higher proportion of cluster F2 and F3 astrocytes (Fig. 3F″). These results highlight regional and disease-severity-related differences in fibrous-like astrocyte clusters.
Examination of gene expression revealed that clusters F0 and F3 had higher expression levels of glutamate transporters SLC1A2 and SLC1A3 (Fig. 4A). Conversely, clusters F1 and F2 showed relatively lower levels. The small cluster F3 showed relatively high expression levels of GLUL and SYNE1 (Fig. 4A). Because cluster F3 was relatively small (less than 200 cells), it was not included in downstream analyses. Cluster F1 expressed metallothionein genes like MT3, MT2A, and MT1E, Clusterin (CLU), and APOE, whereas cluster F2 expressed heat-shock protein genes, as well as multiple genes correlated with CAG-repeats and the integrated lipidomics signature (Fig. 4A).
We next used a supervised analysis to annotate astrocytic clusters with putative functional characteristics. We performed cluster-level gene set enrichment analysis of specific informative gene sets: (1) a previously defined core astrocytic gene set found to be depleted in human and murine HD20, (2) genes associated with quiescent astrocytes21, which are indicators of baseline astrocyte function including regulation of glutamate transport, (3) a “putative” neuroprotective gene signature21, which we defined based on our previous work to be enriched in metallothionein genes encoding proteins rich in cysteine amino acids known to confer protection against oxidative damage54, (4) genes derived from the integrated HD lipidomic signature, which are associated with response to unfolded protein and cell death (Figs. 2), (5) CAG-correlated genes, which are linked to genetic disease-severity (Figs. 1E), and (6) the response to unfolded protein gene ontology (GO:0006986), which is relevant to HD55. We conceptualize the first two gene sets as indicative of homeostatic astrocytic function, the latter three as disease associated, and the putative neuroprotective gene set as potentially compensatory. These gene sets are provided in Supplementary Dataset-2.
Based on this supervised enrichment analysis, we found several cluster-specific patterns of gene expression (Fig. 4B). Across all regions, the quiescent signature was enriched in cluster F0, and the putative neuroprotective signature was enriched in cluster F1. Furthermore, combinations of disease-associated signatures - CAG-correlated, integrated lipidomic signature, and unfolded protein response - were enriched in cluster F2. These results demonstrate that fibrous-like astrocyte clusters show distinct transcriptional states that may be associated with functional variation.
There were several region-specific patterns of gene expression in fibrous-like astrocyte clusters. For example, the core astrocytic signature20 was specifically enriched in cingulate cluster F0, and depleted in caudate cluster F1. Also, in the cingulate cortex but not other regions, cluster F2 showed enrichment of putative neuroprotective genes. Additionally, the CAG-correlated genes were enriched in cluster F0 astrocytes in the caudate, and in cluster F2 in the caudate and nucleus accumbens, but not in the cingulate cortex (Fig. 4B).
Together, these results show a strong regional influence on gene expression and likely functional states of fibrous-like astrocytes. Moreover, the CD44-high, HD-enriched cluster F2 displayed regional heterogeneity with expression of putative neuroprotective genes in the cortex, but CAG-correlated genes in the striatal regions.
Protoplasmic astrocytic clusters
Protoplasmic astrocytes showed notable regional heterogeneity. For example, cingulate protoplasmic astrocytes were most abundant in the most sizeable clusters P0, P2, and P3. Conversely, caudate and accumbens astrocytes were more represented in clusters P1 and P4. Clusters P5 and P6 were composed of cells from the nucleus accumbens and caudate nucleus, respectively (Fig. 3G′). Notably, there was a decrease in the abundance of P1 astrocytes in the HD caudate (log fold change: −1.17 ± 0.48, ANCOMBC p value: 0.014). With the exception that most juvenile-onset HD astrocytes were present in cluster P4 cells, no other discernable correlations were apparent between cluster and grade (Fig. 3G″).
Examination of protoplasmic astrocyte cluster markers showed that the genes associated with baseline/quiescent astrocyte function (e.g., SLC1A2, SLC1A3, ADGRV1), while expressed in most protoplasmic astrocytes, were highest in clusters P0 and P6. Clusters P1, P3 and P5 expressed elevated levels of metallothionein genes, cluster P1 exhibited the highest levels of CHI3L1, and clusters P1, P3 and P6 expressed high levels of CLU. Clusters P4 and P5 showed expression of genes associated with the integrated lipidomics signature and CAG-correlated genes (Fig. 4D). Because clusters P5 and P6 were small (containing less than 200 cells), further interpretation was not pursued.
Next, we interrogated protoplasmic astrocyte clusters using supervised gene set enrichment analysis as we had done for fibrous-like astrocytes (Fig. 4E). Across all regions, cluster P2 displayed enrichment of the core astrocytic gene set, clusters P1 and to a lesser extent P3 were enriched in putative neuroprotective genes, and cluster P4 showed highest scores for the unfolded protein response (GO:0006986). These data suggest that, similar to fibrous-like astrocytes, protoplasmic astrocyte clusters represent distinct states.
There was clear regional heterogeneity in protoplasmic astrocyte clusters. For example, cluster P0 was enriched for the quiescent signature in the cingulate and accumbens, but not in the caudate – where it showed increased enrichment of the unfolded protein response GO. This is expected given the severity of degeneration in the caudate nucleus compared with the cingulate cortex and nucleus accumbens. Also, clusters P1 and P4 showed enrichment of the CAG-correlated gene set in the striatum but not in the cingulate cortex. Similarly, the integrated lipidomic signature was enriched in cluster P3 in the striatum but not in the cortex (Fig. 4E).
Together, these results show that HD protoplasmic astrocytes, similar to fibrous-like astrocytes, exhibit distinct transcriptional states influenced by anatomic localization. Of particular interest, cluster P1, which showed increased enrichment of the putative neuroprotective signature, was enriched in disease-severity-associated gene sets in the striatum and not the cortex.
Differential gene expression analysis highlights regional differences in HD astrocytes
As an additional analytic layer, we determined the DEGs in fibrous-like and protoplasmic astrocytes across the three brain regions (Fig. 4C, F, Supplementary Dataset-6). While some DEGs were shared among the three brain regions, a large proportion of the DEGs were uniquely attributed to individual brain regions, further highlighting the regional diversity of HD pathology. Of the unique DEGs between control and HD astrocytes, those in the nucleus accumbens displayed the largest number of genes with higher expression in HD, and the cingulate cortex displayed the largest number of genes with lower expression in disease (Fig. 4C, F).
To further analyze the DEGs in both protoplasmic and fibrous-like astrocytes across brain regions, we performed GO enrichment analysis (Fig. 4G and Supplementary Dataset-6). As expected, fibrous-like and protoplasmic astrocytes exhibited increased response to unfolded protein and heat stress across all three regions. However, there were significant differences in astrocytic response to injury that were type- and region-specific. For example, the ontology term “metallothionein binding activity” was significantly enriched in DEG sets for protoplasmic and not fibrous-like astrocytes, reflecting larger changes in levels of metallothionein genes in protoplasmic astrocytes. This is consistent with our pseudotime results suggesting a down-regulation of metallothionein-related genes as astrocytes transition from protoplasmic to fibrous-like. The metallothionein related terms were most enriched in cingulate and accumbens protoplasmic astrocytes when compared caudate astrocytes. Conversely, the GO term transport across blood brain barrier was enriched in genes with lower expression in fibrous-like astrocytes in HD – mostly in the cingulate. Finally, the glutamate receptor activity term was most significantly enriched in genes with lower expression in accumbens fibrous-like astrocytes in HD. Altogether, protoplasmic and fibrous-like astrocytes exhibit diverse, region-specific responses to injury in HD.
Examining cell groupings at higher resolution, our cluster-level DEG analysis showed that most DEGs were downregulated in HD compared to control (Fig. S6A). Ontology terms related to transport across the blood brain barrier were enriched in DEGs decreased in cluster F1, and extracellular matrix related ontology terms were enriched in DEGs decreased in clusters F0, F1, P1 and P3. Most of the genes increased in clusters P0, P1, and P3 were enriched for terms related to Huntington’s disease, tau protein binding, and oxidative phosphorylation. Metal-binding related ontologies were enriched in DEGs increased in clusters P1 and P3 (Fig. S6B and Supplementary Dataset-5). As mentioned above, cluster F2 was significantly overrepresented by HD samples, particularly in the accumbens, and very few DEGs were retrieved. Together, these results demonstrate that HD differentially influences astrocytic types and states.
Selective and differential neuronal vulnerability in HD across different brain regions
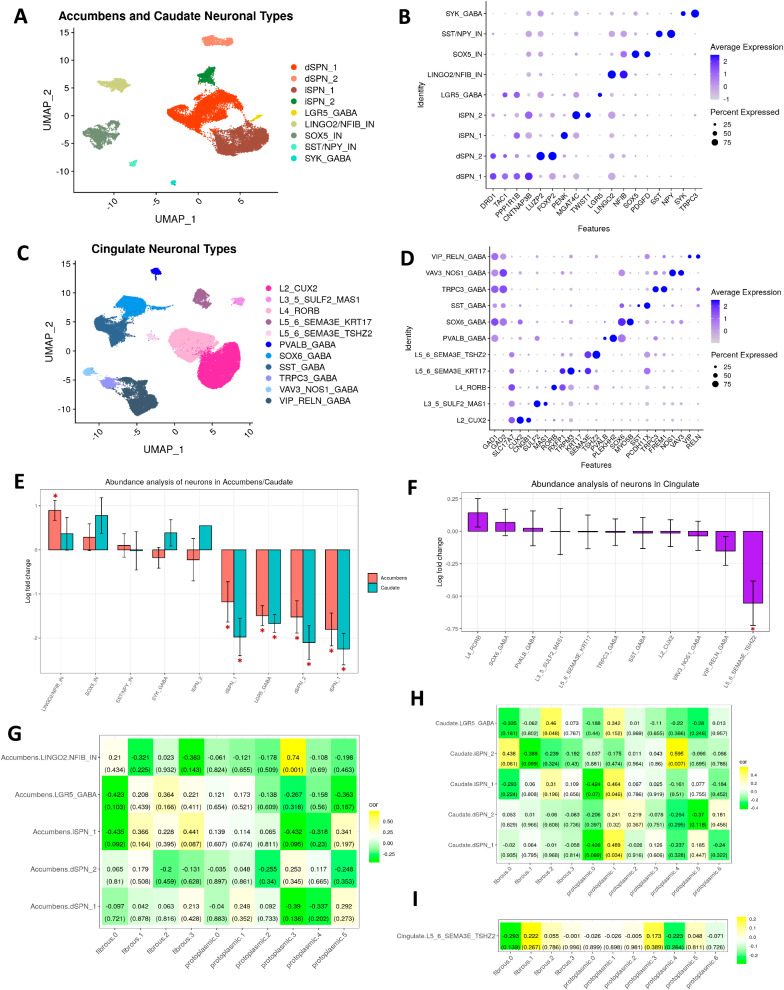
We next turned our attention to neurons and examined differences in their abundance and gene expression across different brain regions. To define neuronal subtypes, we sub-clustered striatal (n = 19,155 nuclei) and cortical neurons (n = 62,253 nuclei) separately. We identified nine major neuronal subtypes in the accumbens and caudate, consisting of direct (d) and indirect (i) pathway SPNs as well as several interneurons/GABAergic cells that expressed canonical markers, like LGR5, LINGO2, SST, NPY, and SYK38,56 (Fig. 5A, B). In the cingulate, we identified a number of layer (L) specific projection/excitatory glutamatergic neurons from L2, L3, L4, L5, and L6 as well as GABAergic neurons as defined by expression of canonical markers (Fig. 5C, D; Supplementary Dataset-8).
Fig. 5. Single nucleus RNAseq analysis and differential abundance analysis of neurons in HD show correlations to astrocytic states.
A UMAP plot of nucleus accumbens and caudate neuronal subtypes. B Dot plot of select gene markers for the striatal neuronal subtypes in (A). C UMAP plot of cingulate neuronal subtypes. D Dot plot of select gene markers for cingulate neuronal subtypes in (C). E Differential abundance analysis comparing the enrichment or depletion of striatal neuronal subtypes in (A) in HD (caudate n = 16, accumbens n = 12) versus control (caudate n = 5, accumbens n = 4). The logFC values from ANCOM-BC linear regression model are shown on the y-axis for each cell type. Stars indicate statistically significant differences (Holm-adjusted p-values < 0.05). Holm-adjusted p-values indicated by stars from left to right are 7.72e−5, 9,76e−3, 3.30e−6, 3.58e−11, 1.99e−16, 3.28e−5, 4.85e−8, 1.33e−6, 2.78e−10. Error bars indicate standard errors. F Similar to E but for cingulate neurons from (C) in HD (n = 19) versus control (n = 9) with Holm-adjusted p-value 1.19e−3. G Heatmap displaying the correlation proportion of fibrous-like and protoplasmic sub-clusters (Fig. 3F, G) – columns, with proportions of select accumbens neuronal sub-clusters – rows. The values in the tile represent the Pearson correlation coefficient and p-values in parentheses. Two-tailed Pearson correlation p values were determined using cor.test(). H Similar to (G) but for select caudate neurons. I Similar to (G) but for a select cingulate neuronal cluster (L5/6 glutamatergic SEMA3E+TSHZ2+) depleted in HD.
To uncover patterns of selective neuronal vulnerability in HD, we performed differential abundance analysis. In the caudate and accumbens we found that dSPN_1, dSPN_2, iSPN_1, and LGR5_GABA were depleted in HD (Fig. 5E). In the accumbens, LINGO2/NFIB_IN was relatively increased, which may reflect an apparent relative increase in proportion resulting from depletion of other neuronal types (Fig. 5E). In the cingulate, only the L5_6_SEMA3E_TSHZ2 neurons were depleted (Fig. 5F). Although SPN loss in the caudate has been described before8,57,58, as well as in the nucleus accumbens59,60, to our knowledge, loss of LGR5+ interneurons has not been documented. Thus, we validated that LGR5 is expressed in striatal neurons and that fewer neurons were LGR5+ in HD versus control (Fig. S7F), confirming the depletion of the LGR5 cluster we observed in the abundance analysis.
Neuronal dysregulation at the snRNAseq level in the striatum has been documented38, and we provide an analysis of the DEGs in the striatal and cortical neurons in the Supplementary Results (Fig. S7 and Supplementary Dataset-8). Because we were interested in how patterns of neuronal loss correlated with astrocytic phenotypes, we quantified the correlations in abundance between neurons and the astrocyte subclusters. Specifically, we sought to determine if certain astrocytic subclusters were correlated with vulnerable neuronal clusters. In the accumbens, the only significant correlation was between protoplasmic cluster P3 and LINGO2/NFIB inter-neuronal cluster (Fig. 5G). No further analysis was pursued due to the small number of cells in this cluster. In the caudate, there were significant positive correlations between protoplasmic cluster P1 with dSPN_1 and iSPN_1, fibrous-like cluster F2 and LGR5_GABA, and the juvenile-onset HD enriched protoplasmic cluster P4 and iSPN_2 (Fig. 5H). In the cingulate cortex, there were no significant correlations (Fig. 5I).
Together, these results validate previously described selective neuronal vulnerability in the HD striatum. Also, the data link the abundance of the metallothionein-high protoplasmic state (cluster P1) to the abundance of vulnerable caudate SPN neurons in HD. Accordingly, since P1 and SPN abundances were positively correlated, the depletion of SPNs in the HD caudate led us to hypothesize that astrocytic P1 will be depleted as well - as seen in the snRNAseq abundance analysis (above). We validated the expected depletion of P1 astrocytes using immunohistochemistry below.
Validation of astrocyte phenotypes in patient samples
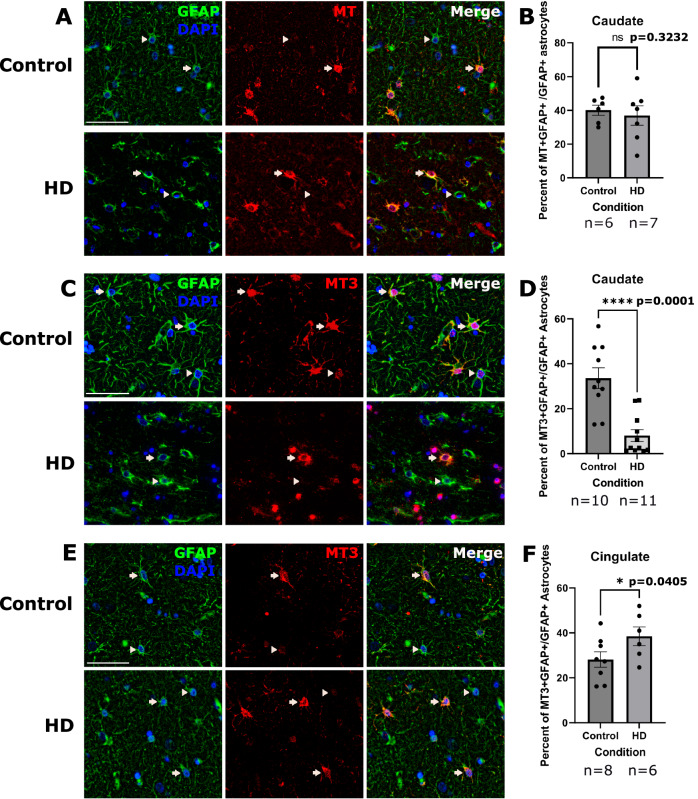
We have shown that astrocytes display an upregulation of a putative neuroprotective signature driven by metallothioneins, including MT3, in multiple brain regions (Fig. 4). We were intrigued by this finding because it was most prominent in protoplasmic rather than fibrous-like astrocytes (Fig. 4G). We had previously shown that cingulate astrocytes upregulate MT1 and MT2 genes in HD21. However, we have not quantified metallothionein protein expression in the severely affected caudate nucleus. As such, we quantified MT1 and MT2A protein levels in caudate nucleus astrocytes (Fig. 6A), and found no significant differences between control and HD astrocytes (Fig. 6B). In contrast, when we examined levels of MT3, which is also expressed in the putative neuroprotective astrocytes (Fig. 6C), we found it to be decreased in parenchymal caudate astrocytes (Fig. 6D). This is consistent with our differential abundance analysis findings showing depletion of cluster P1, which is high in MT3, in the HD caudate. Conversely, we found MT3, like other metallothioneins, to be increased in the cingulate cortex (Fig. 6E, F). These data are consistent with the finding of significant enrichment of metallothionein GO term in the cingulate compared to the caudate (Fig. 4G).
Fig. 6. MT3 expression is increased in the cingulate and decreased in the Caudate of HD brains.
A Immunofluorescent images of the caudate sections labeled for nuclei (DAPI-blue) and GFAP (green) to detect astrocytes (left), and MT (red-middle panel). A merged panel is shown on the right. Arrows indicate DAPI, GFAP and MT positive cells (MT-positive astrocytes) and arrowheads indicate MT negative astrocytes. The antibody detects MT2A and MT1 proteins. Scale bar = 50 μm. B Quantification of the percent of MT-positive astrocytes in the caudate. Unpaired one-tailed t-test with n = 6 for control and 7 for HD. Data are shown as mean ± SEM. P value = 0.3232. C Immunofluorescent images of the caudate labeled for nuclei (DAPI-blue) and GFAP (green) to detect astrocytes (left panel), and MT3 (red-middle panel). A merge of the three channels is shown on the right. Arrows indicate DAPI, GFAP and MT3 positive cells (MT3 positive astrocytes) and arrowheads indicate MT3 negative astrocytes. Scale bar = 50 μm. D Quantification of the percent of astrocytes that were MT3 positive in the caudate. Unpaired one-tailed t-test used with n = 10 for control and 11 for HD. Data are shown as mean ± SEM. P value = <0.0001. E Same as D but for the cingulate. F Quantification of the percent of percent of astrocytes that were MT3 positive in the cingulate. Unpaired one-tailed t-test with n = 8 for control and 6 for HD. Data are shown as mean ± SEM. P value = 0.0405. For all IHC panels, control and HD images are shown on the top and bottom rows, respectively.
To ascertain whether the metallothionein dysregulation is specific to protoplasmic versus fibrous-like astrocytes, we performed multiplex immunofluorescence for GFAP, CD44 (to label fibrous-like astrocytes), and MT3 in caudate samples (Fig. S4A). GFAP+/CD44+ fibrous-like astrocytes did not show a significant change in the proportion of cells that were MT3+ in HD (Fig. S4D). Conversely, MT3 was depleted in HD GFAP+/CD44− protoplasmic astrocytes (Fig. S4E). These data confirm that the metallothionein dysregulation in HD astrocytes is a phenomenon specific to protoplasmic astrocytes.
GWAS analysis identifies a single nucleotide polymorphism in the MT gene locus associated with delayed disease onset
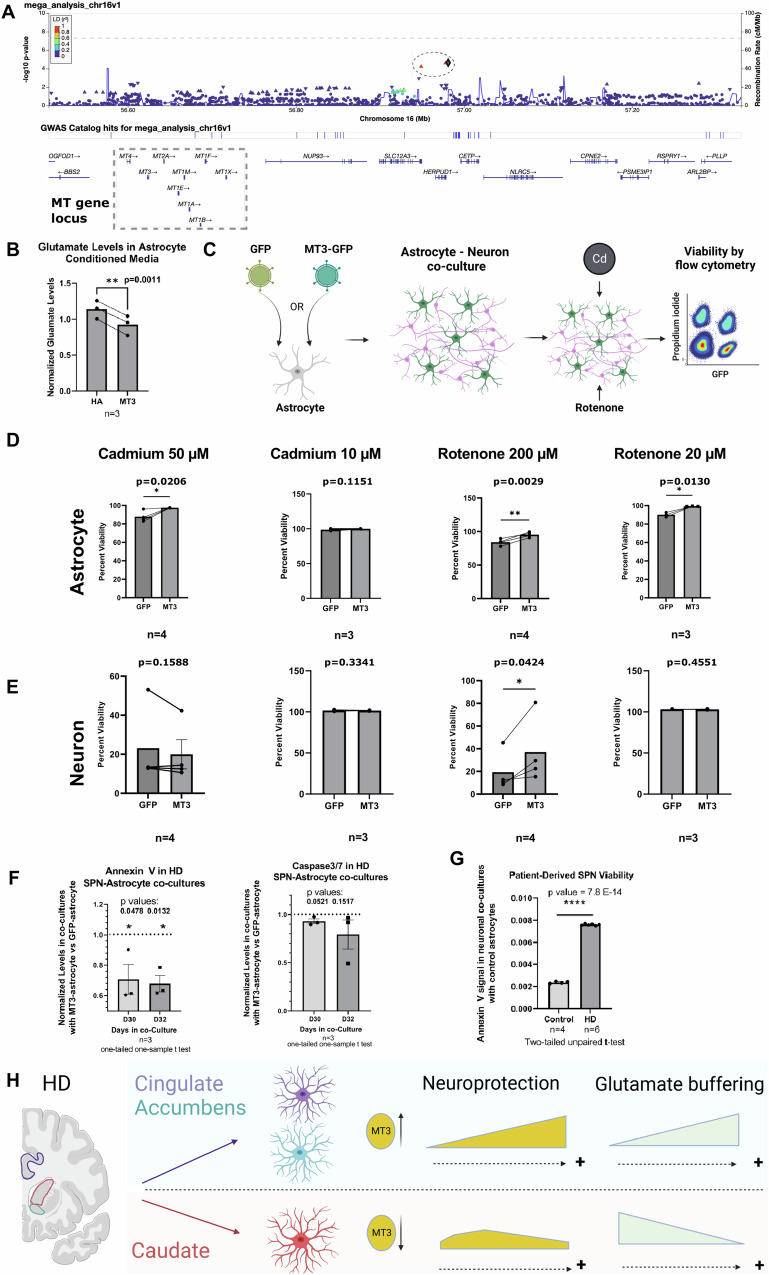
We next asked whether upregulation of metallothioneins in resilient versus vulnerable brain regions is clinically relevant for HD. The age of onset of HD is a clinical phenotype that can be correlated with genetic traits, and is inversely correlated with the CAG length in the HTT gene. Once the effect of the CAG repeat length on age of onset is accounted for, the “residual” age of onset can be treated as a heritable trait. Using this trait, prior genome-wide association studies identified several loci and genes as potential modifiers of the HD age of onset (see61 for review). Here we directed our focus to the metallothionein gene cluster on chromosome 16, utilizing HD patient cohorts from both the GeM-HD consortium and Venezuelan Kindreds. We genotyped the Venezuelan Kindreds with a fine-mapping approach that combined whole genome sequencing and an Illumina CoreExome SNP array across 390 HD patients with corresponding clinical data. Our comprehensive mega-analysis, which combined the genotypic and clinical data from the Venezuelan patients with the GeM-HD consortium data62–65, identified three SNPs that were in linkage disequilibrium (rs3812963, rs74611520, and rs2518054) and were associated with delayed age of onset in HD and the MT gene locus (Fig. 7A and Supplementary Dataset-7). Further, we found that the two SNPs rs3812963 and rs74611520 acted as expression quantitative trait loci (eQTLs) and were associated with increased metallothionein levels in prefrontal cortex66 – see Supplementary Dataset-7.
Fig. 7. Metallothioneins are implicated in GWA studies and promote neuronal viability.
A Genome-wide Association study (GWAS) of residual age of motor onset in HD. This study combines data from the Venezuelan Kindreds and GeM-HD consortium of patients. A LocusZoom plot of the metallothionein (MT) gene cluster (box) on chromosome 16 is shown, depicting the SNPs in the association analysis on chr16 and the log-transformed associated Wald’s p-value (y-axis) measured using a mixed linear model. SNPs are color-coded by the r2 of linkage disequilibrium (LD) with the representative SNP rs74611520 (diamond). Two neighboring SNPs (rs2518054, rs3812963) in high linkage disequilibrium are indicated by red triangles and outlined by dotted ellipse. B Glutamate levels in media conditioned by MT3 astrocytes normalized by levels in control human astrocytes (HA). Paired one-tailed t-test with N = 3 experiments. p = 0.0011. C A cartoon depiction of the design of the astrocyte-neuron co-culture viability experiment. Neurons (GFP−) and astrocytes (GFP+) were separated using flow cytometry-activated sorting (FACS). Cd: Cadmium. D Bar plots of the viability of FACS sorted MT3 overexpressing astrocytes normalized by viability of control GFP astrocytes under the indicated conditions. n = 3–4 independent biological replicates as indicated. Paired one-tailed t-tests. E Bar plots of FACS sorted murine neuronal viability when co-cultured with MT3 overexpressing astrocytes normalized by neuronal viability of sorted neurons co-cultured with control GFP+ astrocytes. Treatment conditions as per D. n = 3–4 independent biological replicates as indicated. Paired one-tailed t-tests. P values are indicated. F Expression of Annexin V and Caspase 3/7 in HD-derived directly reprogrammed SPNs co-cultured with control (GFP) versus MT3 astrocytes at 30 and 32 days in co-culture. The values are expressed as fold change from control. N = 3 biological replicates. The p values are indicated. One-tailed one-sample t-test. G Example of Annexin V signal in control versus HD derived directly reprogrammed SPNs co-cultured with control astrocytes at day 30 demonstrating significant neurodegeneration in HD co-cultures evidenced by the increase in Annexin V signal. N = 4 and 6 technical replicates for control and HD, respectively, two-tailed unpaired t-test, the p values are indicated. For B, D–G, the data are shown as mean ± SEM. H Summary of our understanding of astrocytic states and regional heterogeneity in HD as it relates to neuroprotection. Panels C and H were created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Metallothionein-3 promotes astrocytic and neuronal viability
We next sought to define the functions of the putative neuroprotective signature (Fig. 4). By neuroprotection, we refer to homeostatic astrocytic functions that promote neuronal viability. Thus, we modeled the neuroprotective astrocytes in vitro by overexpressing MT3 in human astrocytes to selectively increase the expression of MT3 but not other metallothionein genes (Fig. S9A). MT3 is increased in astrocytic clusters with enrichment of the putative neuroprotective gene set - cluster P1, P3, and F1 (Fig. S9E, F). We selected MT3 over other metallothioneins because it was decreased in the HD caudate (Fig. 6D). Next, we asked if MT3-overexpressing astrocytes exhibited higher glutamate buffering capacity compared with control astrocytes, which would be an indicator of enhanced homeostatic functions. So, we measured glutamate levels in the conditioned medium of astrocytes cultured for 5–7 days. We found that MT3 astrocyte-conditioned media contained lower levels of glutamate compared to control, suggesting MT3 astrocytes were better able to buffer glutamate (Fig. 7B). Consistent with this finding, MT3 astrocytes increased the expression of glutamate transporter SLC1A2 and glutamine synthetase GLUL in vitro (Fig. S9D), which is consistent with snRNAseq results showing that levels of GLUL were higher in MT3-high astrocytes, and the two genes were correlated (Fig. S9G). In the snRNAseq data, MT3-high astrocytes did not show increased SLC1A2 expression compared with MT3-low astrocytes (Fig. S9G). The discordance between in vivo and in vitro SLC1A2 levels in MT3-high astrocytes may reflect differences between astrocytes in vivo and in vitro. Overall, the results indicate that MT3 expression in astrocytes enhances glutamate buffering – a key astrocyte function that can prevent neurotoxicity of glutamate.
We expected metallothioneins to be neuroprotective, or compensatory, i.e., promote neuronal viability, given that MT1 and MT2 knockout mice exhibit increased vulnerability to neurodegeneration67. To our knowledge, the role of astrocytic MT3 has not been tested. As such, we tested whether MT3 astrocytes could enhance the viability of murine neurons subjected to heavy metal (Cadmium) or Rotenone-induced neurodegeneration (Fig. 7C). We exposed astrocyte-neuronal co-cultures to Rotenone or Cadmium and measured cell viability. We found that MT3-astrocytes protected neurons against Rotenone-induced damage at the higher dose, but not Cadmium-induced damage (Fig. 7E). Conversely, MT3 protected astrocytes from both types of damage (Fig. 7D). Finally, we tested whether MT3 astrocytes could protect directly reprogrammed HD patient-derived SPNs68 (Fig. 7F). Thus, we co-cultured control versus MT3-overexpressing astrocytes with HD-derived SPNs for 30 and 32 days and measured the levels of cell death marker signals (Annexin V and Caspase3/7, n = 3 biological replicates). We found these markers to be decreased in the setting of MT3-astrocyte co-culture (Fig. 7F). HD-derived SPNs showed a significant increase in the Annexin V signal compared to control-derived SPNs (Fig. 7G), supporting the utility of this model to test the effects of MT3 astrocytes on neurodegeneration. Thus, our results overall showed that MT3 expression in astrocytes enhanced neuronal and astrocytic viability, supporting its compensatory functions.
Discussion
We used transcriptomic and lipidomic analyses of three different regions in the human HD brain to extract HD-related disease severity gene signatures. We found that these involve upregulation of glial genes including astrocytic and immune genes and downregulation of genes related to neuronal function. Lipidomic analysis of the HD cortex revealed that poly-unsaturated long-chain fatty acids, which are largely metabolized by astrocytes, were increased in HD. snRNAseq of protoplasmic and fibrous-like astrocytes allowed us to uncover astrocytic states that vary across brain regions and exhibit heterogeneous gene expression profiles. We identified a compensatory state of protoplasmic astrocytes, protoplasmic cluster P1, which upregulated metallothioneins mainly in the cingulate cortex and nucleus accumbens, but was depleted in the caudate nucleus – where HD neurodegeneration was most severe. The depletion of the compensatory astrocytic state was correlated with the depletion of vulnerable SPNs in the caudate nucleus. GWAS analysis incriminated a SNP, rs3812963, in the MT gene locus – which is associated with increased MT levels in the brain - to be associated with delayed disease age of onset. Functional experiments confirmed that astrocytic MT3 expression enhanced neuronal and astrocytic viability, nominating astrocytic MT3 expression as a potential compensatory mechanism to block neurodegeneration in HD.
Regional and type-specific responses to injury in HD
Comparing snRNAseq profiles of astrocytes in the three differentially affected brain regions in HD to those in control individuals, and across different grades of HD severity, revealed regional heterogeneity in astrocytic responses to HD. For example, caudate astrocytes failed to upregulate the metallothioneins 1 and 2, and even downregulated MT3. In contrast, cingulate astrocytes increased metallothioneins 1 and 2, as we previously reported21 and confirmed in this study, and increased MT3 as well. Functional experiments revealed that MT3 overexpression promoted neuronal and astrocytic viability. We interpret this finding as a compensatory or neuroprotective astrocytic state, which will be the subject of future validation studies in animal models.
Metallothioneins are protective in cerebral ischemia, and other contexts of neuronal injury69–72. Most reports investigated MT1 and MT2 as compensatory or neuroprotective molecules, but here, we found that MT3 is also compensatory or neuroprotective. MT3 can regulate neuronal differentiation and promote survivability in vitro73, protect against seizure-induced cell death in vivo74, and is involved in the regulation of lysosomal functions and autophagy in astrocytes75. The function of metallothioneins as chelators of heavy metals is well known76, and given the increases in iron and copper in the HD brain (see review77), the presence of metallothioneins in astrocytes may be protective against oxidative stress. In this respect, the responses of cingulate vs. caudate astrocytes may be crucial for neuronal survival in these brain regions. Our finding that MT3 overexpression in astrocytes increased their ability to buffer glutamate is another potential mechanism by which MT3 enhances viability - given the neurotoxic effects of excessive glutamate78. MT3 also hinders mHTT protein aggregation in vitro79, and may be secreted via unknown non-canonical mechanisms80. Thus, there are multiple ways through which MT3 and other metallothioneins may block neurodegeneration in HD. Future experiments will define the exact mechanism of neuroprotection conferred by MT3-expressing astrocytes.
We separated astrocytes into two major categories, protoplasmic astrocytes and fibrous-like astrocytes, realizing that there is heterogeneity within each group. Fibrous-like astrocytes comprise a diverse group, including the interlaminar cortical astrocytes and white matter astrocytes in the normal human CNS42, as well as protoplasmic astrocytes that have diminished the expression of protoplasmic genes and elevated the expression of other genes such as CD44 and GFAP. Notably, fibrous-like astrocytes showed the highest enrichment of the disease-associated CAG-correlated gene signature (Fig. 4B) and, in contrast to protoplasmic astrocytes, did not significantly alter metallothionein levels in the caudate nucleus (Fig. S4A, D). These results demonstrated astrocyte-type-specific responses to injury in HD.
Our neuropathologic examination of the HD caudate using IHC for CD44 suggested a transition from protoplasmic to CD44+ fibrous-like astrocytes. We have described similar transitions in different pathologies, such as hypoxia and epilepsy42,53. When applying pseudotime analysis to astrocytes as a means to investigate state transitions, the shift from protoplasmic to fibrous-like states was associated with down-regulation of genes related to metallothioneins and upregulation of ontologies related to glutamate receptor activity. This computational analysis requires further validation in astrocyte models.
We previously described a model for astrocytic responses to injury in the HD cortex, which was based on protoplasmic astrocytes21. In it we hypothesized that astrocytes that express quiescence genes at baseline respond to injury by upregulating neuroprotective genes and/or reactive genes like GFAP, then progressively decrease quiescence genes before they decrease neuroprotective genes. At end-stage, only disease-associated genes are present. Here, we see evidence to support this model in our expanded cohort of protoplasmic astrocytes across different brain regions. The addition of other anatomic regions as a dimension of progressive disease severity from cingulate to accumbens to caudate nucleus makes a stronger case than the analysis of only the cingulate. Our model of astrocytic regional heterogeneity in response to HD is illustrated in Fig. 7H. Cingulate and accumbens astrocytes increase metallothioneins, which is a compensatory phenomenon that potentially confers protection to neurons and enhances glutamate buffering. In the caudate at advanced stages of neurodegeneration, astrocytes fail to increase metallothioneins and may be less efficient at glutamate uptake.
Associations between astrocyte responses and CAG repeat length and selective neuronal loss
Given that HD individuals carry variable numbers of CAG repeats, we were interested in associating the different astrocyte clusters with the genes correlated with CAG repeat length. We found stronger CAG-correlated gene scores with astrocytic clusters in the caudate compared to those in the cingulate cortex. This may not be surprising, since the caudate pathology is far more profound than the cortical pathology. Of interest, the compensatory protoplasmic cluster P1 displayed regional heterogeneity whereby it increased the CAG-correlated genes in striatal regions and not the cingulate cortex. This may be attributed to the differences in disease burden, but may also be due to potential inherent differences between human neocortical and striatal astrocytes. This is a subject of great interest to us, and we will investigate it in a separate endeavor.
Correlations between astrocyte pathology and neuronal loss
The known selective neuronal vulnerability in HD8,57,81 was recapitulated in our dataset. Our examination of neuronal populations in HD confirmed the loss the SPNs in the caudate, extended these results to the nucleus accumbens, and discovered the depletion of layer 5/6 SEMAS3E/TSHZ2 neurons in the cingulate cortex and a loss of LGR5+ interneurons in the caudate. We were particularly interested in any relationships between neuronal loss and astrocyte pathology, and so looked for correlations between astrocyte states and neuronal abundance. One of the significant positive correlations was between the compensatory protoplasmic cluster P1 and vulnerable SPNs. Since SPNs are depleted in the caudate, we reasoned that this compensatory astrocyte state might also be depleted, and indeed, our validation studies showed that there was a loss of MT3 in caudate astrocytes. It is possible that the selective neuronal vulnerability in the caudate nucleus is augmented by the lack of robust compensatory responses in caudate protoplasmic astrocytes. Additional studies are needed to validate the pro-survival effect of astrocytic MT3 in mouse models.
Astrocyte-microglial crosstalk
Finally, as described in Supplemental Results, we were interested in examining microglial gene expression in our HD samples and then determining if astrocytes with MT3 overexpression co-cultured with microglia altered microglial gene expression. We indeed found significant changes in tissue microglial gene expression (Fig. S10B–F and Supplementary Dataset-9) and furthermore found that MT3-astrocytes co-cultured with microglia significantly altered microglial gene expression, which included genes related to fatty acid metabolism and lipid transport. These changes were only present in microglia of the cingulate cortex (Fig. S10 and S11), where astrocytes upregulated metallothioneins, and the MT3-primed microglia exhibited increased phagocytic capacity (Fig. S11G).
Open questions
Lipidomic analysis identified an increase in poly-unsaturated fatty acids in HD. Astrocytes are the main cells that metabolize these fatty acids in the brain50–52. The link between fatty acid metabolism and metallothionein expression in astrocytes is yet to be explored. Which astrocytic state, if any, is the source of the increase in potentially toxic poly-unsaturated fatty acids in the HD brain? And are MT3 astrocytes less prone to producing such toxic lipids, or even produce other protective lipids? Future functional experiments are needed to test these hypotheses.
In conclusion, we showed that astrocytes in HD exhibit state-specific and regional heterogeneity in response to HD pathology. We have defined gene signatures associated with CAG repeat length, lipidomic abnormalities, and potentially, neuroprotection. The latter was enriched in an astrocyte state that was depleted in the caudate nucleus and was correlated with the depletion of vulnerable striatal neurons. This compensatory gene signature was characterized by high levels of metallothionein gene expression. Analysis of HD GWAS studies identified a SNP associated with delayed disease onset in the MT gene locus, and functional experiments confirmed that MT expression in astrocytes promoted enhanced viability.
Limitations
First, while we provide metrics on the differences in modal CAG repeat length between blood/cerebellum and each of the regions utilized in our study in Fig. S1C, we do not measure repeat instability or an expansion index as outlined in Lee et al.82. Thus, our CAG-correlated gene signature does not account for changes in somatic instability. Second, we do not define causal relationships between astrocytic phenotypes and differential neuronal vulnerability; instead, we make correlations between an astrocytic state modeled in vitro and enhanced neuronal viability. Further studies are needed to define the causal relationships between these cell states. Also, due to limited sample numbers in HD grade 1 and grade 4 in our cohort, our study does not define the relationships between astrocytic states/clusters and HD grades. Finally, our functional studies focused on MT3, however we did not determine if MT3’s effects were specific to that protein or shared with other metallothionein family proteins - which share sequence homology.
Methods
Brain tissue
All brain tissue used in the study was obtained from the New York Brain Bank. All brains were donated after consent from the next of kin or an individual with legal authority to give consent. The use of postmortem brain tissues for research was approved by the Columbia University Institutional Review Board (IRB protocol # AAAT2895). The Institutional Review Board has determined that performing clinicopathologic studies on de-identified postmortem samples is exempt from Human Subject Research according to exemption # 45 CFR 46.104(d)(2). All studies conducted herein complied with relevant ethical regulations enforced by Columbia University and per IRB protocol # AAAT2895.
Bulk RNA sequencing
Total RNA was extracted from frozen brain tissue samples using an automated Qiagen platform (QiaSymphony). RIN values were measured on an Agilent Bioanalyzer. All samples had RIN values ≥7. The samples were processed using an Illumina® Stranded mRNA Prep kit and were sequenced on an Illumina NOVASEQ 6000 sequencer to produce stranded 100-base pair paired-end libraries at 20 million read depth per sample. For RNAseq of microglial cultures, the samples were processed using an Illumina TruSeq Stranded Total RNA kit and sequenced on an Illumina NextSeq sequencer at a read depth of 10 million reads per sample. Raw reads were aligned to the reference (GRCh38.92) using STAR(version 2.7.1)83. Count matrices were generated from the BAM files using featureCount with default options. Differential gene expression analysis was performed in EdgeR using the glmQLFit method84. Batch was considered in the design formula. Gene set enrichment analysis was performed in enrichR85 and/or gprofiler286.
CAG regression analysis
The gene expression count matrix (20276 genes) was filtered to remove genes with low expression using the “filterByExpr” function of the edgeR84 package (version 3.30.3), with default settings. 15413 genes were retained for subsequent analysis. The matrix was then transformed using the “limma::voom” function (version 3.44.3). Batch effects were visualized with principal component analysis and batch correction performed with the “limma::removeBatcheffects” function. To determine how CAG repeat length affects expression of the retained genes in HD, we performed multiple linear regression with CAG as one of the predictors of the level of gene expression among HD samples across all three regions. We assumed that level of gene expression would also be affected by age, region of the brain, and sex. A regression model was fit separately for each gene using the transformed gene abundance as the response variable, and CAG length, age, brain region, and sex as predictor variables. Regression coefficients were estimated using least squares, and the p-values for the fit models were corrected for multiple hypothesis testing using the Benjamini-Hochberg method. Among the significant gene models (adjusted p-value < 0.01), genes that had a significant regression coefficient estimate for CAG repeat length (p-value < 0.01) were considered as the genes whose expressions were associated with CAG expansion – referred to as “CAG-correlated genes”. For enrichment analysis using EnrichR85, we set an FDR threshold of 0.1 for select genes for the enrichment analysis.
Lipidomics
Total lipids were extracted from frozen 40–70 mg human brain dissected as described above. Lipidomics profiling in fresh frozen brain tissue was performed using Ultra Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC-MSMS). Lipid extracts were prepared from homogenized tissue samples using modified Bligh and Dyer method78, spiked with appropriate internal standards, and analyzed on a platform comprising Agilent 1260 Infinity HPLC integrated to Agilent 6490 A QQQ mass spectrometer controlled by Masshunter v 7.0 (Agilent Technologies, Santa Clara, CA). Glycerophospholipids and sphingolipids were separated with normal-phase HPLC as described before87, with a few modifications. An Agilent Zorbax Rx-Sil column (2.1 × 100 mm, 1.8 µm) maintained at 25 °C was used under the following conditions: mobile phase A (chloroform: methanol: ammonium hydroxide, 89.9:10:0.1, v/v) and mobile phase B (chloroform: methanol: water: ammonium hydroxide, 55:39:5.9:0.1, v/ v); 95% A for 2 min, decreased linearly to 30% A over 18 min and further decreased to 25% A over 3 min, before returning to 95% over 2 min and held for 6 min. Separation of sterols and glycerolipids was carried out on a reverse phase Agilent Zorbax Eclipse XDB-C18 column (4.6 × 100 mm, 3.5 µm) using an isocratic mobile phase, chloroform, methanol, 0.1 M ammonium acetate (25:25:1) at a flow rate of 300 μl/min. Quantification of lipid species was accomplished using multiple reaction monitoring (MRM) transitions under both positive and negative ionization modes in conjunction with referencing of appropriate internal standards: PA 14:0/14:0, PC 14:0/14:0, PE 14:0/14:0, PG 15:0/15:0, PI 17:0/20:4, PS 14:0/14:0, BMP 14:0/14:0, APG 14:0/14:0, LPC 17:0, LPE 14:0, LPI 13:0, Cer d18:1/17:0, SM d18:1/12:0, dhSM d18:0/12:0, GalCer d18:1/12:0, GluCer d18:1/12:0, LacCer d18:1/12:0, D7-cholesterol, CE 17:0, MG 17:0, 4ME 16:0 diether DG, D5-TG 16:0/18:0/16:0 (Avanti Polar Lipids, Alabaster, AL). Lipid levels for each sample were calculated by summing up the total number of moles of all lipid species measured by all three LC-MS methodologies, and then normalizing that total to mol %. The final data are presented as mean mol % with error bars showing mean ± S.E. Statistical comparisons were done using a one-way ANOVA and Tukey’s test for post-hoc analysis. We had previously only presented the DAG results in our paper on oligodendrocytes15.
WGCNA
The “WGCNA” R package (version 1.72) was used to identify modules of co-expressed and co-regulated genes, associated with various clinical and anatomic traits in HD88. Briefly, the gene expression count matrix of both control and HD samples was normalized using the voom function from limma89, batch corrected using combat90. To retain the most varying genes expressed across all samples, we determined the top 6183 genes with the highest median absolute deviation. A signed weighted gene co-expression network was constructed. Adjacencies of the retained genes were calculated using a soft thresholding power of 16 for control and 9 for HD networks. To minimize spurious associations, the adjacencies were transformed to a topological overlap matrix (TOM), and corresponding dissimilarity calculated. The genes were then clustered on the TOM-based dissimilarity. Modules of co-expressed genes were subsequently identified from the dendrogram of clustered genes using hybrid-tree cutting, with a deepsplit parameter set to TRUE and minimum module size of 75. Modules with similar expression profiles (cor > 0.75) were then merged. Significantly associated clinical and anatomic attributes were then determined from the Pearson correlation between module eigengene expression and different traits. Module preservation statistics were generated using the modulePreservation() function with 100 permutations and network type set to signed and quickCor = 0. Otherwise, default parameters were used. Module-trait correlation was performed by Pearson correlation of module eigenvalues per sample and traits. Correlation p values were calculated using the corPvalueStudent function with default parameters.
Dysregulated networks of co-expressed genes in HD
To identify and characterize dysregulated gene expression networks in HD, we first built a reference network using control samples only following similar steps as above, and then evaluated the preservation of these modules in HD using the “modulePreservation” function in WGCNA with 100 permutations, as previously described91. Specifically, several module preservation statistics were generated and used to determine the degree of preservation of each control module in HD. Gene ontology enrichment analysis on the module genes was performed using the moduleGO function from DGCA92 using a hypergeometric test with default parameters. The reference dataset included all genes included in the WGCNA analysis (n = 6183 genes). The plots in Fig. S2E–F were generated using extractModuleGO and plotModuleGO functions. Modules of interest were selected for visualization.
Integration of lipidomics with bulk RNAseq and sparse-PLS discriminant analysis
Integration of bulk RNAseq and lipidomics data was performed using the sparse-partial least squares/projection to latent space (spls) function in the R package mixOmics93. Both the normalized lipidomics and the log-normalized bulk RNAseq matrices were first filtered based on variance (by detecting outlier features with high variance using the isOutlier(nmads = 2, type = “higher”) function from scater94). 5251 genes and 250 lipid species remained from 21 samples. The spls model was first tuned resulting in three components. The network was exported and visualized in cytoscape95. Spls-discriminant analysis (splsda) was achieved after tuning the model using the following command: tune.splsda(X = lipidomics_data, Y = Grade, ncomp = 4, test.keepX = c(1:10, seq(2, 300 10)), validation = “Mfold”, folds = 5, dist =“max.dist”, measure = “BER”, nrepeat = 10). The resulting x and y components were used in the final tuned model, which was used for discriminant analysis. ROC curve for sensitivity and true negative rates were generated using the auroc function from mixOmics.
Single nucleus RNAseq and quality control
The tissue dissection, extraction of nuclei, sequencing, and QC analysis of this dataset presented herein was described previously15. Briefly, ~60 mm3 pieces of fresh frozen post-mortem tissue were dissected and mechanically homogenized to extract nuclei using a Triton X100 and sucrose-containing buffer. 5000–10000 nuclei were processed for GEM generation on a Chromium Controller (10× Genomics) using single Cell 3′ Reagent Kit v2 or v3 (Chromium Single Cell 3′ Library & Gel Bead Kit v2/v3, catalog number PN-1000075; Chromium Single Cell A Chip Kit, 48 runs, catalog number: 120236; 10× Genomics). Sequencing of the snRNAseq libraries was done on Illumina NOVAseq 6000, and alignment to of the fastq files to the reference was done using the CellRanger pipeline (10× Genomics) and the GRCh38.p12 reference). Count matrices were generated using DropEst96 or CellRanger. Low-quality cells were filtered from the based on %mitochondrial reads ≥ 15%, ngenes <200 gene, and logtotalreads<6 using the scater package94. Ambient RNA removal was performed to improve the accuracy of clustering in decontX with default parameters97. Doublets were removed using scran98 with default parameter and by identifying clusters with ambiguous markers.
Clustering of cell types from Huntington’s disease samples
The complete Seurat object was created by aligning multiple datasets through the R package Seurat99,100 (version 4.06). Each sample’s cell type was identified using known markers and further corrected by region for biological consistency. The samples were log-normalized and scaled using Seurat’s NormalizeData function. Coordinates for the PCA were calculated using Seurat’s RunPCA function for a total of 30 principal components. The datasets were merged using Harmony91 (version 1.0) through Seurat’s RunHarmony function. All 30 principal components were used as inputs into RunHarmony. In addition, the integration corrected for three variables, donor, batch, and case numbers (some donors had more than one sample). UMAP coordinates were calculated with Seurat’s RunUMAP function using the Harmony coordinates as input into the dimensional reduction argument. Cluster assignment on the main object followed Seurat’s standard workflow with passing the object’s 30 Harmony coordinates through the FindNeighbors function followed by the FindClusters function with all default parameters. The coordinates of t-SNE were produced using runTSNE function in scater94 (version 1.22.0). Each t-SNE plot was created using the first three coordinates through the package plotly (version 4.10.0).
Sub-clustering of all astrocyte samples
After clustering the complete object which comprised various cell types, the astrocytes were subset into a separate Seurat object for reclustering. The samples were first log-normalized and scaled using Seurat’s NormalizeData function with default inputs. Afterwards, PCA was performed using Seurat’s RunPCA function for a total of 30 principal components. All samples were then corrected for donor and batch through Seurat’s RunHarmony function91. The inputs consisted of the first 30 principal components, while all other arguments were set to default settings. UMAP coordinates were calculated with Seurat’s RunUMAP function using the Harmony coordinates as input into the dimensional reduction argument. Cluster assignment on the main object followed Seurat’s standard workflow by passing the object’s 30 Harmony coordinates through the FindNeighbors function followed by the FindClusters function with all default parameters. Cluster assignments were evaluated iteratively for any contamination by looking at standard markers for neuronal and glial cell types. Samples which consistently expressed certain cell markers were removed to ensure all samples were true astrocytes. To further sub-cluster astrocytes into fibrous-like and protoplasmic, we partitioned the astrocyte samples into those that expressed CD44 (fibrous-like) and those that did not (protoplasmic). These sub-types were then split into their own respective Seurat objects and subjected to the same procedure of clustering as described above. Subclusters that contained under 200 cells were excluded from downstream analysis; a total of 278 cells were excluded.
Differential gene expression analysis – on the single cell level
An initial counts filter was applied to all DGE analysis for every cell type. The genes were filtered by minimum expression and whether genes were robustly expressed. Any genes with raw counts less than 4 were not considered. In order to consider the most robustly expressed genes for downstream analysis, the average expression of each gene was calculated for all cells in a cell-type or cluster, (i.e., row average), and a gene is retained if 5% of cells in a given type/cluster have an expression level greater than or equal to the overall average expression in that cluster/type.
Significantly differentially expressed genes between HD and control were identified with the voom function89 in a regression formula to correct for donor, batch, sex, and age. The analysis was followed with the eBayes function to apply statistics and to finally extract the top 1000 genes with an FDR of 5%.
Gene set enrichment analysis and Gene ontology enrichment analysis
The counts matrix for gene set enrichment analysis was prepared by pseudo-bulking the astrocyte snRNAseq data. The pseudo-bulking procedure was performed by aggregating the counts from all donors within its respective UMAP cluster. All donor-cluster samples were then transferred into a Seurat object (version 4.06) and were normalized using Seurat’s NormalizeData function followed by the SCTransform function to correct for any covariates. All normalization functions were applied with default parameters. Gene set enrichment analysis was done using the GSVA101 package (version 1.44), using the pseudo-bulked matrix and the gene sets as input to the gsva function. The arguments of the function mx.diff were set to false and the method set to “gsva”. From the output, we normalized the individual gene set scores into a z-score for each pseudo-bulk sample by applying the base R scale function. Finally, the gene set scores were averaged across region, grade, and cluster. For Preranked gsea (Fig. S11), the DEGs increased in HMC3 cells co-cultured with MT3-astrocytes versus those co-cultured with control astrocytes was used as a gene set. DEGs from HD versus control microglia in different brain regions were ranked based on log fold change and the enrichment was measured using the fgsea package (version 1.28.0) in R. Normalized enrichment score, p values and adjusted p values are indicated in the figure. Gene ontology analysis was performed on significant DEGs only using gprofiler2 package (version 0.2.3) with the multi_query option set to true, with default parameters.
Pseudotime analysis on astrocyte sub-populations
We created PHATE102 (version 1.0.4) embeddings for all astrocytes. All raw counts matrices for each group were normalized using the sqrt function. We corrected for donors in the embedding by providing the phate function with a vector of all donors using fastmnn function, and used default parameters to create the final embeddings. Pseudotime analysis was performed with slingshot103 (version 2.8.0) on the astrocytes in PHATE reduction. Initial clusters were defined using the Mclust package (version 6.0). The origin point was defined by the cluster with the highest expression levels of SLC1A2. Cluster annotations and PHATE embeddings were provided to the slingshot function with default parameters. Downstream start vs end point differential expression was performed using the tradeSeq104 package (version 1.14.0).
Differential abundance analysis
A matrix of cell types by donor for each brain region was created as input into the differential abundance analysis. Any donor with less than 10 cells from any particular cluster was considered an outlier and was not included in the analysis. An otu table from the phyloseq package (version 1.42.0) was created for each matrix as required for running ANCOM-BC105 (version 2.02). The ancombc function was used to run the differential abundance analysis with an input formula of “Age + Sex + Batch + Condition + Region”. This ensured our results were corrected for covariates. The prv_cut parameter was set to 0, neg_lb = TRUE, struc_zero = TRUE, conserve = FALSE, and all other parameters were set to their default settings. Log fold change values from the “Condition” column were used to compare whether specific cell types were enriched or depleted in HD. Further, log fold change values for each “Region” column were referenced to determine enrichment of astrocyte clusters in various regions. In addition, standard error values were extracted to place onto the final bar plot. Statistically significant comparisons were determined using the corrected p-value provided from ANCOM-BC.
Correlation of astrocyte and neuron proportion analysis
Using the significantly differentially abundant cells from the differential abundance analysis, we correlated the proportions of significant neuronal types to the proportions of astrocyte sub-clusters for each brain region. The correlation was performed using the cor.test function from the stats package (version 3.6.2). All default parameters were used. The Pearson correlation coefficients and respective p-value were extracted from the output of cor.test and plotted as heatmaps.
Immunohistochemistry and multiplex immunofluorescence
Five-to-seven micron-thick sections of formalin-fixed and paraffin-embedded human brain tissue were processed on an automated Leica© Bond RXm autostainer according to the manufacturers’ instructions. For chromogenic DAB stains, generic IHC protocols were employed as per manufacturer protocols. Standard deparaffinization and rehydration steps preceded antigen retrieval in Leica ER2 (cat# AR9640) antigen retrieval buffer for 10–20 min according to manufacturer protocols. For multiplex immunofluorescence, 1 h incubation in a 10% donkey serum containing PBS-based blocking buffer preceded labeling with primary antibodies for 1 h at room temperature. Three wash steps in Leica wash buffer (Ref#AR9590) preceded labeling with species-appropriate Alexa fluor conjugated secondary antibodies (Invitrogen). After three washes, a DAPI containing mounting solution (Everbright TrueBlack Hardset Mounting Medium with DAPI Cat#23018) was used to label nuclei and quench autofluorescence prior to cover-slipping. All steps were conducted at ambient temperature.
Alternatively, multiplex Immunofluorescence of paraffin-embedded blanks was done manually. First, the slides were rehydrated (xylene, 100% EtOH, 95% EtOH, 70% EtOH, 50% EtOH, ddH2O) and then washed in PBS-Tween-based wash buffer. Slides were submerged in pre-heated 1×Trilogy for 20 min for antigen retrieval, cooled to room temperature, and washed in PBS-T. Samples were outlined with a hydrophobic marker to contain antibody solutions on the tissue. Slides were blocked with 1:10 dilution of blocking serum to AB diluent. Antibodies were diluted, applied, and slides were left to incubate 4 °C overnight. On Day 2, slides were washed in PBS-T, species-appropriate secondary antibodies were diluted (1:500), applied, and slides were left to incubate at room temperature in the dark for 2 h, and then washed in PBS-T. Slides were incubated with 0.1% Sudan Black B diluted in 70% EtOH for 30 min to quench the tissue’s autofluorescence, and then washed in PBS-T. Finally, slides were mounted with DAPI media to stain nuclei.
The following primary antibodies were used: Rabbit ALDH1L1 (1:100, EnCor, Cat#RPCA-ALDH1L1), Chicken GFAP (1:1000, Abcam, Cat#4674), Goat Clusterin (1:200, Thermo fisher, cat#PA5-46931), Rabbit MT3 (1:200, Novus Biologicals, cat#NPP1-89772), Mouse CD44 (1:50, Millipore, cat#SAB1405590), Rabbit CD44 (1:100, Abcam, Cat#ab101531), Rabbit MT3 (1:100, Millipore, Cat#HPA004011), Mouse anti Metallothionein (Detects MT1 and MT2 proteins – Abcam, Ab12228, 1:100). Secondary antibodies conjugated to fluorophores: anti-mouse Alexa Fluor 488, 568, and 633, anti-rabbit Alexa Fluor 488, 594, anti-chicken Alexa Fluor 488 and 647, and anti-goat Alexa Fluor 488, 568, 633; all from goat or donkey (1:500, ThermoFisher Scientific, Eugene, OR).
Imaging and image analysis
All brightfield images and immunofluorescent images were taken on the Leica Thunder Imager DMi8. Images were acquired at 20x air or 40× oil immersion objectives using a Leica K5 camera or a Leica DMC5400 color camera. Leica Biosystems LAS X software was used for image capture. Tiles covering the region of interest were taken and stitched. Leica Thunder instant computational clearing was used to remove out of focus light.
All observers were blind to experimental conditions. Tiff files were used for analysis in Qupath 0.4.2. Annotations delineating the dorsal caudate nucleus or the entirety of the cingulate cortex at the level of the caudate nucleus head were created. Next, to detect cells, we used the “cell detection” function (analysis tab), and the DAPI channel was set as the Detection Channel. Next, we trained an object classifier to classify the detections for the different channels. Training data was created from each image, for each channel, to mark the cells positive for a specific antigen. One classifier per channel was trained by calling the “train object classifier” function under the analysis tab > classify with the following parameters: type = Random Trees, measurements = Cell: Channel X standard deviation, mean, max, and min measurements for the channel in question. To increase the accuracy of the classifier, additional training annotations were created on the image in question until the classification results matched the impression of the observer. A composite classifier was created from the individual trained classifiers using the “create composite classifier” function. Training images were included from all slides quantified, and classifiers were re-trained for each image separately as appropriate. To perform a morphological analysis of GFAP-positive cells, 12 GFAP-positive cells per case were quantified. The morphology was analyzed by counting the number of processes per cell and determining the average length of those processes. A two-tailed t-test was performed comparing these parameters between the HD and control group with N = 3 for both groups.
Genome-wide genotyping and imputation of Venezuelan patients, and data acquisition from GeM-HD consortium
DNA was extracted from EBV-transformed lymphoblastoid cell lines established from blood samples of the Venezuelan individuals using the Qiagen Blood and Tissue DNeasy Kit. Genotyping on these DNA samples was performed at the New York Genome Center on a HiScan Illumina machine with the Illumina CoreExome Array. The genotypes were then imputed using the IMPUTE2 software package (version 2.3.2) and the publicly available 1000 Genomes reference panel, along with whole genome sequences from select Venezuelan patient samples. For the GeM-HD consortium patients62, genotyping array data was downloaded from dbGaP (Study Accession: phs000222.v4.p2). The IMPUTE2 software package was used to impute these downloaded genotypes, guided by the 1000 Genomes reference panel.
Genome-wide association study of residual age of onset
For the comprehensive mega-analysis, the genotypic and clinical data from the Venezuelan patients were combined with the data from the GeM-HD consortium. A quantitative genome-wide association (GWA) test was carried out using the GENESIS software package (version 2.30). A linear mixed model regression was implemented using the residual age at motor onset as a phenotype, allowing for adjustment of empirical pairwise relatedness, as well as other covariates such as sex and population structure. The residual age of onset was calculated using the motor age of onset and CAG repeat length. To subtract the effects of CAG repeats from the age of onset, a previously developed phenotype model was utilized. To mitigate skewness in the distribution of residual age at onset and to model a theoretical normal distribution, a Box-Cox power transformation was applied to the distribution prior to its usage as a phenotype for association testing. The resulting significance values from the GWA were uploaded to the LocusZoom browser and plotted for the rs3812963 locus – the y-axis indicates a p value threshold of 10−6. Although the SNPs in the MT locus did not meet the traditional cut-off for GWAS (10−8), we proceeded with examining the association of the SNPs of interest with MT gene levels in the xQTL database – see main text.
Real-time quantitative PCR
Total RNA was extracted from brain specimens using RNAeasy minikit (Qiagene©). RNA concentration and purity were determined using NanoDrop (Thermo Scientific™, MA). RNA was converted to cDNA using High-capacity RNA-to-cDNA kit (Thermo Fisher Scientific, Applied Biosystems™, MA). The following Taqman assays were used (GLUL cat #4453320, SLC1A2 cat# 44488952 and GADPH cat#4448484). The reaction volumes were 10–15 µl per reaction. TaqMan™ Multiplex Master Mix (Thermo Fisher Scientific cat# 4461881) was used. All reactions included 2–5 ng of cDNA. Thermal cycling parameters were conducted per manufacturer’s standard recommendations. The qPCR plates were read on a QuantStudio™ 5 Real-time PCR system (Thermo Fisher Scientific, Applied Biosystems™, MA). The reactions were done in triplicates. Relative gene expression was calculated using the delta delta Ct method with GAPDH or RPL13A as a reference genes. N = three biological replicates. For MT genes, qPCR was preformed using PowerTrack SYBR Green Master mix for qPCR kit (Thermofisher, cat#A46109). Primers: MT1E (Invitrogen cat#10336022), MT1A (Invitrogen cat#10336022), MT1X (Invitrogen cat#10336022), MT2 (Invitrogen cat#10336022), MT3 (Invitrogen cat#10336022) and RPL13A (Invitrogen cat#10336022).
Cell culture and transduction of human astrocytes
Human astrocytes (Sciencell© cat#1800) were cultured in Astrocyte culture medium (Sciencell © cat#1801) according to vendor’s protocols on poly L-Lysine coated cell culture plates. The following Lentiviruses were obtained from VectorBuilder ™; Lentivirus transduction for mGFP (pLenti-C-mGFP-P2A-Puro – Origene Cat# RC211875L4V), CLU_mGFP (CLU(mGFP–tagged)-human clusterin(CLU) - Origene Cat# PS100093V), and MT3 (pLV[Exp]-EGFP:T2A:Puro-EF1A > hMT3 VectorBuilder cat# VB900003-8937eud). Glutamate levels in culture media of GFP or MT3 transduced astrocytes were measured using Cayman glutamate colorimetric/Fluorometric assay (cat# 702330) according to manufacturer suggested protocol using a glutamate oxidase-based conversion of glutamate to alpha-ketoglutarate, ammonia, and H2O2 followed by a horseradish peroxidase-based reaction generating a fluorescent substrate excitable at 530 nm. Fluorescence was collected at 590 nm on a Varioskan Lux plate reader in a 96 well-plate.
Patient-derived neuronal cultures
Striatal spiny projection neurons (SPNs) were directly converted from the fibroblasts of healthy controls and HD patients using brain-enriched microRNAs, miR-9/9* and miR-124 and subtype-defining transcription factors CTIP2, DLX1/2, and MYT1L as previously described68,106. Specifically, three independent cell lines were used: ND30013 - male, AG04194 - female, and AG04230 - male, with CAG repeats of 43, 46, 45, respectively. P2A (GFP-transduced) astrocytes and MT3 astrocytes were co-cultured with Control SPNs (Ctrl-SPNs – n = 1 cell line) and HD-SPNs (n = 3 cell lines) at 1:1 ratio on post-induction day 22 (PID22). Reprogrammed cells were treated with Incucyte Caspase-3/7 Green Reagent and Annexin V Red Reagent on PID27. Four-to-six technical replicates per condition were used. Imaging scheduling, collection and data analyses were performed with the Incucyte Live-Cell Analysis System. Ctrl-SPNs and HD-SPNs were imaged every 24 h for 8 days (PID28 to 35). Images were analyzed for the number of green or red objects per well of 96 well plates. For the apoptotic index, the number of green or red objects divided by phase area (μm2) per well was quantified.
Co-cultures of astrocyte with murine neurons
Murine Neuro 2a (N2A – Sigma cat # 89121404) were cultured in DMEM (11995073, Gibco) containing 10% fetal bovine serum (FBS, A3840301, Gibco) and 1% penicillin/streptomycin at 37 °C and 5% CO2. The cells were seeded onto a cell culture treated 24 well-plate at 3.5 × 104 cells/well in for 24 h. The next day, the media was changed and switched to a DMEM differentiation media containing 2% FBS and 20 µM of Retinoic acid. Cells were allowed to differentiate for 24 h before adding control GFP or MT3 overexpressing astrocytes at 3.5 × 104 cells/well in astrocyte culture medium (Sciencell©). The cells were co-cultured for 24 h before adding Cadmium at 50 µM or 10 µM, or Rotenone at 200 nM or 20 nM (HY-B1756, MedChemExpress), versus DMSO. 24 h later, the cells were stained with Propidium Iodide (Invitrogen cat# P3566) at 1:500 for 30 min prior to washing, trypsinization, and analysis by flowcytometry using BD Bioscience LSRII flowcytometer. After gating on the live singlet cells, the percentage of positive FITC± that were PI± were quantified by FCS express 7 (De Novo Software). The experiment was replicated 4 times.
Astrocyte-microglia co-culture for microglia RNAseq and phagocytosis assay
HMC3 cells (ATCC cat#CRL-3304) were seeded in 24 well-plate at 5 × 104 each well in 0.5 ml culture medium (DMEM with 10% FBS medium) and cultured overnight before co-culture with control, CLU overexpressing, or MT3 overexpressing astrocytes in a transwell assay. The astrocytes were seeded in the upper chamber of a 24 well-plate 0.4 µm polycarbonate membrane inserts (costar cat#3413) pre-coated with Poly L-Lysine, at 5 × 104 cells per insert in 0.1 ml of astrocyte growth medium (Sciencell™) 3 h before transferring the insert to the wells containing the microglia. The two cell types were co-cultured for 24 h before the astrocyte inserts were removed for collecting microglial RNA for RNAseq or conducting the phagocytosis assay. For the latter, the media was changed into fresh HMC3 media containing fluorescent latex beads (1:500, latex –beads-rabbit IgG-FITC complex—Cayman chemical cat#500290). The cells were incubated for 30 min at 37 °C before being washed as the manufacturer protocol, trypsinized, and analyzed by flowcytometry using BD Bioscience LSRII flowcytometer. The percentage of positive FITC+ cells was evaluated by FCS express 7 (De Novo Software) after gating on the live singlet cells. The experiment was replicated 4 times.
Murine neuronal cultures and viability studies
N2A cells were cultured as above but with the following changes. Cells were seeded at 20,000 per well in 96-well plates (3340, Corning). After 24 h of culture in DMEM with 10% FBS, the culture medium was removed. The N2A cells were then cultured for an additional 24 h in DMEM with 2% FBS and 20 µM retinoic acid (R2625, Sigma) to induce neuronal differentiation. To assess the cytotoxicity of Dihomo-γ-linolenic acid (DGLA, HY-A0143, MedChemExpress) on neurons, differentiated N2a cells were cultured with DGLA at 20, 100, and 200 µg/ml or equivalent weight/volume control ethanol (1:5000, 1:1000, or 1:500, respectively) in the absence or presence of 20 µM Rotenone or DMSO.
After 24 h of treatment, the viability of the neurons was determined using PrestoBlue™ HS Cell Viability Reagent (P50201, Invitrogen). Specifically, the culture medium was removed, and 100 µL of culture medium was added to each well containing 10% PrestoBlue reagent. The plates were then returned to the incubator for 20 min and fluorescence was measured using VarioSkan Lux (VLBLATGD1, ThermoFisher) at 560/590 nm (excitation/emission). Viability values above 100% are explained by well-to-well variability in control-normalized fluorescence values. However, these values were not significantly different from 100% when a one-sample t-test comparing the values per group to a hypothetical mean of 100% (two tailed p values: 0.9994, 0.9332, 0.6901, 0.1100 for the groups shown in Fig. 2F, respectively).
Data analysis and statistics
Unpaired t-tests were used unless otherwise stated. Paired tests were used for data shown in Fig. S11G and Fig. 7D, E, as the data was derived from three/four experiments each with condition and control. At least 3 technical replicates were averaged per experiment. The average fold-change from control per experiment experiments was used as a biological replicate. Since there was a batch effect related to experiments being done on separate plates and separate days, we used pairing to control for this confounding effect. A one-tailed t-test was used when we had a prior hypothesis, and when we assigned equal weights to results of the alternative hypothesis (no differences or differences opposite to what was expected), because both equally invalidate the driving hypothesis107. In Fig. S11G, a two-tailed test was used because there was no prior hypothesis regarding the effects of MT3 on phagocytosis. In analyses in Fig. 6B, D, F, Fig. S7F, and Fig. 7F, we use one-tailed unpaired t-tests because our transcriptomic analysis allowed us to formulate a priori hypotheses: MT3 was decreased in caudate astrocytes (Fig. 6D), MT3 was increased in cingulate astrocytes (Fig. 6F), LGR5 was reduced in caudate neurons (S7F), and MT3 could increase viability (Fig. 7F) - based on predicted function of metallothioneins. For Fig. S3, one-way anova followed by Tuckey’s test for multiple comparisons was done. The n and p values are indicated in the respective figures and legends. For qPCR tests to verify overexpression of CLU and MT3 in CLU and MT3 overexpressing astrocytes, respectively, a one-sample one-tailed t-test was used. Figures 7H and S11A were created in BioRender.com.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
O.A. was supported previously through funding from the Huntington Disease Society of America and the Hereditary Disease Foundation, and Currently by the American brain tumor association, NIA ADRC REC program Grant Number P30AG066462. O.A., F.P., J.E.G. and V.M. are supported by NIH R21 AG075754. This research was supported by the Digital Computational Pathology Laboratory in the Department of Pathology and Cell Biology at Columbia University Irving Medical Cancer, and by the Biomarkers Core Laboratory at the Irving Institute for Clinical and Translational Research, home to Columbia University’s Clinical and Translational Science Award. We thank Nancy Wexler for all her support throughout this project.
Author contributions
F.P., A.Y., J.E.G., V.M., and O.A. designed the study. O.A., A.T,. K.O., N.M., H.L., J.L., J.S.K., A.M. performed the experiments. J.S.K. and A.Y. perform the HD patient neuronal cultures experiments. C.W.N., A.B., D.E.H., C.M., M.Z. conducted the GWAS analysis. J.V.P. and R.H. provided the brain tissue and performed neuropathologic studies. H.L. and S.X. performed cell culture and flowcytometry analysis. SMD performed lipidomic analysis. J.L., N.M., A.T. performed the sequencing and IHC experiments. K.O. performed the WGCNA analysis. O.A., F.P., K.J., N.M., V.M., J.S.K., A.T., J.E.G., C.N., S.M.D. analyzed the data. All authors read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The human snRNAseq and bulk RNAseq dataset have been deposited in the GEO database under accession GSE242198. The lipidomics dataset is provided in the supplementary information. Source data are provided with this paper.
Code availability
The customized code and object used for analysis in this manuscript is provided here: https://github.com/Al-Dalahmah-lab/HD_astrocytes_paper/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vilas Menon, Email: vm2545@cumc.columbia.edu.
Osama Al-Dalahmah, Email: oa2298@cumc.columbia.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50626-0.
References
- 1.Hirano, M. et al. Clinicopathological differences between the motor onset and psychiatric onset of Huntington’s disease, focusing on the nucleus accumbens. Neuropathology39, 331–341 (2019). 10.1111/neup.12578 [DOI] [PubMed] [Google Scholar]
- 2.Hebb, M. O., Denovan-Wright, E. M. & Robertson, H. A. Expression of the Huntington’s disease gene is regulated in astrocytes in the arcuate nucleus of the hypothalamus of postpartum rats. FASEB J.13, 1099–1106 (1999). 10.1096/fasebj.13.9.1099 [DOI] [PubMed] [Google Scholar]
- 3.DiFiglia, M. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science277, 1990–1993 (1997). 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- 4.Mangiarini, L. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell87, 493–506 (1996). 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- 5.Roos, R. A., Pruyt, J. F., de Vries, J. & Bots, G. T. Neuronal distribution in the putamen in Huntington’s disease. J. Neurol. Neurosurg. Psychiatry48, 422–425 (1985). 10.1136/jnnp.48.5.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosas, H. D. et al. Cerebral cortex and the clinical expression of Huntington’s disease: complexity and heterogeneity. Brain131, 1057–1068 (2008). 10.1093/brain/awn025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vonsattel, J. P. et al. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol.44, 559–577 (1985). 10.1097/00005072-198511000-00003 [DOI] [PubMed] [Google Scholar]
- 8.Reiner, A. et al. Differential loss of striatal projection neurons in Huntington disease. Proc. Natl Acad. Sci. USA85, 5733–5737 (1988). 10.1073/pnas.85.15.5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy, J. C. L. et al. Somatic CAG expansion in Huntington’s disease is dependent on the MLH3 endonuclease domain, which can be excluded via splice redirection. Nucleic Acids Res.49, 3907–3918 (2021). 10.1093/nar/gkab152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonitel, R. et al. DNA instability in postmitotic neurons. Proc. Natl Acad. Sci. USA105, 3467–3472 (2008). 10.1073/pnas.0800048105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelbourne, P. F. et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet.16, 1133–1142 (2007). 10.1093/hmg/ddm054 [DOI] [PubMed] [Google Scholar]
- 12.Wang, N. et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat. Med.20, 536–541 (2014). 10.1038/nm.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swami, M. et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet.18, 3039–3047 (2009). 10.1093/hmg/ddp242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuccato, C. et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science293, 493–498 (2001). 10.1126/science.1059581 [DOI] [PubMed] [Google Scholar]
- 15.Lim, R. G. et al. Huntington disease oligodendrocyte maturation deficits revealed by single-nucleus RNAseq are rescued by thiamine-biotin supplementation. Nat. Commun.13, 7791 (2022). 10.1038/s41467-022-35388-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vonsattel, J. P., Keller, C. & Del Pilar Amaya, M. Neuropathology of Huntington’s disease. Handb. Clin. Neurol.89, 599–618 (2008). 10.1016/S0072-9752(07)01256-0 [DOI] [PubMed] [Google Scholar]
- 17.Lievens, J. C. et al. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol. Dis.8, 807–821 (2001). 10.1006/nbdi.2001.0430 [DOI] [PubMed] [Google Scholar]
- 18.Jiang, R., Diaz-Castro, B., Looger, L. L. & Khakh, B. S. Dysfunctional calcium and glutamate signaling in striatal astrocytes from Huntington’s disease model mice. J. Neurosci.36, 3453–3470 (2016). 10.1523/JNEUROSCI.3693-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faideau, M. et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum. Mol. Genet.19, 3053–3067 (2010). 10.1093/hmg/ddq212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz-Castro, B., Gangwani, M. R., Yu, X., Coppola, G. & Khakh, B. S. Astrocyte molecular signatures in Huntington’s disease. Sci. Transl. Med.11, eaaw8546 (2019). [DOI] [PubMed]
- 21.Al-Dalahmah, O. et al. Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol. Commun.8, 19 (2020). 10.1186/s40478-020-0880-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford, J. et al. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc. Natl Acad. Sci. USA106, 22480–22485 (2009). 10.1073/pnas.0911503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradford, J. et al. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J. Biol. Chem.285, 10653–10661 (2010). 10.1074/jbc.M109.083287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao, H. Y., Chen, Y. C., Chen, H. M., Tu, P. H. & Chern, Y. A critical role of astrocyte-mediated nuclear factor-kappaB-dependent inflammation in Huntington’s disease. Hum. Mol. Genet.22, 1826–1842 (2013). 10.1093/hmg/ddt036 [DOI] [PubMed] [Google Scholar]
- 25.Wood, T. E. et al. Mutant huntingtin reduction in astrocytes slows disease progression in the BACHD conditional Huntington’s disease mouse model. Hum. Mol. Genet.28, 487–500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benraiss, A. et al. Cell-intrinsic glial pathology is conserved across human and murine models of Huntington’s disease. Cell Rep.36, 109308 (2021). 10.1016/j.celrep.2021.109308 [DOI] [PubMed] [Google Scholar]
- 27.Yu, X. et al. Reducing astrocyte calcium signaling in vivo alters striatal microcircuits and causes repetitive behavior. Neuron99, 1170–1187.e1179 (2018). 10.1016/j.neuron.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes-Ortiz, A. M. et al. Single-nuclei transcriptome analysis of Huntington disease iPSC and mouse astrocytes implicates maturation and functional deficits. iScience26, 105732 (2023). 10.1016/j.isci.2022.105732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meunier, C., Merienne, N., Jolle, C., Deglon, N. & Pellerin, L. Astrocytes are key but indirect contributors to the development of the symptomatology and pathophysiology of Huntington’s disease. Glia64, 1841–1856 (2016). 10.1002/glia.23022 [DOI] [PubMed] [Google Scholar]
- 30.Gangwani, M. R. et al. Neuronal and astrocytic contributions to Huntington’s disease dissected with zinc finger protein transcriptional repressors. Cell Rep.42, 111953 (2023). 10.1016/j.celrep.2022.111953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benraiss, A. et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat. Commun.7, 11758 (2016). 10.1038/ncomms11758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong, X. et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci.17, 694–703 (2014). 10.1038/nn.3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arregui, L., Benitez, J. A., Razgado, L. F., Vergara, P. & Segovia, J. Adenoviral astrocyte-specific expression of BDNF in the striata of mice transgenic for Huntington’s disease delays the onset of the motor phenotype. Cell. Mol. Neurobiol.31, 1229–1243 (2011). 10.1007/s10571-011-9725-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giralt, A. et al. BDNF regulation under GFAP promoter provides engineered astrocytes as a new approach for long-term protection in Huntington’s disease. Gene Ther.17, 1294–1308 (2010). 10.1038/gt.2010.71 [DOI] [PubMed] [Google Scholar]
- 35.Oliveira, A. O., Osmand, A., Outeiro, T. F., Muchowski, P. J. & Finkbeiner, S. alphaB-Crystallin overexpression in astrocytes modulates the phenotype of the BACHD mouse model of Huntington’s disease. Hum. Mol. Genet.25, 1677–1689 (2016). 10.1093/hmg/ddw028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labadorf, A. et al. RNA sequence analysis of human huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS ONE10, e0143563 (2015). 10.1371/journal.pone.0143563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodges, A. et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet.15, 965–977 (2006). 10.1093/hmg/ddl013 [DOI] [PubMed] [Google Scholar]
- 38.Lee, H. et al. Cell type-specific transcriptomics reveals that mutant Huntingtin leads to mitochondrial RNA release and neuronal innate immune activation. Neuron 107, 891–908.e8 (2020). [DOI] [PMC free article] [PubMed]
- 39.Garcia, F. J. et al. Single-cell dissection of the human brain vasculature. Nature603, 893–899 (2022). 10.1038/s41586-022-04521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renelt, M., von Bohlen und Halbach, V. & von Bohlen und Halbach, O. Distribution of PCP4 protein in the forebrain of adult mice. Acta Histochem.116, 1056–1061 (2014). 10.1016/j.acthis.2014.04.012 [DOI] [PubMed] [Google Scholar]
- 41.Tyagi, E., Fiorelli, T., Norden, M. & Padmanabhan, J. Alpha 1-antichymotrypsin, an inflammatory protein overexpressed in the brains of patients with Alzheimer’s disease, induces tau hyperphosphorylation through c-Jun N-terminal kinase activation. Int J. Alzheimers Dis.2013, 606083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Dalahmah, O. et al. The Matrix Receptor CD44 is present in astrocytes throughout the human central nervous system and accumulates in hypoxia and seizures. Cells13, 129 (2024). [DOI] [PMC free article] [PubMed]
- 43.Block, R. C., Dorsey, E. R., Beck, C. A., Brenna, J. T. & Shoulson, I. Altered cholesterol and fatty acid metabolism in Huntington disease. J. Clin. Lipido.4, 17–23 (2010). 10.1016/j.jacl.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreilaus, F., Spiro, A. S., Hannan, A. J., Garner, B. & Jenner, A. M. Brain cholesterol synthesis and metabolism is progressively disturbed in the R6/1 mouse model of Huntington’s disease: a targeted GC-MS/MS sterol. Anal. J. Huntingt. Dis.4, 305–318 (2015). 10.3233/JHD-150170 [DOI] [PubMed] [Google Scholar]
- 45.Kreilaus, F., Spiro, A. S., McLean, C. A., Garner, B. & Jenner, A. M. Evidence for altered cholesterol metabolism in Huntington’s disease post mortem brain tissue. Neuropathol. Appl. Neurobiol.42, 535–546 (2016). 10.1111/nan.12286 [DOI] [PubMed] [Google Scholar]
- 46.Phillips, G. R. et al. Cholesteryl ester levels are elevated in the caudate and putamen of Huntington’s disease patients. Sci. Rep.10, 20314 (2020). 10.1038/s41598-020-76973-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valenza, M. et al. Dysfunction of the cholesterol biosynthetic pathway in Huntington’s disease. J. Neurosci.25, 9932–9939 (2005). 10.1523/JNEUROSCI.3355-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valenza, M. et al. Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation. Hum. Mol. Genet.16, 2187–2198 (2007). 10.1093/hmg/ddm170 [DOI] [PubMed] [Google Scholar]
- 49.Valenza, M. et al. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington’s disease. Neurobiol. Dis.28, 133–142 (2007). 10.1016/j.nbd.2007.07.004 [DOI] [PubMed] [Google Scholar]
- 50.Lee, J. A., Hall, B., Allsop, J., Alqarni, R. & Allen, S. P. Lipid metabolism in astrocytic structure and function. Semin. Cell Dev. Biol.112, 123–136 (2021). 10.1016/j.semcdb.2020.07.017 [DOI] [PubMed] [Google Scholar]
- 51.Garcia Corrales, A. V., Haidar, M., Bogie, J. F. J. & Hendriks, J. J. A. Fatty acid synthesis in glial cells of the CNS. Int. J. Mol. Sci.22, 8159 (2021). [DOI] [PMC free article] [PubMed]
- 52.Aizawa, F. et al. Astrocytes release polyunsaturated fatty acids by lipopolysaccharide stimuli. Biol. Pharm. Bull.39, 1100–1106 (2016). 10.1248/bpb.b15-01037 [DOI] [PubMed] [Google Scholar]
- 53.Sosunov, A. A. et al. Phenotypic heterogeneity and plasticity of isocortical and hippocampal astrocytes in the human brain. J. Neurosci.34, 2285–2298 (2014). 10.1523/JNEUROSCI.4037-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isani, G. & Carpene, E. Metallothioneins, unconventional proteins from unconventional animals: a long journey from nematodes to mammals. Biomolecules4, 435–457 (2014). 10.3390/biom4020435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalathur, R. K. et al. The unfolded protein response and its potential role in Huntington’s disease elucidated by a systems biology approach. F1000Res4, 103 (2015). 10.12688/f1000research.6358.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran, M. N. et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron109, 3088–3103.e3085 (2021). 10.1016/j.neuron.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glass, M., Dragunow, M. & Faull, R. L. The pattern of neurodegeneration in Huntington’s disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington’s disease. Neuroscience97, 505–519 (2000). 10.1016/S0306-4522(00)00008-7 [DOI] [PubMed] [Google Scholar]
- 58.Deng, Y. P. et al. Differential loss of striatal projection systems in Huntington’s disease: a quantitative immunohistochemical study. J. Chem. Neuroanat.27, 143–164 (2004). 10.1016/j.jchemneu.2004.02.005 [DOI] [PubMed] [Google Scholar]
- 59.Hedreen, J. C. & Folstein, S. E. Early loss of neostriatal striosome neurons in Huntington’s disease. J. Neuropathol. Exp. Neurol.54, 105–120 (1995). 10.1097/00005072-199501000-00013 [DOI] [PubMed] [Google Scholar]
- 60.Morton, A. J., Nicholson, L. F. & Faull, R. L. Compartmental loss of NADPH diaphorase in the neuropil of the human striatum in Huntington’s disease. Neuroscience53, 159–168 (1993). 10.1016/0306-4522(93)90294-P [DOI] [PubMed] [Google Scholar]
- 61.Donaldson, J., Powell, S., Rickards, N., Holmans, P. & Jones, L. What is the Pathogenic CAG Expansion Length in Huntington’s Disease? J. Huntingt. Dis.10, 175–202 (2021). 10.3233/JHD-200445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genetic Modifiers of Huntington’s Disease, C. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell162, 516–526 (2015). 10.1016/j.cell.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moss, D. J. H. et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol.16, 701–711 (2017). 10.1016/S1474-4422(17)30161-8 [DOI] [PubMed] [Google Scholar]
- 64.Genetic Modifiers of Huntington’s Disease Consortium. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell178, 887–900.e814 (2019). [DOI] [PMC free article] [PubMed]
- 65.Lee, J. M. et al. Genetic modifiers of Huntington disease differentially influence motor and cognitive domains. Am. J. Hum. Genet109, 885–899 (2022). 10.1016/j.ajhg.2022.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng, B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci.20, 1418–1426 (2017). 10.1038/nn.4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murakami, S., Miyazaki, I., Sogawa, N., Miyoshi, K. & Asanuma, M. Neuroprotective effects of metallothionein against rotenone-induced myenteric neurodegeneration in parkinsonian mice. Neurotox. Res.26, 285–298 (2014). 10.1007/s12640-014-9480-1 [DOI] [PubMed] [Google Scholar]
- 68.Victor, M. B. et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci.21, 341–352 (2018). 10.1038/s41593-018-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trendelenburg, G. et al. Serial analysis of gene expression identifies metallothionein-II as major neuroprotective gene in mouse focal cerebral ischemia. J. Neurosci.22, 5879–5888 (2002). 10.1523/JNEUROSCI.22-14-05879.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung, R. S. et al. Neuron-glia communication: metallothionein expression is specifically up-regulated by astrocytes in response to neuronal injury. J. Neurochem88, 454–461 (2004). 10.1046/j.1471-4159.2003.02193.x [DOI] [PubMed] [Google Scholar]
- 71.Stankovic, R. K., Chung, R. S. & Penkowa, M. Metallothioneins I and II: neuroprotective significance during CNS pathology. Int. J. Biochem. cell Biol.39, 484–489 (2007). 10.1016/j.biocel.2006.09.010 [DOI] [PubMed] [Google Scholar]
- 72.Michael, G. J. et al. Up-regulation of metallothionein gene expression in parkinsonian astrocytes. Neurogenetics12, 295–305 (2011). 10.1007/s10048-011-0294-5 [DOI] [PubMed] [Google Scholar]
- 73.Uchida, Y., Gomi, F., Masumizu, T. & Miura, Y. Growth inhibitory factor prevents neurite extension and the death of cortical neurons caused by high oxygen exposure through hydroxyl radical scavenging. J. Biol. Chem.277, 32353–32359 (2002). 10.1074/jbc.M111263200 [DOI] [PubMed] [Google Scholar]
- 74.Erickson, J. C., Hollopeter, G., Thomas, S. A., Froelick, G. J. & Palmiter, R. D. Disruption of the metallothionein-III gene in mice: analysis of brain zinc, behavior, and neuron vulnerability to metals, aging, and seizures. J. Neurosci.17, 1271–1281 (1997). 10.1523/JNEUROSCI.17-04-01271.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koh, J. Y. & Lee, S. J. Metallothionein-3 as a multifunctional player in the control of cellular processes and diseases. Mol. Brain13, 116 (2020). 10.1186/s13041-020-00654-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klaassen, C. D., Liu, J. & Diwan, B. A. Metallothionein protection of cadmium toxicity. Toxicol. Appl. Pharmacol.238, 215–220 (2009). 10.1016/j.taap.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rivera-Mancia, S. et al. The transition metals copper and iron in neurodegenerative diseases. Chem. Biol. Interact.186, 184–199 (2010). 10.1016/j.cbi.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 78.Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol.37, 911–917 (1959). 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- 79.Hands, S. L., Mason, R., Sajjad, M. U., Giorgini, F. & Wyttenbach, A. Metallothioneins and copper metabolism are candidate therapeutic targets in Huntington’s disease. Biochem Soc. Trans.38, 552–558 (2010). 10.1042/BST0380552 [DOI] [PubMed] [Google Scholar]
- 80.Chung, R. S. et al. Redefining the role of metallothionein within the injured brain: extracellular metallothioneins play an important role in the astrocyte-neuron response to injury. J. Biol. Chem.283, 15349–15358 (2008). 10.1074/jbc.M708446200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sieradzan, K. A. & Mann, D. M. The selective vulnerability of nerve cells in Huntington’s disease. Neuropathol. Appl. Neurobiol.27, 1–21 (2001). 10.1046/j.0305-1846.2001.00299.x [DOI] [PubMed] [Google Scholar]
- 82.Lee, J. M. et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol.4, 29 (2010). 10.1186/1752-0509-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44, W90–W97 (2016). 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed]
- 87.Chan, R. B. et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem.287, 2678–2688 (2012). 10.1074/jbc.M111.274142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform.9, 559 (2008). 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47–e47 (2015). 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics8, 118–127 (2007). 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 91.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKenzie, A. T., Katsyv, I., Song, W. M., Wang, M. & Zhang, B. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst. Biol.10, 106 (2016). 10.1186/s12918-016-0349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rohart, F., Gautier, B., Singh, A. & Le Cao, K. A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol.13, e1005752 (2017). 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics33, 1179–1186 (2017). 10.1093/bioinformatics/btw777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Petukhov, V. et al. dropEst: pipeline for accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Genome Biol.19, 78 (2018). 10.1186/s13059-018-1449-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang, S. et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol.21, 57 (2020). 10.1186/s13059-020-1950-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420 (2018). 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e3529 (2021). 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma.14, 7 (2013). 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuchroo, M. et al. Multiscale PHATE identifies multimodal signatures of COVID-19. Nat. Biotechnol.40, 681–691 (2022). 10.1038/s41587-021-01186-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics19, 477 (2018). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Van den Berge, K. et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun.11, 1201 (2020). 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin, H. & Peddada, S. D. Analysis of compositions of microbiomes with bias correction. Nat. Commun.11, 3514 (2020). 10.1038/s41467-020-17041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oh, Y. M. et al. Age-related Huntington’s disease progression modeled in directly reprogrammed patient-derived striatal neurons highlights impaired autophagy. Nat. Neurosci.25, 1420–1433 (2022). 10.1038/s41593-022-01185-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruxton, G. D. & Neuhäuser, M. When should we use one-tailed hypothesis testing? Methods Ecol. Evol.1, 114–117 (2010). 10.1111/j.2041-210X.2010.00014.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The human snRNAseq and bulk RNAseq dataset have been deposited in the GEO database under accession GSE242198. The lipidomics dataset is provided in the supplementary information. Source data are provided with this paper.
The customized code and object used for analysis in this manuscript is provided here: https://github.com/Al-Dalahmah-lab/HD_astrocytes_paper/.