Abstract
Enterovirus infection and persistence have been implicated in the pathogenesis of certain chronic muscle diseases. In vitro studies suggest that persistent enteroviruses mutate, evolving into forms that are less lytic and display altered tropism, but it is less clear whether these mechanisms operate in vivo. In this study, persistent coxsackievirus RNA from the muscle of mice afflicted with chronic inflammatory myopathy (CIM) was characterized and compared with RNA from a virus that had established a persistent infection of G8 mouse myoblasts for 30 passages in vitro. Competitive strand-specific reverse transcription-PCR and susceptibility to RNase I treatment revealed that plus- and minus-strand viral RNAs were present at nearly equivalent levels in muscle and that they persisted in a double-stranded conformation. All regions of the viral genome persisted and were amplified as a series of seven overlapping fragments. Restriction endonuclease fingerprinting coupled with sequencing indicated that there was no evolution of the viral genome associated with its persistence in muscle. This contrasted with the productive persistent infection that was established in myoblast cultures, where plus-strand RNA predominated and persistent virus developed distinct mutations. In vitro persistence proceeded by a carrier culture mechanism and was completely dependent on production of infectious virus, since persistent viral RNA was not detected in cultures subjected to antibody-mediated curing. These experiments demonstrate that persistence of coxsackievirus RNA in muscle is not facilitated by distinct genetic changes in the virus that give rise to replication-defective forms but occurs primarily through production of stable double-stranded RNA that is produced as the acute viral infection resolves. The data suggest a mechanism for coxsackievirus persistence in myofibers and perhaps other nondividing cells whereby cells that survive infection could harbor persistent viral RNA for extended times without producing detectable levels of infectious virus.
Coxsackievirus-induced chronic inflammatory myopathy (CIM) is an experimental model in which viral infection initiates a chronic postviral immunopathic muscle disease. Infection of newborn mice with the Tucson strain of coxsackievirus B1 (CVB1T) initially causes an acute myotropic infection that lasts for 2 weeks (37, 40). As the acute infection resolves, CIM develops and is manifest as chronic mononuclear cell infiltration of the muscle accompanied by ongoing myofiber degeneration and regeneration. The histopathology resembles that found in human inflammatory myopathies such as polymyositis and dermatomyositis. CIM expression is T-cell dependent and does not occur in T-cell-depleted or nude mice (50, 51). Levels of CIM peak around 1 month after initial infection and show a slow steady decline thereafter. Using standard techniques of homogenization and plating on BGMK cells, infectious virus cannot be recovered when CIM is prominent, even though long-term persistence of viral RNA in muscle has been documented by both in situ hybridization and reverse transcription-PCR (RT-PCR) (44, 45). Persistent viral RNA is readily detected at 1 month after infection and declines in parallel with CIM expression, but it has been detected as long as 12 months after the initial infection (44). Persistent viral RNA is also more prevalent in strains that are susceptible to CIM and correlates positively with the increased CIM severity seen in mice which possess the H-2d haplotype (41). These observations suggest that viral RNA persistence is linked to the expression of CIM and that continued presentation of viral epitopes may be involved in promoting the immunopathic response against muscle.
Enteroviruses such as coxsackievirus, poliovirus, and echovirus are small nonenveloped viruses that belong to the picornavirus family. They possess a single-stranded RNA genome of approximately 7.4 kb that acts directly as mRNA in infected cells. Persistent enterovirus RNA has been found in patients with neuropathic and muscular diseases including chronic fatigue syndrome (7, 17), idiopathic inflammatory myopathy (8), sporadic motor neuron disease (49), dilated cardiomyopathy (2, 3, 9), and postpolio syndrome (27). These and other reports have led to the hypothesis that persistent enteroviruses might be involved in certain chronic medical conditions with unexplained pathogenesis (16), although negative results from some laboratories have spawned a debate regarding their significance (32, 33). Experimental models have also implicated enterovirus persistence as a contributing factor in the development of postinfectious pathology. In addition to being found in mice with CIM, persistence of coxsackievirus RNA has been described in mouse models of diabetes and myocarditis (4, 26, 46). The most direct evidence that persistent coxsackievirus RNA can exert pathologic effects on tissue was recently found in experiments with transgenic mice that express defective CVB3 in heart muscle. In this model, viral RNA synthesis alone, in the absence of infectious virus, is sufficient to cause abnormalities in excitation-contraction coupling and dilated cardiomyopathy (48).
The mechanisms by which enteroviruses persist are incompletely understood. Viral clearance is highly dependent on the production of antiviral antibody, and persistence of infectious virus is generally observed only in immunocompromised hosts such as patients with agammaglobulinemia (30) or during in vitro infection of certain primary cell cultures or cell lines (10, 12, 13, 18, 29). Single-stranded RNA viruses such as enteroviruses evolve rapidly due to high error rates and lack of a DNA template for use in correcting mismatches (22). As a result, microvariants or quasispecies that possess altered replication and host range characteristics may arise. Production of replication-defective variants has been proposed as a likely molecular mechanism for enterovirus persistence in vivo and is based in part on experimental data which show that mutated forms arise during persistent infection of cell cultures (5, 31). Synthesis of equal amounts of genomic (plus-strand) and template (minus-strand) viral RNAs in vivo has been observed in the muscle of patients with postviral fatigue syndrome (15) and in heart muscle of mice afflicted with coxsackievirus-induced chronic myocarditis (1, 21). These studies have been interpreted as a sign that enterovirus RNA persistence results from mutations that impair replication, but such mutations have not been mapped. In the following study, we analyzed the nature of persistent coxsackievirus RNA at the molecular level to test the hypothesis that enterovirus persistence proceeds by a mechanism involving evolution of the viral genome. For purposes of comparison, a persistent infection of mouse myoblasts was established and evaluated in parallel with persistent viral RNA amplified from muscle. These data are the first to evaluate the genetic basis for persistence of coxsackievirus RNA in muscle, and they illustrate important differences between the molecular mechanisms which underlie viral persistence in vivo and in vitro.
MATERIALS AND METHODS
Viruses.
The parental stock of CVB1T was propagated in BGMK cells as described previously (45). MP1 is a plaque-purified variant derived from parental CVB1T that induces CIM and hindlimb weakness which are comparable to those induced by the parental virus (42). MP1/M represents the viral RNA that persists in muscle at 1 month after infection. MP1/G8 is a preparation of MP1 derived from persistently infected G8 (PI-G8) cells passaged 30 times as described below. The pNI4 infectious cDNA clone of CVB1 was provided by A. Nomoto (23).
Persistent infection of G8 cells.
G8 mouse myoblasts (American Type Culture Collection [ATCC], Bethesda, Md.) were cultured on collagen-coated plates in Dulbecco’s minimal essential medium (DMEM) with high glucose containing 10% fetal bovine serum (FBS) and 10% horse serum. To establish persistent infection, 106 G8 cells were infected with 105 PFU of MP1. The initial culture was incubated for 1 week at 37°C in a humidified atmosphere containing 5% CO2. Thereafter, persistently infected G8 (PI-G8) cells were serially passaged at a 1/20 dilution when they reached 80 to 100% confluence (once or twice per week).
Infectious-center assay.
PI-G8 cells were removed from culture dishes by using trypsin-EDTA, washed twice in Hanks balanced salt solution, (HBSS), and resuspended at 106 cells/ml in HBSS containing a 1/1,000 dilution of horse anti-CVB1 neutralizing antibody (ATCC). The cells were incubated on ice for 30 min and washed three times in HBSS to remove antibody and any extracellular virus. The cells were resuspended in MEM containing 5% FBS and subjected to 10-fold dilutions in the same medium. A 1-ml volume of cells was combined with an agar overlay containing 2 ml of modified Eagle medium (MEM), 5% FBS, and 2.7% Bacto Agar at 50°C. This cellular overlay was plated onto a confluent monolayer of BGMK cells in a 100-mm petri dish. The agar was allowed to solidify, and then 7 ml of a nutrient overlay consisting of MEM, 5% FBS, and 0.9% Bacto Agar was added. The plates were incubated for 5 days at 37°C in a humidified atmosphere containing 5% CO2 and stained with 0.16% neutral red in phosphate-buffered saline. Infected cells were quantitated as PFU.
Antibody-mediated curing of PI-G8 cells.
At passage 30, replicates of PI-G8 cells were plated at 7 × 105 cells per 10 ml of medium in a 100-mm petri dish with the addition of either horse anti-CVB1 serum or horse preimmune serum (ATCC) at a final dilution of 1/1,000. The cells were passaged twice a week and maintained in either the immune or preimmune serum. At each passage, the cell-free supernatant was tested for infectious virus by being plated onto BGMK monolayers and scored for cytopathic effects. Half of the cells were disrupted by a single freeze-thaw step and tested for virus-induced cytopathic effects, while the other half was subjected to RNA extraction and amplification by RT-PCR with viral primers 2C.4365 and 3D.6693. Complete curing of a culture was confirmed by performing two additional sequential passages without the addition of horse serum and testing for infectious virus and viral RNA as described above.
RNA extraction.
MP1 RNA was prepared from infected BGMK supernatants by polyethylene glycol precipitation and acidic guanidine extraction as previously described (11, 44). For amplification of virus from muscle, RNA was extracted from frozen pulverized hamstring muscle at 1 month after infection, as previously described, by a modification of the acidic guanidine method (43). For studies of virus persistence in G8 cells, cell pellets were solubilized in guanidine and processed the same as for muscle. RNA extracted from muscle or cell cultures was immediately frozen in aliquots and stored at −70°C to minimize annealing of complementary strands.
Primers.
Oligonucleotide primers used in this study are shown in Table 1. The numbering system is based on the published sequence for CVB1 described by Iizuka et al. (23), which was also used to design some of the primers. However, as the study progressed, we determined that sufficient sequence heterogeneity was present between CVB1T and the pNI4 sequence to compromise amplification efficiency. Subsequently, regions of CVB1T were sequenced and used to design CVB1T-specific primers as indicated in Table 1.
TABLE 1.
Oligonucleotide primers used in this study
| Primer namea | Genomic location (nucleotides) | Nucleotide sequenceb (5′ to 3′) |
|---|---|---|
| Strand-specific and competitive template primers | ||
| T7.5UTR.1 | 1–20 | TAATACGACTCACTATAGGGTTAAAACAGCCTGTGGGTTG |
| T7.3UTR.7359 | 7341–7359 | TAATACGACTCACTATAGGGGCACCGTTATCTAGTTCGG |
| 2C.4365*4774 | 4365–4387* | CATACAGTTCAAGTCCAAATGCC* |
| 4774–4798 | ATCGAGGTTATCTCCATGTACAGCC | |
| 3UTR.7359 | 7341–7359 | GCACCGTTATCTAGTTCGG |
| REF primers (sense/antisense pairs) | ||
| 5UTR.1 | 1–20 | TTAAAACAGCCTGTGGGTTG |
| 5UTR.603 | 584–603 | TCAATTGTCACCATAAGCAG |
| 5UTR.450 | 450–474 | CCGGCCCCTGAATGCGGCTAATCCT |
| 1C.2154 | 2134–2154 | TGCGCCTGGTGGTGAGTATGC |
| 1C.2093 | 2093–2120 | TGTTCTGTGGGTCTGCAATGGCCACAG |
| 2B.4022 | 3994–4022 | GCCATTGGAATCCCATAATATTGGGACAC |
| 2A.3307 | 3307–3338 | GGGGCTGTCTACGTAGGCAACTACAGAGTGGT |
| 2B.4417 | 4393–4417 | CGTGCAAGAGACAGCATACAGGTTC |
| 2C.4365 | 4365–4387 | CATACAGTTCAAGTCCAAATGCC |
| 3C.5552 | 5524–5552 | CCATCCTTGTCCACCAACTCTTTAGCGTC |
| 3C.5479 | 5479–5507 | CCTGGGCCAACCATCCTGATGAATGACCA |
| 3D.6693 | 6669–6693 | GTTGGTTTCCTTGTGCGAGTATCCG |
| 3D.6693.82T-C | 6669–6693 | GTTGGTTTCCTCGTGCGAGTATCCGc |
| 3D.6255 | 6255–6279 | CCCGTATGTCGCATTAGGTATCAAG |
| Tag.3DT.C | 7389–poly(A) | CGATCTGAGTCATGTCACTGTTTTTTTTTTTTTTC |
With the exception of Tag.3DT.C, virus-specific primer names include the viral gene designation and the number of the first nucleotide on the 5′ end of the primer. In primer 2C.4365*4774, the location of a deletion that was engineered into the primer is indicated (*).
Primers 5UTR, 5UTR.603, 5UTR.450, 1C.2154, 2B.4022, and 2B.4417 were based on the published sequence of pNI4. All remaining primer sequences were based on sequence information obtained from CVB1T (unpublished data). The Tag sequence in primer Tag.3DT.C is a nonviral sequence appended to the 5′ end to facilitate amplification of the 3′ end of the virus and is italicized. T7 promoter sequences in T7.5UTR.1 and T7.3UTR.7359 are underlined.
This primer has the same sequence as 3D.6669-93 but with a T-to-C transition at nucleotide 6682 (shown in boldtype).
Strand-specific RT-PCR.
Control plus- or minus-strand viral RNAs were synthesized from a nearly full-length viral amplicon generated by primers T7.5UTR.1 and 3UTR.7359 or primers 5UTR.1 and T7.3UTR.7359, respectively. Control RNAs were synthesized by T7 transcription (MegaScript; Ambion) followed by two rounds of DNase treatment and extraction with guanidinium isothiocyanate to remove the transcription template. Transcripts were quantitated on denaturing agarose gels by densitometric scanning of the negative image (14) followed by analysis with the public domain NIH Image 1.6 program (developed at the U.S. National Institutes of Health and available on the Internet [33a]). The RT reaction mixture contained 200 U of Moloney murine leukemia virus reverse transcriptase (SuperScript II; GIBCO BRL, Gaithersburg, Md.) and 20 U of RNasin (Promega, Madison, Wis.) in a 20-μl volume. The reaction mixture was preheated to 42°C for 2 min, the reverse transcriptase was added, and the mixture was incubated at 42°C for 1 h followed by 70°C for 15 min. The minus strand was primed with 2A.3307, and the plus strand was primed with 3UTR.7359. Following RT, the primer was removed by centrifugal diafiltration through a 30K membrane (Pall Filtron, Northborough, Mass.). For amplification, 25 pmol each of primers 2C.4365 and 3D.6693 were used in a hot-start PCR with 2 U of rTth DNA polymerase XL (PE Applied Biosystems, Foster City, Calif.) and reaction conditions as recommended by the manufacturer to yield a 2.3-kb amplicon (nucleotides 4365 to 6693). The cycling conditions consisted of 94°C for 30 s, 50°C for 1 min, and 72°C for 1 min for 32 cycles. The final extension was at 72°C for 10 min. To determine whether the RNA was single or double stranded, the samples were treated with 0.2 U of RNase I (Epicenter Technologies, Madison, Wis.) for 30 min at 37°C and inactivated by heating at 70°C for 20 min immediately prior to RT. The double-stranded viral RNA control was prepared by annealing the plus and minus control strands as described for RNase I mismatch analysis in the manufacturer’s protocol.
The amounts of plus- and minus-strand viral RNA were determined by RT-PCR with a competitive template constructed with the sense primer 2C.4365*4774. This primer consisted of a 5′ sequence of viral nucleotides 4365 to 4387 and a 3′ sequence to anneal at nucleotides 4774 to 4798. Amplification by primers 2C.4365*4774 and 3D.6693 yielded a 1.9-kb amplicon, which could then be reamplified by primers 2C.4365 and 3D.6693. For strand-specific quantitation, fourfold dilutions of the 1.9-kb gel-purified competitive template were spiked with a constant amount of viral cDNA and amplified by 32 cycles of PCR. Test samples were examined by using an input of 4 μg of total RNA or, when the target was more abundant, 0.5 μg of total RNA. The amplicons were electrophoresed through 1.5% agarose gels (50:50 mixture of SeaKem and SeaPlaque; FMC, Rockland, Maine). Mass amounts of the 2.3-kb viral target and the 1.9-kb competitive template amplicon produced in each reaction were determined by densitometry as described above. The number of copies of virus was calculated at the point of equivalence, corrected for the difference in size between the target and the competitive template, and expressed per microgram of total input RNA.
REF.
Amplicons for restriction endonuclease fingerprinting (REF) analysis were generated by RT-PCR as described above with some modifications. Random hexamers were used for RT at a final concentration of 2.5 μM except when the extreme 3′ end of the virus was to be amplified, in which case 2.5 pmol of primer Tag.3DT.C was used (Table 1). Primer 3D.6693.82T-C was identical to primer 3D.6693 except for a single-base T-to-C transition at position 6682. The PCR product produced by this primer and primer 3C.5479 was used as a single-base mutation detection control for REF. The RT reaction mixtures were incubated for 10 min at 25°C (random hexamers only), 1 h at 42°C, and 15 min at 70°C. A 5-μl volume of each RT reaction mixture was then amplified by PCR in a final reaction volume of 100 μl by using the cycling profile described above. Muscle samples taken 1 month after infection were first evaluated individually for the presence of persistent viral RNA by RT-PCR amplification of a 1.7-kb product with primers 5UTR.450 and 1C.2154. Eight muscle samples that were positive for persistent viral RNA were pooled, aliquoted into individual 4-μg amounts, and stored as ethanol precipitates at −70°C.
REF was used to screen for mutations in persistent viral RNA as described by others (24, 28) with minor modifications. The starting material consisted of amplified subgenomic viral cDNA fragments that were gel purified and radiolabeled as follows. RT-PCR products were electrophoresed through low-melting-point agarose gels (SeaPlaque; FMC) in 1× Tris-acetate-EDTA (TAE). A gel slice containing the amplicon was excised and melted, and 5 μl was transferred to a tube for radiolabeling by PCR (52). The 60-μl hot-start PCR mixture contained 1.1 mM magnesium acetate, 0.02 mM each deoxyribonucleoside triphosphate, 15 pmol of each primer originally used to synthesize the amplicon, 5 μCi of [α-32P]dCTP (Amersham, Arlington Heights, Ill.), and 1 U of rTth DNA polymerase XL. Twelve cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 3 min were followed by a final extension of 72°C for 5 min. The amount of incorporated radioisotope was determined by binding to DE-81 filters (38) and was found to average 57%. Fragments were digested with at least four different combinations of restriction enzymes that had 4-base recognition sequences, including MnlI, BsaJI, MseI, MboI, RsaI, and HaeIII. A total of 7.5 × 105 cpm of product was restricted in a 20-μl volume. A 3-μl volume of restricted amplicon was added to 2 μl of denaturing loading buffer, heated to 80°C for 5 min, and quick-chilled on ice. The same volume of labeled and restricted DNA was also prepared in a nondenaturing loading buffer (PE Applied Biosystems), heated to 60°C, and chilled on ice. This sample was run in parallel to ensure that restriction was complete and to provide a reference point for the REF bands. For gel analysis, 3 μl of each sample was loaded onto a 1× GeneAmp acrylamide gel prepared as specified by the manufacturer (Perkin-Elmer) in a 38-cm by 50-cm by 0.4-mm sequencing cell (Bio-Rad, Hercules, Calif.). The running buffer was 0.5× Tris-borate-EDTA (TBE) (pH 8.3), and the gels were electrophoresed at 800 V at either 4°C or room temperature. Nondenatured samples to evaluate the restriction component of the assay were loaded and electrophoresed for 30 min. Denatured samples used for single-strand conformation polymorphism analysis were loaded 30 min later, and the gel was run for an additional 2 h. The gel was then transferred to 3MM paper, covered with plastic wrap, and autoradiographed at −70°C with Hyperfilm-MP (Amersham).
Sequencing.
Sequencing reaction mixtures contained 0.5 to 1.0 μg of gel-purified cDNA amplicon and 3.2 pmol of primer with cycle-sequencing components, and the cycling conditions used were those recommended by the manufacturer (ABI Prism Cycle Sequencing Kit; PE Applied Biosystems). The sequence was determined by using either an ABI Prism 310 Genetic Analyzer or a 370A DNA Sequencer (PE Applied Biosystems). Sequence information presented in this report was derived from both strands of cDNA and was analyzed by using GCG version 9.0 (Genetics Computer Group, Madison, Wis.) and GeneWorks version 2.45 (IntelliGenetics, Inc., Campbell, Calif.). Manual editing was performed by using parameters for automated sequencing described by Parker et al. (34).
RESULTS
Single- versus double-stranded nature of persistent viral RNA.
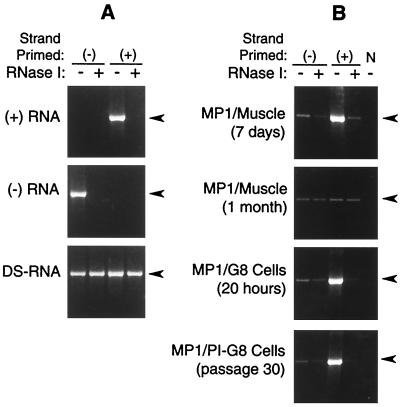
The specificity of the strand-specific RT-PCR procedure was evaluated by using viral RNAs transcribed in vitro. Both plus- and minus-strand viral RNAs were amplified only after RT with the appropriate complementary primer (Fig. 1A), indicating that the RT-PCR procedure was strand-specific with no detectable mispriming of the opposite strand. Also, no self-priming was detected in reactions without added primer (results not shown). These single-stranded RNAs were susceptible to RNase I treatment, whereas RNase I had little or no effect on amplification of the target sequence from the annealed double-stranded template. Thus, strand-specific RT-PCR, in concert with RNase I, could be used to determine whether persistent viral RNA was single or double stranded. RNA samples from infected muscle were evaluated by the same method during acute and persistent infection (Fig. 1B). At 7 days after infection, plus-strand viral RNA was present in abundance with respect to the minus strand, with low levels persisting as a double-stranded moiety. In contrast, the viral RNA that persisted at 4 weeks appeared to be present at roughly equivalent levels of plus and minus strands annealed in a double-stranded form. Viral RNA in both freshly infected G8 cells at 20 h after infection and persistently infected G8 myoblasts at passage 30 resembled that found in acutely infected muscle, with the plus strand present in excess of the minus strand and most of the minus strand complexed as double-stranded RNA. Lack of band production in the control reactions, which did not contain a primer during RT, indicates that neither self-priming nor cDNA contamination occurred in these samples.
FIG. 1.
Strand-specific characterization of viral RNAs produced during acute and persistent infection. The presence of plus (+)- and minus (−)-strand viral RNAs was determined by strand-specific RT-PCR and evaluated in combination with RNase I treatment to determine whether the RNA existed in a single- or double-stranded form. Production of a 2.3-kb amplicon which includes viral nucleotides 4365 to 6693 is shown (arrowhead) for control T7-transcribed single-stranded and annealed double-stranded RNAs (A) and viral RNAs produced during acute or persistent infection of muscle or G8 myoblasts (B). The strand that was primed during the RT reaction is indicated by (+) or (−). In panel B, an RT reaction without any added primer (N) served as a negative control.
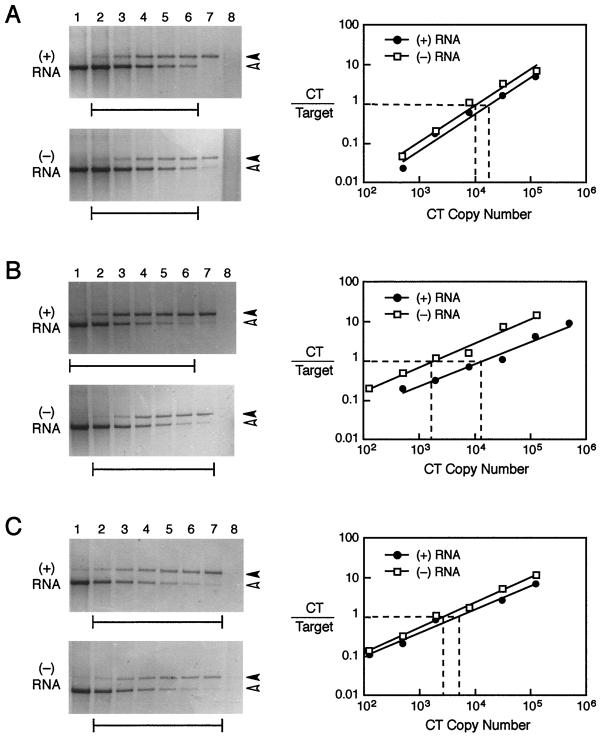
Quantitation of plus- and minus-strand viral RNAs was performed using strand-specific RT-PCR in the presence of a competitive template. Selected experimental results are shown in Fig. 2, and the compiled data are displayed in Table 2. As shown in Fig. 2A, an input of 4 × 104 copies of either the plus or minus control transcript gave output values of 1.1 × 104 copies of minus strand and 1.8 × 104 copies of plus strand. In multiple trials, the efficiency of detection varied from 45 to 62% for the plus strand and 25 to 40% for the minus strand with means of 45% and 28%, respectively. Since amplification efficiencies should be similar for either strand, this most probably reflects differences in the efficiency of the RT step or slight variations in the original quantitation of control transcripts. Acutely infected muscle tested at an input of 0.5 μg of total muscle RNA for the plus strand and 4 μg of total muscle RNA for the minus strand yielded an average of 4.5 × 104 copies of plus strand per μg and 600 copies of minus strand per μg with a 75:1 ratio of plus to minus strands (Fig. 2B). By 1 month, the amount of plus strand had diminished and was similar to that of the minus strand. In the experiment shown (Fig. 2C), the ratio of plus to minus strand was 2:1. In contrast, G8 myoblasts consistently showed an excess of plus strand over minus strand during both acute and persistent infection (Table 2). The plus-strand-to-minus-strand ratio was slightly lower in the PI-G8 cells, and the absolute levels of plus strand averaged 70-fold lower than in freshly infected G8 cells. This paralleled the lower titers of infectious virus that were found in PI-G8 compared to freshly infected G8 cells. Thus, PI-G8 cells produce less virus and correspondingly lower levels of viral RNA but still maintain the excess of plus strand that is characteristic of a productive infection.
FIG. 2.
Quantitation of plus (+)- and minus (−)-strand viral RNAs during acute and persistent infection. Competitive, strand-specific RT-PCR was used to measure the levels of plus and minus strands of viral RNA. The solid arrowhead marks the position of the target amplicon, and the open arrowhead marks the competitive template. Brackets underneath each gel indicate which lanes were used for quantitation, as shown in the adjacent plot. Representative data are shown, and a summary of data from all experiments is presented in Table 2. (A) Control plus and minus transcripts were input at 4 × 104 copies per reaction and used to evaluate the efficiency of the strand-specific competitive RT-PCR procedure. (B) Acutely infected muscle at 7 days postinfection showed a 61-fold excess of plus strand with 1.3 × 104 copies per 0.5 μg of input RNA for the plus strand and 1.7 × 103 copies per 4 μg of input RNA for the minus strand. (C) At 1 month after infection, the plus strand was present at 5.1 × 103 copies per 4 μg of input RNA while the minus strand was at 2.6 × 103 copies per 4 μg, yielding a final plus-strand-to-minus-strand ratio of 2.0.
TABLE 2.
Comparison of plus- and minus-strand viral RNA levels in acute and persistently infected samples
| Sample | Virus titera | No. of copies viral RNAb
|
Viral RNA strand ratio (plus:minus)c | |
|---|---|---|---|---|
| Plus strand | Minus strand | |||
| MP1/M (7 days) | 2.1 × 108 | 4.5 × 104 (2.6 × 104–6.6 × 104) | 6.0 × 102 (4.2 × 102–7.5 × 102) | 75:1* |
| MP1/M (1 mo) | NDd | 2.0 × 103 (1.3 × 103–2.8 × 103) | 5.2 × 102 (3.9 × 102–6.5 × 102) | 4:1 |
| MP1/G8 (20 h) | 1.1 × 106 | 4.6 × 105 (0.47 × 105–7.1 × 105) | 7.7 × 103 (6.9 × 103–9.0 × 103) | 60:1* |
| MP1/PI-G8 (passage 30) | 3.4 × 104 | 6.3 × 103 (4.4 × 103–9.3 × 103) | 1.5 × 102 (1.1 × 102–2.3 × 102) | 42:1* |
Virus titers are expressed as PFU per gram of muscle for muscle samples (M) or PFU per 106 cells for G8 and PI-G8 cells.
When quantitated by competitive RT-PCR, in vitro-transcribed control transcripts input at 4 × 104 copies gave mean values of 2.1 × 104 copies of plus-strand RNA (range, 1.8 × 104 to 2.5 × 104 copies) and 1.2 × 104 copies of minus-strand RNA (range, 1.0 × 104 to 1.6 × 104 copies). Values shown in the table are the mean number of copies of the plus or minus strand expressed per microgram of total RNA from muscle or G8 cells. Values were determined from three independent samples, and the range is shown in parentheses.
*, The number of copies of the plus strand was significantly different from that of the minus strand by the paired t test (α = 0.05).
ND, none detected.
Sequence changes linked to viral RNA persistence.
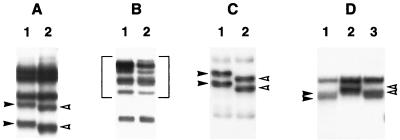
Each of the seven overlapping amplicons generated from viral RNA that persisted in muscle or in PI-G8 cells was evaluated by REF at least eight times under conditions which included a minimum of four different restriction digests and two different temperatures for electrophoresis. A control amplicon, engineered with a single-base transition at nucleotide 6682, showed altered migration of the appropriate bands under 9 of 10 conditions tested. An example of the shift in migration of the two bands which contained this mutation is shown in Fig. 3A. Despite a similarly rigorous analysis of MP1/M, no distinct band shifts were observed. The only evident change occurred in the region from 5508 to 6668, where there appeared to be a broadening of the banding pattern rather than a complete shift in mobility (Fig. 3B). Since this phenomenon was reproducible and occurred in different digests, the region was sequenced. Sequencing revealed a dimorphism at nucleotide 6249, with adenine and guanine peaks of equal height in two trials each for the forward and reverse sequencing reactions, that could not be resolved by manual editing (Table 3). The dimorphism occurred in the third position of an alanine codon and did not alter the amino acid sequence compared to parental MP1. In contrast, MP1/G8 showed a number of distinct genomic changes in five of the seven regions examined. The region from 5508 to 6668 contained a uracil-to-cytosine transition at position 5787, which was a silent mutation in the third position of a glycine codon (Fig. 3C). The mutation at position 269 in the untranslated region of MP1/G8 was identified as a cytosine-to-uracil transition. Sequencing of this region for MP1/M did not reveal any mutations, in agreement with what was observed on REF gels.
FIG. 3.
Detection of sequence changes in persistent viral RNA by REF analysis. The open arrowheads indicate the positions of bands whose migration was altered relative to those of parental MP1, which are marked by the solid arrowheads. (A) BsaJI digest of fragment 5479 to 6693 for the detection control showing MP1 (lane 1) and the altered migration of two bands caused by a T-to-C transition engineered into the primer at nucleotide 6682 (lane 2). (B) HaeIII-MboI digest of fragment 5479 to 6693 from MP1 (lane 1) compared to persistent MP1/M (lane 2). Broadening of the banding pattern in MP1/M resulting from an A-to-G dimorphism is indicated by the brackets. (C) MnlI-BsaJI digest of fragment 5479 to 6693 from MP1 (lane 1) and altered migration of two bands caused by a U-to-C transition at position 5787 in MP1/G8 (lane 2). (D) DdeI-HaeIII digest of fragment 4365 to 5552 from MP1 (lane 1), MP1/M from a mouse which contained a U-to-C transition at nucleotide 5157 (lane 2), and MP1/M from a second mouse which did not contain the mutation (lane 3).
TABLE 3.
Mutations that occurred during persistence of MP1 in muscle or in PI-G8 myoblasts
| Genomic region (nucleotides)a | Mutations detected in persistent viral RNAb
|
|||
|---|---|---|---|---|
| MP1/M
|
MP1/G8
|
|||
| REF | Sequencing | REF | Sequencing | |
| 21–583 | − | − | + | 269 (C-U) |
| 475–2133 | − | NS | − | NS |
| 2121–3993 | − | NS | + | NS |
| 3339–4392 | − | NS | − | NS |
| 4388–5523 | − | NS | + | NS |
| 5508–6668 | + | 6249 (GCA-GCRc) (Ala-Ala) | + | 5787 (GGU-GGC) (Gly-Gly) |
| 6280–7388 | − | NS | + | NS |
Nucleotide numbers indicate the genomic region screened and exclude the primer-binding region of the amplicon. The numbering system is based on alignment of partial-sequence information for MP1 with the complete sequence of CVB1 published for the pNI4 clone.
Results of REF analysis indicate whether altered migration either was (+) or was not (−) observed for restriction fragments within a given genomic region. Summarized sequence information indicates that there were no sequence changes detected within a region (−) or that the region was not sequenced (NS). Mutations are reported as the nucleotide position of the mutation followed by the base change and the codon change for coding sequences.
R = A or G.
An additional REF analysis of MP1/M was performed individually on muscle from two infected mice at 1 month postinfection by using the following three combinations of restriction digests: MnlI-BsaJI, RsaI-MseI-HaeIII, and DdeI-HaeIII. A mutation in the region from 4388 to 5523 was detected in one mouse and mapped as a uracil-to-cytosine transition at nucleotide 5157 (Fig. 3D). This change represented a silent mutation of CCU to CCC in a proline codon. When this region was amplified in an independent RT-PCR, the mutation was still present. No mutations were detected in MP1/M from the other mouse. These results confirmed that there was no consistent mutation of the virus associated with in vivo persistence and that the mutations which occurred were silent. REF analyses of persistent virus covered the entire 7.4-kb viral genome except the 20 nucleotides at the 5′ end and the terminal nucleotide at the 3′ end.
Frequency of infection and curing of PI-G8 myoblasts.
Infectious-center assays were performed to determine the frequency of infected cells in PI-G8 cultures. The proportion of productively infected PI-G8 cells ranged from 10% at passage 10 to 14% at passage 20 and 8% at passage 30. Virus obtained from these supernatants was fully cytopathic when used to infect BGMK cells. Three experiments with G8 cells that were freshly infected at a multiplicity of infection of 10 and evaluated at 20 h after infection yielded values ranging from 15 to 21%, indicating that not all G8 cells are productively infected during the initial exposure to virus. In controls prepared by adding 2 × 106 MP1 to 1 × 106 G8 cells in the presence of neutralizing antibody and immediately proceeding with the infectious-center assay, an average of 0.0005% of the input virus was detected as PFU, indicating that antibody neutralization was effective. To control for the removal of neutralizing antibody and the efficiency of intracellular virus detection, 106 G8 cells were pretreated with antibody and washed and 102 PFU of MP1 was added. In multiple trials, at least 96% of the input virus was detected as infectious centers in these controls.
To evaluate whether PI-G8 cells could be cured of their infection, horse anti-CVB1 neutralizing antibody was added to cultures at the beginning of passage 30. By the time the cells were passaged several days later, the cell-free supernatant and the cell extract were negative for both infectious virus and viral RNA, indicating that all of the cell-associated virus must have been released and neutralized by the time the cells were subjected to passage 31 (Table 4). Continued passage in the presence of antibody yielded similarly negative results for virus. When antibody treatment was discontinued, no reactivation of infectious virus was observed and no viral RNA was detected by RT-PCR, indicating that the culture had been completely cured of virus and that viral RNA did not persist. A second experiment showed that the net result was the same regardless of whether antibody treatment was maintained for two or five passages.
TABLE 4.
PI-G8 myoblasts are completely cured of persistent infection by anti-CVB1 treatment
| Horse serum | Serum added | CPE/viral RNA detected at passagea:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | ||
| Preimmune | p30-p35 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| Anti-CVB1 | p30-p35 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| Preimmune | p30-p31 | +/+ | +/+ | +/+ | +/+ | +/+ | NAb | NA | NA |
| Anti-CVB1 | p30-p31 | −/− | −/− | −/− | −/− | −/− | NA | NA | NA |
Results indicate that cultures were consistently positive (+) or negative (−) for virus-induced CPE or viral RNA (detected by RT-PCR) at a given passage. Uninfected G8 cells were uniformly negative when run as a negative control for detection of CPE and viral RNA.
NA, not assayed for CPE or viral RNA at these passages.
DISCUSSION
Coxsackieviruses infect and replicate in susceptible hosts by a mechanism that is primarily cytolytic. However, the association of persistent viral RNA with various disease states has spurred interest in understanding how these viruses persist and whether persistence of the viral genome is pathogenic. In this study, we have explored the fate of coxsackievirus RNA as it enters the persistent state in diseased muscle and compared it with viral persistence in a myoblast cell line. During acute productive infection, muscle contained on average a 75-fold excess of plus-strand compared to minus-strand RNA. By 1 month after infection, which is a time when infectious virus can no longer be recovered, the level of plus-strand RNA in muscle had diminished to a fourfold excess over minus-strand RNA. Given the slightly higher efficiency for RT-PCR of the plus-strand RNA, actual levels of plus- and minus-strand RNAs were nearly equal. Moreover, the RNA appeared to persist as a double-stranded complex, and absolute levels of minus-strand RNA were similar to those which were present during acute infection. Taken together, these observations suggest that persistence is characterized by reduced plus-strand RNA synthesis, a result which could occur as RNA polymerase activity subsides, leading to a corresponding lack of strand displacement and formation of the double-stranded replicative form. Since single-stranded intracellular viral RNAs decay within hours (19), this double-stranded form may lend stability to and protect the RNA from degradation, thereby promoting long-term persistence. It may also contribute to pathogenicity. Using mutagenized coxsackievirus cDNA to prevent maturation cleavage, Wessely et al. demonstrated that viral RNA alone, in the absence of infectious virus, can exert direct pathologic effects on cultured cardiac myocytes and on cardiac muscle (47, 48).
In CVB1T-induced CIM, active viral replication proceeds for about 2 weeks, after which time infectious virus cannot be recovered. Viral RNA persists in most mice at 1 month and gradually decays thereafter, although it can still be detected in a low percentage of samples after 1 year (44). The second objective of this study was to determine whether the transition from active to persistent infection occurred in concert with genetic changes in the virus. Error rates are high during replication of RNA viruses, since RNA-dependent RNA polymerases lack proofreading capabilities. Assuming that the mutations are not deleterious, RNA viruses may evolve rather quickly (22). We sought to determine if there was a uniform mechanism of genetic change associated with the transition to a persistent state. Premature termination, disrupted maturation cleavage, or changes in the untranslated region controlling viral replication were some distinct possibilities. Using persistently infected muscle samples pooled from eight mice, we identified only one region that contained an apparent genetic change. This mutation, at nucleotide 6249 in the 3D polymerase region, was silent. Moreover, it was sequenced as a dimorphism and probably represents a mutation that occurred in at least one but not all of the samples used to prepare the pooled sample. Conserved silent mutations in coding regions, which occur during in vitro persistence of poliovirus (6), have led to speculation that such mutations may exert effects through alteration of RNA secondary structure, but this has not been demonstrated experimentally. Because MP1/G8 contained a mutation in the region from 21 to 583, this region was sequenced for MP1/M, and the results confirmed that there were no changes or ambiguities in MP1/M that went undetected by the REF screening. The possibility that nonconserved mutations with similar functional effects occurred in individual mice but were diluted out and therefore not detected in the pooled sample was also considered. Individual analysis of muscle from two additional mice revealed a single silent mutation at nucleotide 5157 in the 3A proteinase region that was not present in viral RNA from the other mouse. For single-stranded RNA genomes, large amounts of genomic plus-strand RNA are derived from the minus strand template. Thus, only mutations in the minus strand, generated early in the replication cycle, would be uniformly reproduced and detected in persistent viral RNA. The 3A proteinase mutation was reproduced in a second amplification reaction and thus was not an artifact of RT-PCR. Nonetheless, its lack of effect on the coding sequence further supports the conclusion that viral RNA does not undergo specific genetic mutations which facilitate its persistence.
The characteristics described for MP1/M persistence are in sharp contrast to those of MP1/G8. A number of in vitro models of persistent enterovirus infection have been described, and despite the use of different viruses and host cells, they share certain key features. Among them, persistence often proceeds by a carrier culture mechanism in which only a small percentage of cells are infected (39). Our model of persistent infection of G8 myoblasts also behaved as a carrier culture, with less than 15% of the cells supporting production of infectious virus. PI-G8 cultures could be completely cured of infectious virus by antibody treatment, and once they were cured, virus could not be reactivated. Despite the presence of low levels of double-stranded RNA in PI-G8 cells, there was no production of a stable persistent form of viral RNA that survived the elimination of infectious virus by antibody treatment. Plus-strand-to-minus-strand ratios, although slightly lower than those of freshly infected G8 cells, were shifted toward an excess of plus-strand RNA production, as would be expected for an active infection and similar to that described for persistent infection of RD cells by coxsackievirus B5 (18). Unlike MP1/M, MP1/G8 clearly evolved into a genetically altered virus. Changes were detected in five of the seven overlapping regions evaluated by REF. Of the two specific mutations that were identified by sequencing, one was silent and the other occurred at nucleotide 269 in the 5′ untranslated region, where it could potentially affect viral replication. Viral mutations which facilitate attachment, entry, and replication have been described for persistent poliovirus (35) and are likely to be selected during serial passage in the relatively homogeneous cultured cell environment.
The observation that MP1 persistence in G8 cells does not exemplify the process in muscle may in part reflect a difference in the state of host cell differentiation. G8 cells are actively growing cells, while myofibers are end-stage nondividing cells. Myofibers that sustain localized cytopathic damage or rupture may survive and be restored by processes of regeneration and reinnervation (36). Viral RNA could then persist in the remaining portion of the myofiber. It is unlikely that persistence is contingent upon evolution of MP1, since the only mutations which occurred were generated in positions of redundancy. This implies that the virus does not mutate to evade the host immune system or promote its survival in muscle. It also suggests that reduced RNA synthesis is not mediated by changes in viral proteins. The mechanism of enterovirus replication is not completely understood, but it is possible that downregulation of RNA polymerase activity occurs in damaged myofibers through changes in cellular proteins that participate as elements of the viral replication complex, resulting in production of double-stranded persistent RNA. As a potent inducer of interferon and other cell mediators, double-stranded RNA may itself be pathogenic (25). Activation of interferon-inducible protein kinase has multiple effects on cellular proteins, which, in addition to inhibiting viral replication, include upregulating the transcription of cytokine genes through activation of NFκB. In virus-induced myopathies, tissue injury could result directly from inducible nitric oxide synthase produced by muscle or through production of cytokines and other mediators such as those that have been implicated in the pathogenesis of fatigue syndromes (20). Equivalent levels of plus and minus strands of enterovirus RNA have been observed in patients with chronic fatigue syndrome (15), and what we have found in mouse muscle may now provide a basis for understanding the processes which are at work in human diseases (33). Whether pathogenicity in CIM is derived directly from the presence of viral RNA or requires some degree of translation is not known. The lack of deleterious mutations and the fact that all regions of the viral genome were amplified leaves open the possibility that under appropriate but as yet unknown conditions, persistent coxsackievirus RNA possesses the capacity to produce viral proteins or infectious virus, thereby promoting an ongoing immunopathic response in muscle.
ACKNOWLEDGMENTS
We thank Akio Nomoto for providing the pNI4 clone and Christoph Eggert and Andy Schmidt for excellent technical assistance.
This work was supported by Public Health Service grant AI36223 from the National Institutes of Health, the National Chapter of the Arthritis Foundation, and the Graduate School of the University of Minnesota.
REFERENCES
- 1.Andreoletti L, Hober D, Becquart P, Belaich S, Copin M C, Lambert V, Wattre P. Experimental CVB3-induced chronic myocarditis in two murine strains: evidence of interrelationships between virus replication and myocardial damage in persistent cardiac infection. J Med Virol. 1997;52:206–214. doi: 10.1002/(sici)1096-9071(199706)52:2<206::aid-jmv15>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 2.Arbustini E, Grasso M, Porcu E, Bellini O, Diegoli M, Fasani R, Banchieri N, Pilotto A, Morbini P, Dal Bello B, Campana C, Gavazzi A, Vigano M. Enteroviral RNA and virus-like particles in the skeletal muscle of patients with idiopathic dilated cardiomyopathy. Am J Cardiol. 1997;80:1188–1193. doi: 10.1016/s0002-9149(97)00638-3. [DOI] [PubMed] [Google Scholar]
- 3.Archard L C, Bowles N E, Cunningham L, Freeke C A, Olsen E G, Rose M L, Meany B, Why H J, Richardson P J. Molecular probes for detection of persisting enterovirus infection of human heart and their prognostic value. Eur Heart J. 1991;12(Suppl. D):56–59. doi: 10.1093/eurheartj/12.suppl_d.56. [DOI] [PubMed] [Google Scholar]
- 4.Berger M M, See D M, Aymard M, Lina B. Demonstration of persistent enterovirus in the pancreas of diabetic mice by in situ polymerase chain reaction. Clin Diagn Virol. 1998;9:141–143. doi: 10.1016/s0928-0197(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 5.Borzakian S, Couderc T, Barbier Y, Attal G, Pelletier I, Colbere G F. Persistent poliovirus infection: establishment and maintenance involve distinct mechanisms. Virology. 1992;186:398–408. doi: 10.1016/0042-6822(92)90005-a. [DOI] [PubMed] [Google Scholar]
- 6.Borzakian S, Pelletier I, Calvez V, Colbere-Garapin F. Precise missense and silent point mutations are fixed in the genomes of poliovirus mutants from persistently infected cells. J Virol. 1993;67:2914–2917. doi: 10.1128/jvi.67.5.2914-2917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowles N E, Bayston T A, Zhang H Y, Doyle D, Lane R J, Cunningham L, Archard L C. Persistence of enterovirus RNA in muscle biopsy samples suggests that some cases of chronic fatigue syndrome result from a previous, inflammatory viral myopathy. J Med. 1993;24:145–160. [PubMed] [Google Scholar]
- 8.Bowles N E, Dubowitz V, Sewry C A, Archard L C. Dermatomyositis, polymyositis, and Coxsackie-B-virus infection. Lancet. 1987;i:1004–1007. doi: 10.1016/s0140-6736(87)92271-9. [DOI] [PubMed] [Google Scholar]
- 9.Bowles N E, Richardson P J, Olsen E G, Archard L C. Detection of Coxsackie-B-virus-specific RNA sequences in myocardial biopsy samples from patients with myocarditis and dilated cardiomyopathy. Lancet. 1986;i:1120–1123. doi: 10.1016/s0140-6736(86)91837-4. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Schnurr D P. Persistent infection of YAC-1 cells by coxsackievirus B3. J Gen Virol. 1988;69:59–65. doi: 10.1099/0022-1317-69-1-59. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Colbere-Garapin F, Duncan G, Pavio N, Pelletier I, Petit I. An approach to understanding the mechanisms of poliovirus persistence in infected cells of neural or non-neural origin. Clin Diagn Virol. 1998;9:107–113. doi: 10.1016/s0928-0197(98)00009-9. [DOI] [PubMed] [Google Scholar]
- 13.Conaldi P G, Serra C, Mossa A, Falcone V, Basolo F, Camussi G, Dolei A, Toniolo A. Persistent infection of human vascular endothelial cells by group B coxsackieviruses. J Infect Dis. 1997;175:693–696. doi: 10.1093/infdis/175.3.693. [DOI] [PubMed] [Google Scholar]
- 14.Correa-Rotter R, Mariash C N, Rosenberg M E. Loading and transfer control for Northern hybridization. BioTechniques. 1992;12:154–158. [PubMed] [Google Scholar]
- 15.Cunningham L, Bowles N E, Lane R J, Dubowitz V, Archard L C. Persistence of enteroviral RNA in chronic fatigue syndrome is associated with the abnormal production of equal amounts of positive and negative strands of enteroviral RNA. J Gen Virol. 1990;71:1399–1402. doi: 10.1099/0022-1317-71-6-1399. [DOI] [PubMed] [Google Scholar]
- 16.Fohlman J, Friman G, Tuvemo T. Enterovirus infections in new disguise. Lakartidningen. 1997;94:2555–2560. [PubMed] [Google Scholar]
- 17.Galbraith D N, Nairn C, Clements G B. Evidence for enteroviral persistence in humans. J Gen Virol. 1997;78:307–312. doi: 10.1099/0022-1317-78-2-307. [DOI] [PubMed] [Google Scholar]
- 18.Gow J W, Behan W M, Cash P, Simpson K, Behan P O. Genomic and template RNA transcription in a model of persistent enteroviral infection. J Neurovirol. 1997;3:76–82. doi: 10.3109/13550289709015796. [DOI] [PubMed] [Google Scholar]
- 19.Graeber I, Tischer J, Heinrich J, Hachula G, Lopez-Pila J M. Persistence of heterologous nucleic acids after uptake by mammalian cells. DNA Cell Biol. 1998;17:945–949. doi: 10.1089/dna.1998.17.945. [DOI] [PubMed] [Google Scholar]
- 20.Hickie I, Lloyd A. Are cytokines associated with neuropsychiatric syndromes in humans? Int J Immunopharmacol. 1995;17:677–683. doi: 10.1016/0192-0561(95)00054-6. [DOI] [PubMed] [Google Scholar]
- 21.Hohenadl C, Klingel K, Mertsching J, Hofschneider P H, Kandolf R. Strand-specific detection of enteroviral RNA in myocardial tissue by in situ hybridization. Mol Cell Probes. 1991;5:11–20. doi: 10.1016/0890-8508(91)90033-g. [DOI] [PubMed] [Google Scholar]
- 22.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 23.Iizuka N, Kuge S, Nomoto A. Complete nucleotide sequence of the genome of coxsackievirus B1. Virology. 1987;156:64–73. doi: 10.1016/0042-6822(87)90436-3. [DOI] [PubMed] [Google Scholar]
- 24.Iwahana H, Yoshimoto K, Itakara M. Detection of point mutations by SSCP of PCR-amplified DNA after endonuclease digestion. BioTechniques. 1992;12:64–66. [PubMed] [Google Scholar]
- 25.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 26.Klingel K, Stephan S, Sauter M, Zell R, McManus B M, Bultmann B, Kandolf R. Pathogenesis of murine enterovirus myocarditis: virus dissemination and immune cell targets. J Virol. 1996;70:8888–8895. doi: 10.1128/jvi.70.12.8888-8895.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leparc-Goffart I, Julien J, Fuchs F, Janatova I, Aymard M, Kopecka H. Evidence of presence of poliovirus genomic sequences in cerebrospinal fluid from patients with postpolio syndrome. J Clin Microbiol. 1996;34:2023–2026. doi: 10.1128/jcm.34.8.2023-2026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Sommer S S. Restriction endonuclease fingerprinting (REF): a sensitive method for screening mutations in long, contiguous segments of DNA. BioTechniques. 1995;18:470–477. [PubMed] [Google Scholar]
- 29.Matteucci D, Paglianti M, Giangregorio A M, Capobianchi M R, Dianzani F, Bendinelli M. Group B coxsackieviruses readily establish persistent infections in human lymphoid cell lines. J Virol. 1985;56:651–654. doi: 10.1128/jvi.56.2.651-654.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney R E, Jr, Katz S L, Wilfert C M. Chronic enteroviral meningoencephalitis in agammaglobulinemic patients. Rev Infect Dis. 1987;9:334–356. doi: 10.1093/clinids/9.2.334. [DOI] [PubMed] [Google Scholar]
- 31.McLaren J, Argo E, Cash P. Evolution of coxsackie B virus during in vitro persistent infection: detection of protein mutations using two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1993;14:137–147. doi: 10.1002/elps.1150140122. [DOI] [PubMed] [Google Scholar]
- 32.Melchers W, Zoll J, van Kuppeveld F, Swanink C, Galama J. There is no evidence for persistent enterovirus infections in chronic medical conditions in humans. Rev Med Virol. 1994;4:235–243. [Google Scholar]
- 33.Muir P, Archard L C. There is evidence for persistent enterovirus infections in chronic medical conditions in humans. Rev Med Virol. 1994;4:245–250. [Google Scholar]
- 33a.National Institutes of Health. 7 June 1999, revision date. Image analysis software. [Online.] http://rsb.info.nih.gov/nih-image/. [8 October 1999, last date accessed.]
- 34.Parker L T, Zakeri H, Deng Q, Spurgeon S, Kwok P-Y, Nickerson D A. AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. BioTechniques. 1996;21:694–699. doi: 10.2144/96214rr02. [DOI] [PubMed] [Google Scholar]
- 35.Pelletier I, Duncan G, Pavio N, Colbere-Garapin F. Molecular mechanisms of poliovirus persistence: key role of capsid determinants during the establishment phase. Cell Mol Life Sci. 1998;54:1385–1402. doi: 10.1007/s000180050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rantanen J, Ranne J, Hurme T, Kalimo H. Denervated segments of injured skeletal muscle fibers are reinnervated by newly formed neuromuscular junctions. J Neuropathol Exp Neurol. 1995;54:188–194. doi: 10.1097/00005072-199503000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Ray C G, Minnich L L, Johnson P C. Selective polymyositis induced by coxsackievirus B1 in mice. J Infect Dis. 1979;140:239–243. doi: 10.1093/infdis/140.2.239. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schnurr D P, Schmidt N J. Persistent infection of mouse fibroblasts with Coxsackievirus. Arch Virol. 1984;81:91–101. doi: 10.1007/BF01309299. [DOI] [PubMed] [Google Scholar]
- 40.Strongwater S L, Dorovini-Zis K, Ball R D, Schnitzer T J. A murine model of polymyositis induced by coxsackievirus B1 (Tucson strain) Arthritis Rheum. 1984;27:433–442. doi: 10.1002/art.1780270411. [DOI] [PubMed] [Google Scholar]
- 41.Tam P E, Messner R M. Genetic determinants of susceptibility to coxsackievirus B1-induced chronic inflammatory myopathy: effects of host background and major histocompatibility complex genes. J Lab Clin Med. 1996;128:279–289. doi: 10.1016/s0022-2143(96)90029-3. [DOI] [PubMed] [Google Scholar]
- 42.Tam P E, Messner R P. Coxsackievirus-induced chronic inflammatory myopathy: virus variants distinguish between acute cytopathic effects and pathogenesis of chronic disease. Virology. 1997;233:199–209. doi: 10.1006/viro.1997.8592. [DOI] [PubMed] [Google Scholar]
- 43.Tam P E, Schmidt A M, Messner R P. Modified tissue pulverization technique and evaluation of dihydrofolate reductase amplification as a pan-tissue RT-PCR control. PCR Methods Applic. 1993;3:71–72. doi: 10.1101/gr.3.1.71. [DOI] [PubMed] [Google Scholar]
- 44.Tam P E, Schmidt A M, Ytterberg S R, Messner R P. Duration of virus persistence and its relationship to inflammation during the chronic phase of coxsackievirus B1-induced murine polymyositis. J Lab Clin Med. 1994;123:346–356. [PubMed] [Google Scholar]
- 45.Tam P E, Schmidt A M, Ytterberg S R, Messner R P. Viral persistence during the developmental phase of coxsackievirus B1-induced murine polymyositis. J Virol. 1991;65:6654–6660. doi: 10.1128/jvi.65.12.6654-6660.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vella C, Brown C L, McCarthy D A. Coxsackievirus B4 infection of the mouse pancreas: acute and persistent infection. J Gen Virol. 1992;73:1387–1394. doi: 10.1099/0022-1317-73-6-1387. [DOI] [PubMed] [Google Scholar]
- 47.Wessely R, Henke A, Zell R, Kandolf R, Knowlton K U. Low-level expression of a mutant coxsackieviral cDNA induces a myocytopathic effect in culture: an approach to the study of enteroviral persistence in cardiac myocytes. Circulation. 1998;98:450–457. doi: 10.1161/01.cir.98.5.450. [DOI] [PubMed] [Google Scholar]
- 48.Wessely R, Klingel K, Santana L F, Dalton N, Hongo M, Jonathan Lederer W, Kandolf R, Knowlton K U. Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J Clin Investig. 1998;102:1444–1453. doi: 10.1172/JCI1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodall C J, Riding M H, Graham D I, Clements G B. Sequences specific for enterovirus detected in spinal cord from patients with motor neurone disease. Br Med J. 1994;308:1541–1543. doi: 10.1136/bmj.308.6943.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ytterberg S R, Mahowald M L, Messner R P. Coxsackievirus B1-induced polymyositis. Lack of disease expression in nu/nu mice. J Clin Investig. 1987;80:499–506. doi: 10.1172/JCI113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ytterberg S R, Mahowald M L, Messner R P. T cells are required for coxsackievirus B1 induced murine polymyositis. J Rheumatol. 1988;15:475–478. [PubMed] [Google Scholar]
- 52.Zintz C B, Beebe D C. Rapid reamplification of PCR products purified in low melting point agarose gels. BioTechniques. 1991;11:158–162. [PubMed] [Google Scholar]