Abstract
Recombinant bovine viral diarrhea virus (BVDV) nonstructural protein 5B (NS5B) produced in insect cells has been shown to possess an RNA-dependent RNA polymerase (RdRp) activity. Our initial attempt to produce the full-length BVDV NS5B with a C-terminal hexahistidine tag in Escherichia coli failed due to the expression of insoluble products. Prompted by a recent report that removal of the C-terminal hydrophobic domain significantly improved the solubility of hepatitis C virus (HCV) NS5B, we constructed a similar deletion of 24 amino acids at the C terminus of BVDV NS5B. The resulting fusion protein, NS5BΔCT24-His, was purified to homogeneity and demonstrated to direct RNA replication via both primer-dependent (elongative) and primer-independent (de novo) mechanisms. Furthermore, BVDV RdRp was found to utilize a circular single-stranded DNA as a template for RNA synthesis, suggesting that synthesis does not require ends in the template. In addition to the previously described polymerase motifs A, B, C, and D, alignments with other flavivirus sequences revealed two additional motifs, one N-terminal to motif A and one C-terminal to motif D. Extensive alanine substitutions showed that while most mutations had similar effects on both elongative and de novo RNA syntheses, some had selective effects. Finally, deletions of up to 90 amino acids from the N terminus did not significantly affect RdRp activities, whereas deletions of more than 24 amino acids at the C terminus resulted in either insoluble products or soluble proteins (ΔCT179 and ΔCT218) that lacked RdRp activities.
The Flaviviridae family currently comprises three genera of single-stranded positive-sense RNA viruses: flaviviruses, pestiviruses, and hepaciviruses (26). Bovine viral diarrhea virus (BVDV) is a prototype virus in the genus Pestivirus, which also includes classical swine fever virus and border disease virus. One remarkable property of pestiviruses is the existence of two biotypes (noncytopathic and cytopathic) in each strain. The molecular basis for this distinct biotype pair is due to mechanically novel cellular gene insertions or viral genome rearrangements, which result in either enhanced cleavage at the junction site between nonstructural protein 2 (NS2) and NS3 or increased expression of NS3 (18). This unique property makes BVDV an ideal molecular tool to study RNA recombination and the associated process, RNA replication. Both RNA replication and recombination are poorly understood at the mechanistic level in comparison to their DNA counterparts.
The RNA genome of BVDV is one of the largest (12.5 kb) among members of the Flaviviridae family (4). Similar to the hepatitis C virus (HCV) genome, it consists of a long 5′ untranslated region which contains an internal ribosomal entry site (IRES) for translation of viral proteins (3, 8, 25). The single large open reading frame encodes a polyprotein of approximately 3,900 amino acids (4, 18, 26) that is processed into at least 12 functional proteins (Npro-C-Erns-E1-E2/p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B) by both host and viral proteases (6, 26, 32). Unique to pestiviruses, the first virally encoded protein is Npro, a papain-like cysteine protease responsible for the cleavage between Npro and the capsid protein (C) (29). BVDV Erns (formerly E0) possesses RNase activity which is believed to play a role during viral RNA replication (28).
As one of the best-characterized members of the Flaviviridae family, BVDV provides a good model system for HCV, a major etiologic agent for non-A, non-B hepatitis. Three features shared between BVDV and HCV making the BVDV model a better one than a flavivirus (such as yellow fever virus) are (i) IRES-mediated translation of viral proteins (3, 8, 15); (ii) NS4A cofactor requirement by NS3 serine protease (31), and (iii) polyprotein processing within the nonstructural region, especially at the NS5A and NS5B junction site (32). Studies by Frolov et al. (8) on the functional substitution of the IRES elements between BVDV and HCV or encephalomyocarditis virus further confirm that these positive-sense RNA viruses have similar strategies for viral translation and replication (9). It is likely that elucidation of the molecular mechanisms for BVDV replication will add to our knowledge of HCV replication. The lack of an efficient cell culture system for HCV makes BVDV a very attractive model system.
NS5B of BVDV, a key enzyme essential for viral replication, has been shown to possess an RNA-dependent RNA polymerase (RdRp) activity (33). A near-dimer-size product was synthesized predominantly from the 3′ end of the RNA template via a copy-back mechanism. A similar copy-back RNA synthesis activity was observed for the HCV NS5B (2, 16). However, such a copy-back mode of RNA synthesis can be demonstrated only by in vitro assays and has not been described for other single-stranded positive-sense RNA viruses in vivo. Poliovirus, the best characterized single-stranded positive-sense RNA virus, has developed a unique de novo protein priming mechanism to initiate the RNA synthesis (22). It is likely that de novo initiation is also the mode of replication in vivo for flaviviruses. This notion is supported by a recent report that BVDV can initiate RNA synthesis in a primer-independent fashion (14). In this report, Kao et al. described a de novo initiation assay in which a synthetic RNA template with a 3′-terminal dideoxynucleotide (abolishing self-priming) was used to direct RNA synthesis. A predominant monomer-size product synthesized by the BVDV RdRp was suggested to represent the full-length complementary copy of the input RNA, which can only be the result of de novo initiation in the presence of ribonucleotide triphosphates (NTPs) (14). Whether de novo and elongative RNA syntheses have similar requirements has not been addressed.
In the present report, a soluble recombinant BVDV NS5B lacking 24 amino acids at the C terminus was expressed in Escherichia coli and purified to homogeneity. The resulting protein, NS5BΔCT24-His, was analyzed for the following activities: (i) primer-dependent (elongative) RNA synthesis; (ii) primer-independent (de novo) RNA synthesis; and (iii) template preference and specificity. Optimal RNA polymerization assay conditions were determined through a scintillation proximity assay (SPA). In addition, extensive site-directed mutagenesis and deletion analysis confirmed the importance of six conserved motifs for RNA synthesis, including the characteristic polymerase motifs A, B, C, and D and two new motifs identified in this study.
MATERIALS AND METHODS
Cells, oligonucleotides, and plasmids.
E. coli JM109(DE3) and XL1-Blue cells were purchased from Promega (Madison, Wis.) and Stratagene (La Jolla, Calif.), respectively. DNA oligonucleotides were purchased from Life Technologies (Gaithersburg, Md.). RNA oligonucleotides were purchased from Oligos Etc. Inc. (Wilsonville, Oreg.). Expression vector pET-28a was purchased from Novagen Inc. (Madison, Wis.). The full-length cDNA clone of BVDV, pVVNADL, was kindly provided by Ruben Donis, University of Nebraska.
Construction of BVDV-NS5B expression plasmids.
cDNAs encoding NS5B of the cytopathic BVDV NADL strain (pVVNADL) was generated by using a standard PCR method. NcoI and BglII sites were engineered in the PCR primers so that the PCR products could be cloned directly into the NcoI and BamHI sites of pET-28a. The 5′ PCR primer also contained an additional methionine codon to initiate translation. Additional codons coding for a polyhistidine tag at the C terminus, GSHHHHHH, was engineered to facilitate the purification of NS5B. NS5B lacking the C-terminal 24 amino acids, NS5BΔCT24, was constructed by a similar strategy (Fig. 1B). The sequences of all clones were confirmed by dideoxynucleotide sequencing, using a model ABI 377 automated sequencer from Perkin-Elmer (Foster City, Calif.).
FIG. 1.
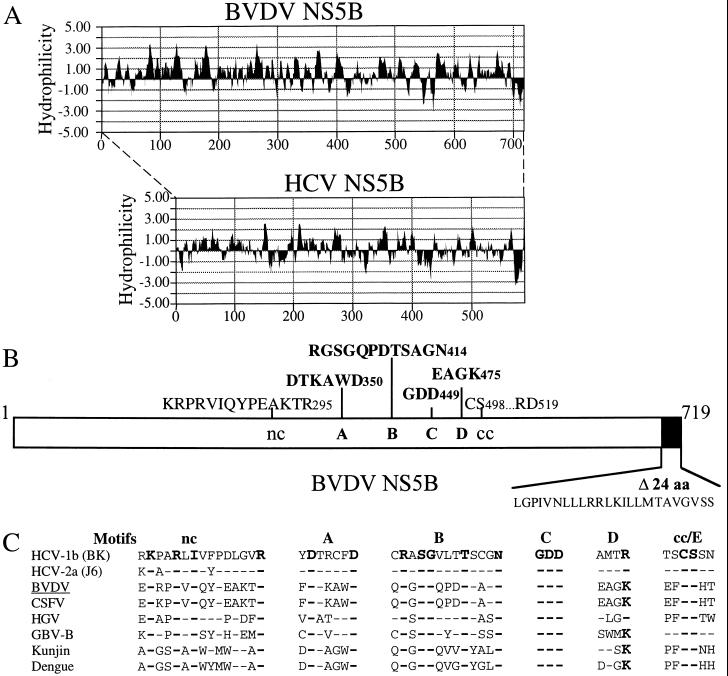
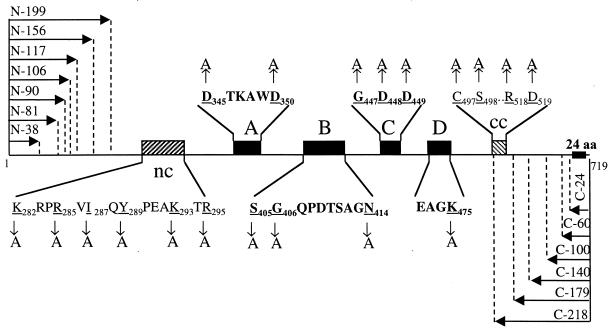
(A) Hydropathy profile comparison between BVDV and HCV NS5B proteins. Both NS5Bs contained a highly hydrophobic region at the C terminus. (B) Motif organization of BVDV NS5B depicting the positions of six conserved motifs (nc, A, B, C, D, and cc) as well as the C-terminal hydrophobic domain (solid bar containing 24 amino acids). The position of each motif is labeled with a number according to its amino acid position in HCV NS5B. (C) Motif alignments among various members of the Flaviviridae family. Amino acids which are 100% conserved are in bold type with a larger font. A short dash represents an identical amino acid compared to the lead sequence derived from HCV-1b BK strain. Motif cc is likely to be motif E, based on secondary structure prediction. The overall homology between viruses of different genera is rather poor, about 20%. CSFV, classical swine fever virus; HGV, hepatitis G virus.
Site-directed mutagenesis and deletional analysis.
The plasmid encoding NS5BΔCT24-His was used as the parental clone for all subsequent manipulations. Site-directed mutagenesis was carried out by using a QuickChange mutagenesis kit (Stratagene). N-terminal and C-terminal deletions were first generated by PCR and then subcloned into the pET-28a vector as described above. All of the mutations and deletions were verified by dideoxynucleotide sequencing (ABI 377 automated sequencer).
Expression and purification of BVDV NS5B proteins.
Expression of NS5B protein in JM109(DE3) cells, grown to an optical density at 600 nm of 0.6, was induced by isopropylthio-β-d-galactoside (IPTG) at a concentration of 0.2 mM. After a 4-h induction at 24°C, the cells were harvested and ruptured with a microfluidizer (Microfluidics Corp., Newton, Mass.). Purification of NS5B protein was conducted under conditions similar to those described previously (7). Briefly, the soluble cell lysates were batch-adsorbed onto Probond resin (a nickel-chelated affinity resin, Ni-nitrilotriacetic acid [NTA], from Invitrogen, Carlsbad, Calif.) for 1 h at 4°C. The bound material was then washed with 15 column volumes of a buffer containing 1 M NaCl to remove most of the contaminating RNA and DNA. The His-tagged fusion proteins were then eluted from the column with a buffer containing 0.35 M imidazole. The eluted materials were dialyzed in a storage buffer (50 mM Tris [pH 7.5], 5 mM dithiothreitol [DTT], 500 mM NaCl, 20% glycerol, 300 nM antipain, 200 nM leupeptin) and stored at −80°C. A portion of the truncated NS5B protein (NS5BΔCT24-His) was further purified by passage through a Superdex-200 gel filtration column (Amersham-Pharmacia Biotech, Arlington Heights, Ill.).
Western blot analysis.
Proteins were separated on a 10 to 20% polyacrylamide gradient gel and electrotransferred onto a nitrocellular membrane as described previously (12). The anti-His6 tag monoclonal antibody (Qiagen Inc., Santa Clarita, Calif.) was used as the primary antibody, and alkaline phosphatase-conjugated anti-mouse immunoglobulin G antibody (Promega) was used as the secondary antibody.
RNA-dependent RNA polymerization assay.
An SPA was developed to optimize the BVDV NS5B RdRp activity as described previously (7). This assay measures the dose-dependent incorporation of [3H]NMP in RNA products captured by the streptavidin-coated SPA beads. The optimized assay conditions were as follows: 20 mM Tris [pH 7.5], 6 mM MnCl2, 0.2 mM MgCl2, 50 μg of bovine serum albumin per ml, 2 mM DTT, 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS), 25 mM NaCl, 1 U of RNasin, and 25% glycerol in a 50-μl reaction volume for 3 h at room temperature. Unless specified, 50 nM NS5B RdRp, 300 ng of RNA homopolymers (polyC), and 25 ng of biotinylated primers [oligo(G)12] were used in a standard assay. The reaction was terminated by adding 100 mM EDTA in phosphate-buffered saline (pH 7.4). The captured RNA products were quantified by using a TopCounter (Packard Instrument Company, Meriden, Conn.).
De novo initiation of RNA synthesis.
A synthetic 22-base RNA, designated (−)21g, was used as the template for the de novo initiation assay. The sequence of (−)21g, derived from the 3′-terminal 21 nucleotides of BVDV minus-strand RNA, was 3′-(diH-g)CAUAUGCUCUUAAUCUUUUCC-5′ (14). The 3′-terminal guanylate was added to the BVDV sequence and modified to have a dideoxyribose (diH) so that it would prevent any RNA synthesis via self-priming. The reaction mixture consisted of 5 pmol of template (−)21g, 20 ng of BVDV NS5B protein, 20 mM sodium glutamate, 4 mM MgCl2, 1 mM MnCl2, 12.5 mM DTT, 0.5% (vol/vol) Triton X-100, 200 μM ATP, 200 μM UTP, 500 μM GTP, and 250 nM [α-32P]CTP in a 40-μl reaction volume. The reaction was carried out at 25°C for 1 h. The labeled RNA products was extracted with phenol-chloroform and precipitated with ethanol in the presence of 0.4 M ammonium acetate and 5 μg of glycogen. The final products were separated on a 20% denaturing polyacrylamide gel containing 8 M urea. The gel was dried and exposed to an X-ray film at −80°C. The radioactivities of the RNA products were quantified by using a PhosphorImager from Molecular Dynamics (Sunnyvale, Calif.).
RNA bandshift assay.
A synthetic RNA template with a stable tetraloop at the 3′ end, 5′-UUUUUUUUUUUUUUUUUUUUUUUUUUUUGGACUUCGGUCC-3′, was used as the probe for RNA bandshift assay. The RNA was end labeled with [γ-33P]ATP by T4 polynucleotide kinase (Amersham Pharmacia Biotech) according to the manufacturer’s protocol. After the labeling reaction, the RNA oligomer was extracted with phenol-chloroform and precipitated by ethanol in the presence of 0.5 M ammonium acetate and 20 μg of glycogen. In each binding assay, 1 pmol of labeled RNA probe and 400 ng of BVDV NS5BΔCT24-His were incubated in a buffer containing 20 mM HEPES (pH 7.3), 7.5 mM MnCl2, 7.5 mM DTT, 5% glycerol, 125 mM NaCl, 100 μg of bovine serum albumin per ml, 1 U of RNase inhibitor, and increasing amounts of unlabeled competitors [0, 0.1, 1, 10, or 100 pmol of poly(U), poly(A), poly(C), or poly(G)]. The binding reactions were performed at room temperature for 30 min in a 10-μl reaction volume. The protein-RNA complexes were analyzed on a native 6% polyacrylamide gel. The gel was dried prior to autoradiography.
Denaturing gel electrophoresis.
RNA products from RdRp reactions were resuspended in 1× denaturing loading buffer (45% deionized formamide, 1.5% glycerol, 0.04% bromophenol blue, 0.04% xylene cyanol) and denatured at 90°C for 3 min. The products were separated by electrophoresis on a 5 or 15% denaturing (8 M urea) polyacrylamide gel, which was then wrapped in plastic film and exposed to X-ray film at −80°C. Product bands were quantified by using a PhosphorImager (Molecular Dynamics).
Preparation of circular single-stranded DNA template.
Bacteriophage MP19 DNA was prepared by polyethylene glycol precipitation according to published protocols (27). The virions were treated with 1 mg of protease K per ml in the presence of 50 mM Tris (pH 7.4), 250 mM NaCl, 2 mM EDTA, and 0.5% sodium dodecyl sulfate followed by extraction with phenol-chloroform and precipitation with 70% ethanol. The DNA pellet was resuspended to a concentration of 20 ng/μl for use in the RNA synthesis assay.
RESULTS
Removal of the C-terminal hydrophobic domain resulted in production of soluble BVDV NS5B protein.
Our initial attempt to produce soluble full-length BVDV NS5B in E. coli failed due to the expression of insoluble protein. Although a recent report by Zhong et al. (33) demonstrated that full-length BVDV NS5B expressed in insect cells can be solubilized by high concentrations of detergent, salt, and glycerol, the solubility was rather poor upon biophysical characterizations using techniques such as ultracentrifugation. This is reminiscent of our experience with the full-length HCV NS5B protein (7). Hydropathy profile comparison between BVDV and HCV NS5Bs in Fig. 1A revealed, similar to findings for HCV, a highly hydrophobic domain of 24 amino acids at the C terminus of BVDV NS5B (Fig. 1B). Amino acid alignments among various members (Fig. 1C) in the Flaviviridae family identified several polymerase motifs present in BVDV NS5B which are highly conserved. Interestingly, the Flavivirus genus lacks the hydrophobic domain at the C terminus (data not shown). This finding suggests that unlike those conserved polymerase motifs, the hydrophobic C-terminal domain may not participate directly in the nucleotidyl transfer reaction, making it a candidate for deletion to improve solubility.
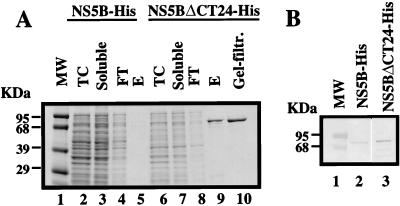
A deletion of 24 amino acids at the C terminus of BVDV NS5B was engineered. To facilitate the purification and immunoblotting analysis, a polyhistidine peptide (GSHHHHHH) was added to both the full-length and C-terminally truncated (ΔCT24) NS5B proteins. Parallel expression and purification were performed; the results are shown in Fig. 2A. Very little if any of the full-length NS5B protein was recovered from the Ni-NTA affinity purification (lane 5), whereas the truncated NS5BΔCT24-His was soluble and readily purified with a good yield and purity (∼85%) (lane 9). The expression levels of both proteins in total cell lysates were comparable, as determined by Western blot analysis (Fig. 2B, lanes 2 and 3) using an anti-His6 tag monoclonal antibody. These results indicate that the hydrophobic domain at the C terminus decreases the solubility of BVDV NS5B protein and its removal significantly improves solubility. The NS5BΔCT24-His protein was further purified through a Superdex-200 gel filtration column. The purity was improved to about 95% (Fig. 2A, lane 10). This 95% pure NS5BΔCT24-His protein was used for the subsequent optimization experiments of RNA synthesis. Mutational analysis of the BVDV NS5B was based on the cDNA encoding NS5BΔCT24-His as the starting construct.
FIG. 2.
(A) Expression and purification of the full-length and C-terminally truncated BVDV NS5Bs. Lanes 2 to 5 contain samples from the full-length NS5B-His; lanes 6 to 10 represent those from the C-terminally truncated NS5BΔCT24-His. Lane 1, molecular weight (mw) markers; lanes 2 and 6, total cell (TC) lysates; lanes 3 and 7, soluble fractions of the cell lysates; lanes 4 and 8, flowthrough (FT) unbound fractions; lanes 5 and 9, eluate (E) from Ni-NTA column; lane 10, the purified protein after passage through a gel filtration column. (B) Western blot analysis of protein expression levels in total cell lysates from full-length (NS5B-His; lane 2) and C-terminally truncated (NS5BΔCT24-His; lane 3) NS5Bs.
Optimal conditions for RNA synthesis by BVDV NS5B.
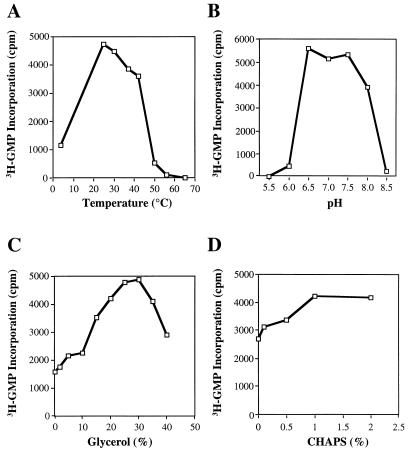
To measure the RdRp activity of the BVDV NS5BΔCT24-His protein, we developed an SPA similar to that for HCV RdRp (7) (see Materials and Methods). We first examined the effects of temperatures, pH, glycerol, and detergents (Fig. 3). The preferred temperature for the RdRp assay was room temperature (∼22°C) (Fig. 3A), as described for HCV NS5B (2, 7, 16, 17), suggesting that BVDV and HCV RdRps may be less stable at temperatures higher than 30°C. BVDV RdRp has comparable activities within a rather broad pH range, from 6.5 to 7.5 (Fig. 3B). In contrast to HCV RdRp, the presence of glycerol (25 to 30% [vol/vol]) and the zwitterionic detergent CHAPS (1% [vol/vol]) in the reaction mixture enhanced the RdRp activity of BVDV (Fig. 3C and D).
FIG. 3.
Optimization of assay conditions for RNA synthesis by the BVDV NS5B. The C-terminally truncated NS5B, NS5BΔCT24-His (50 nM), was used in the optimization experiments. Poly(C) was the template, and oligo(G) was used as a primer. (A) Effect of temperatures; (B) effect of pH; (C) effect of glycerol; (D) effect of a zwitterionic detergent, CHAPS.
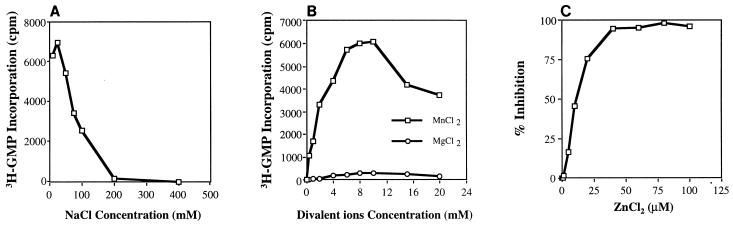
Requirements for monovalent cations and divalent cations were also determined. Monovalent salts such as NaCl (or KCl) at concentrations higher than 25 mM were inhibitory to BVDV RdRp (Fig. 4A). The divalent ion Mn2+ was required for RdRp activity, with an optimal concentration between 6 and 10 mM (Fig. 4B). Surprisingly, Mg2+ ions were not preferred and had minimal effect on RdRp activity (Fig. 4B). The RdRp of tomato spotted wilt virus also has a strict requirement for Mn2+ (1). Several possibilities can be proposed to explain this preference for Mn2+ over Mg2+: (i) presence of His tag; (ii) removal of the C-terminal domain; or (iii) use of a poly(C) template. It was shown that poliovirus 3D polymerase also preferred Mn2+ when poly(C) was used as the template (data not shown). Similar to HCV RdRp (7), BVDV NS5B was inhibited by Zn2+ ions with a 50% inhibitory concentration of approximately 10 μM.
FIG. 4.
Effects of salts on BVDV NS5B RdRp activity. (A) Effects of different concentrations of NaCl; (B) effects of different concentrations of divalent salts, MnCl2 and MgCl2; (C) inhibition of NS5B RNA synthesis by Zn2+ ions.
Template preference of BVDV NS5B.
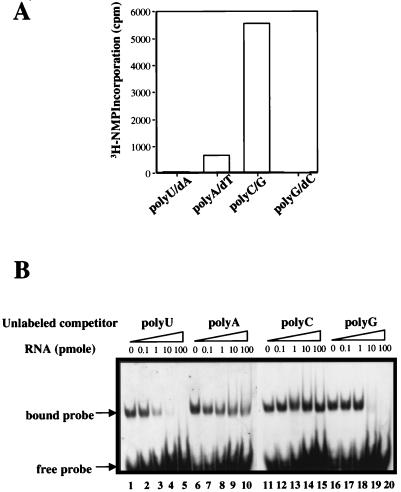
Four homopolymeric RNA template-primer pairs, poly(U)-oligo(dA), poly(A)-oligo(dT), poly(C)-oligo(G), and poly(G)-oligo(dC), were analyzed for the ability to direct RNA synthesis by the BVDV RdRp under the optimized assay conditions described above. As shown in Fig. 5A, the template-primer pair preferred by the BVDV RdRp is poly(C)-oligo(G), followed by poly(A)-oligo(dT). Both poly(G)-oligo(dC) and poly(U)-oligo(dA) were extremely inefficient in supporting the BVDV RdRp activity. Similar template preference was described previously for HCV RdRp (16). To demonstrate further whether this template preference in RNA synthesis inversely correlated with protein-template binding affinity, as in the case of HCV RdRp, an RNA bandshift assay was developed. This assay detected the formation of protein-RNA complexes in solution that could be separated from the unbound RNA on a native polyacrylamide gel. A synthetic RNA of 40 bases with a stable stem-loop at the 3′ end (see Materials and Methods for sequence) was chosen as the radiolabeled probe because of its high binding affinity to BVDV RdRp. While direct binding between polymerase and other RNAs with weaker affinities was difficult to detect, a competition experiment with unlabeled RNAs provided a more quantitative assessment of the binding efficiencies of different RNA templates. NS5BΔCT24-His formed stable complexes with 1 pmol of 33P-labeled probes (Fig. 5B, lanes 1, 6, 11, and 16). Complex formation was inhibited by various competitors with different efficiencies. When increasing amounts of each template (0.1, 1, 10, and 100 pmol) were added, poly(U) was the most efficient competitor in inhibiting the interaction between BVDV RdRp and the radiolabeled RNA probe (lanes 2 to 5). Poly(G) was also a very effective competitor and inhibited most of the binding at 10 pmol (lane 19). Poly(A) competed inefficiently at higher concentrations (10 and 100 pmol), whereas poly(C) did not compete at all. Altogether, these results suggest that the template preference by BVDV NS5B for RNA synthesis is inversely correlated with the binding affinity of the same template, a feature consistent with those reported for HCV and poliovirus RdRps (16, 19).
FIG. 5.
RNA templates preferred by the BVDV NS5B for RNA synthesis and binding. (A) Template preference for RNA synthesis. Four RNA template-primer pairs, poly(U)-oligo(dA), poly(A)-oligo(dT), poly(C)-oligo(G), and poly(G)-oligo(dC), were used to measure primer-dependent RNA synthesis by SPA. (B) RNA bandshift assay to measure the relative affinities of different RNAs bound by BVDV NS5B.
RNA synthesis from a circular single-stranded DNA template.
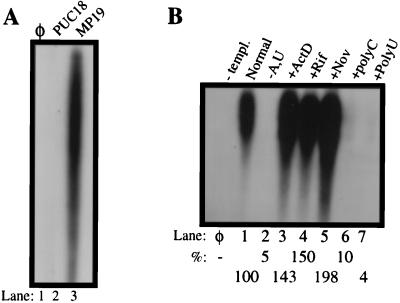
Previous reports demonstrated that the BVDV RdRp was able to utilize DNA template, although less efficiently, to direct RNA synthesis for both primer-dependent and primer-independent modes of replication (14, 33). In contrast, the HCV NS5B RdRp was reported to not use DNA as a template for RNA synthesis (5). To investigate further the template specificity of BVDV RdRp, a circular single-stranded DNA (ssDNA template, MP19(M13), as well as a supercoiled plasmid DNA (pUC18) were tested for RNA synthesis activity (Fig. 6A). We observed that the BVDV RdRp could utilize the circular ssDNA, but not the double-stranded DNA, for RNA synthesis. The RNA synthesis is specific for the BVDV RdRp because mutant BVDV NS5B proteins that were defective for synthesis from RNA did not produce any products from MP19 (data not shown). The RNA products produced by BVDV NS5B entered poorly into the 5% denaturing gel and were heterogeneous in size (longer than 300 bases), suggesting that the polymerase either initiated or terminated at different positions. These results demonstrated that the initiation of RNA synthesis did not require a free terminus on the template. At present, it is not clear whether this synthesis is de novo.
FIG. 6.
RNA synthesis from circular single-stranded MP19 templates, using the wild-type NS5BΔCT24-His. All reactions were performed as described in Materials and Methods for de novo synthesis. (A) Comparison of RdRp products synthesized from double-stranded pUC18 and single-stranded MP19 templates. (B) Effects on RNA synthesis upon addition of 100 μM actinomycin D (ActD), 100 μM rifampin (Rif), 300 μM novobiocin (Nov), 600 ng of poly(U), or 600 ng of poly(C) to 40 μl of RdRp reaction. Lane ø, minus-template control; lane 2, RdRp reaction mixture from which ATP and UTP were omitted. The total RNA products were quantified by using a PhosphorImager, and the relative percentage of synthesis normalized to the wild-type level is shown at the bottom.
RNA synthesis from the ssDNA MP19 was further characterized. The RNA synthesis observed was specific for MP19, not from an endogenous contaminating RNA since no synthesis was detected in the absence of MP19 (Fig. 6B). Also, the radiolabeled products were not produced by terminal labeling since removal of one or more of the NTPs abolished RNA synthesis, as shown in lane 2 of Fig. 6B (removal of ATP and UTP from the reaction mixture abolishes RNA synthesis completely). Poly(U) and poly(C) RNAs present at 1:1 ratio reduced the synthesis from MP19 to less than 20%, suggesting that RNA was the preferred template that competed efficiently for interaction with NS5B (Fig. 6B). Last, product formation from MP19 was not affected by the presence of rifampin and actinomycin D, inhibitors of DNA-dependent RNA polymerases. A somewhat surprising finding is that novobiocin, which was previously shown to inhibit RNA synthesis by brome mosaic virus replicase (50% inhibitory concentration of 75 μM), had no effect on BVDV RdRp-directed RNA synthesis (30). It is possible that novobiocin targets a protein in the brome mosaic virus replicase complex other than RdRp.
Mutational analysis of BVDV NS5B RdRp.
Amino acid sequence alignments between BVDV NS5B and other flavivirus RdRp revealed six conserved sequence motifs, four of which (A, B, C, and D) were characteristic polymerase motifs. The other two have not been previously described for this class of viral RdRps; one (named nc) is N terminal to motif A, and the other is (named cc) is C terminal to motif D (Fig. 1B and C). The nc motif is rich in lysine (K) and arginine (R) and thus is likely to play a role in interacting with RNA template/primer and/or nucleotide. The nc motif is highly conserved (more conserved that motif D), suggesting that it may play a direct and important role in the nucleotidyl transfer reaction. The cc motif consists of an cysteine-serine pair invariable among flavivirus RdRp, which is likely to be part of the motif E as described by Hansen et al. for the poliovirus 3D polymerase (11). However, the cc motif is not well conserved between flavivirus RdRp and that of picornaviruses, making it hard to identify this motif across virus families.
To further characterize the BVDV RdRp activities, a panel of alanine substitutions was introduced by site-directed mutagenesis in all six conserved motifs, with an emphasis on motifs nc and cc. All catalytically important amino acids in motifs A to D are numbered in Fig. 7; the importance of these amino acids had been previously described or hypothesized (11). Mutations created in motifs nc and cc were novel and unique to RdRp. All of the mutant proteins were purified similarly to that of the parental protein, NS5BΔCT24-His, and analyzed for both primer-dependent RdRp [elongative synthesis using poly(C)-oligo(G) as the substrate] (Fig. 5A) and primer-independent RdRp (de novo RNA synthesis) (Fig. 8) activities. The results are summarized in Table 1. For motif A, substitution of the first aspartate residue (D345A) abolished elongative synthesis and significantly reduced de novo synthesis, consistent with its proposed function for binding to the catalytic divalent metal ion (Mg2+ or Mn2+) (11, 13). The aspartate (D350) residue and the asparagine (N414) in motif B are believed to be involved in the selection of ribonucleotide versus deoxyribonucleotide (10, 11). Substitution of D350 with alanine (D350A) did not affect primer-dependent synthesis and somewhat (>2-fold) enhanced de novo synthesis. The N414A substitution, however, reduced both RdRp activities significantly. Neither mutant D350A nor mutant N414A incorporated dNTPs above the background level (data not shown). Two additional substitutions, S405A and G406A, were made for motif B. The invariable glycine residue (G406), was suggested to coordinate template and/or primer positioning (24), and we found it important for both RdRp activities. On the other hand, the S405A mutation had a less severe effect on both RdRp activities.
FIG. 7.
Schematic diagram depicting all alanine substitutions and deletion mutations in BVDV NS5B. Underlined letters represent residues that were changed to alanine by site-directed mutagenesis (vertical arrows); numbers next to the letters represent the positions of these amino acids in unmodified NS5B. Horizontal arrows represent deletions at either the N- or C-terminal end of NS5B.
FIG. 8.
Effects of various alanine substitutions and deletions on de novo RNA synthesis. The results shown are representative; each was reproduced a minimal of three independent times with two repetitions. The product of de novo RNA synthesis is 21 bases. Lane ø represents reaction without template (−)21g. WT, wild type.
TABLE 1.
Summary of elongative and de novo RNA synthesis activities of various NS5B mutants derived from alanine substitutions and deletions
| Mutant | Mean synthesis ± SDa
|
|
|---|---|---|
| Elongative (n = 2) | De novo (n = 2) | |
| Motif nc | ||
| K282A | 190.0 ± 10.0 | 115.6 ± 19.7 |
| R285A | 5.5 ± 0.0 | 1.3 ± 0.9 |
| R295A | 3.5 ± 0.1 | 0 |
| I287A | 14.5 ± 0.5 | 1.3 ± 1.2 |
| Y289A | 54.2 ± 23.5 | 41.3 ± 14.4 |
| K293A | 60.0 ± 35.0 | 30.0 ± 9.0 |
| Motif A | ||
| D345A | 0 | 10.0 ± 3.6 |
| D350A | 98.0 ± 16.0 | 273.7 ± 143.3 |
| Motif B | ||
| S405A | 21.0 ± 7.0 | 76.0 ± 36.1 |
| G406A | 7.4 ± 1.4 | 1.0 ± 0.8 |
| N414A | 23.5 ± 1.5 | 1.7 ± 2.4 |
| Motif C | ||
| G447A | 12.0 ± 2.0 | 13.0 ± 4.5 |
| D448A | 0 | 0 |
| D449A | 0 | 0 |
| Motif D (K475A) | 6.9 | ND |
| Motif cc | ||
| C497A | 17.5 ± 1.5 | 1.0 ± 0.8 |
| S498A | 11.0 ± 1.0 | 2.3 ± 0.5 |
| R518A | 44.0 ± 13.0 | 4.7 ± 0.5 |
| D519A | 80.5 ± 7.5 | 36.7 ± 0.5 |
| Deletions | ||
| N-38 (Δ1-38) | 54.0 ± 5.0 | 23.0 ± 7.0 |
| N-81 (Δ1-81) | 136.0 ± 17.0 | 51.7 ± 1.2 |
| N-90 (Δ1-90) | 156 | ND |
| N-106 (Δ1-106) | 6 | ND |
| N-117 (Δ1-117) | 3.2 ± 0.7 | 2.7 ± 0.5 |
| N-156 (Δ1-156) | NT | NT |
| N-199 (Δ1-199) | NT | NT |
| C-60 (Δ660-719) | NT | NT |
| C-100 (Δ620-719) | NT | NT |
| C-140 (Δ580-719) | NT | NT |
| C-179 (Δ541-719) | 0.9 ± 0.9 | 0 |
| C-218 (Δ502-719) | 0 | 0 |
Percentage of the value for NS5BΔCT24-His (wild type). NT, not tested due to insolubility and unpurifiability; ND, not done.
Motif C is the signature motif for RdRps and is highly conserved in RdRps, although variations in the glycine have been reported (20). Both aspartate residues (D448 and D449) are essential for coordinating divalent cations used in binding NTPs (13). Substitutions (D448A and D449A) at these two positions were lethal to RdRp activities. Substitution of the glycine residue (G447A) was also detrimental to both RNA syntheses, reducing the RNA synthesis to around 15% of the wild-type level. Motif D is less defined by the sequence alignment; the substitution of lysine residue (K475A) reduced primer-dependent synthesis drastically, confirming that the motif assignment may be correct.
Motifs nc and cc were previously uncharacterized, and mutations in these motifs revealed interesting results. In motif nc, two arginine residues (R285 and R295) are conserved. Substitutions at these positions abolished both forms of RNA syntheses, making these two arginine residues candidates for interacting with RNA template and/or primer, or with nucleotide. Interestingly, the two conserved lysines, K282 and K293, were less critical for RNA synthesis (K282A and K293A). In fact, K282A reproducibly exhibited higher activity in elongative synthesis than the wild type (200%). Another conserved amino acid in motif nc is isoleucine (I287). Mutation at this position reduced both activities to the background level. The tyrosine residue (Y289) is less critical since its substitution (Y289A) had lesser effects on RdRp activities.
Alanine substitutions in motif cc revealed that the cysteine and serine residues (C497 and S498) were important for RdRp. Substitutions at these positions reduced the elongation to about 10 to 15% and almost completely abolished de novo synthesis, suggesting that they may be part of the motif E which is less conserved among various viral RdRps (Table 1). The other two conserved amino acids among flavivirus RdRps, R518 and D519, were less important, although R518A seems to have a more profound effect on de novo synthesis, reducing it to the background level. The analyses above demonstrate that while most of the mutations have similar effects on both modes of RNA synthesis, several had selective effects on elongative or de novo modes of RNA synthesis.
A minimal active domain of BVDV NS5B.
To map the minimal domain required for NS5B enzymatic activities, we constructed a series of N- and C-terminal truncations (Fig. 7). Since the N-terminal domain of BVDV NS5B is about 130 amino acids longer than that of HCV NS5B (Fig. 1A), it is desirable to delete this sequence. It has been shown that deletions at the N terminus are detrimental to the RdRp activities of HCV (16) and poliovirus (23). Deletional analysis from the N terminus of BVDV NS5B revealed that up to 90 amino acids could be removed without significantly affecting the RdRp activities. Removal of an additional 16 or more residues (N-106 and N-117) abolished both elongative and de novo RNA syntheses (Table 1). Further truncations (N-156 and N-199) resulted in insoluble products. The N-terminal deletional analysis demonstrates that BVDV can tolerate some degree of truncation from the N terminus.
Deletions from the C terminus revealed, unexpectedly, that only the hydrophobic 24 amino acids could be removed (in NS5BΔCT24-His) without reducing RNA synthesis. Deletions of 60, 100, or 140 amino acids from the C terminus resulted in insoluble products, possibly due to the misfoldings of the truncated products. Further deletions of 179 and 219 amino acids yielded soluble products that, however, lacked RdRp activities (Table 1). These results are in contrast to those of HCV RdRp, in which 63 amino acids can be removed without affecting RdRp activity (7).
DISCUSSION
In this work, we have (i) performed extensive mutational and deletional characterizations of the BVDV RdRp which provide insights to the structure and activity of this class of polymerases; (ii) identified of two additional motifs (nc and cc) important for activity; (iii) examined the correlation between two modes of RNA syntheses (elongative and de novo); and (iv) optimized the conditions for BVDV RdRp activity, allowing comparisons with other related viral RdRps. These results significantly contribute to our knowledge on BVDV viral RNA replication.
Based on sequence comparisons, RdRps have been proposed to contain a number of sequence motifs (11, 24). Mutations in motifs A to D are largely consistent with those of the HCV NS5B protein (16, 17) and those made for numerous RNA viruses (20). We predict that the cc motif is likely to be motif E (11), although motif E is not well conserved between flavivirus RdRp and that of poliovirus. It is interesting that RNA viruses also contain an arginine/lysine-rich nc motif at similar positions in the linear sequences of their RdRps; this well-conserved motif is important for RdRp activities, and we speculate that it may interact with template and/or primers or with nucleotides. Unfortunately, the corresponding region in the poliovirus RdRp is disordered in the crystal structure of poliovirus 3Dpol (11). A higher-resolution structure, preferably in complex with RNA, is required to confirm our prediction.
BVDV NS5B is a large protein (719 amino acids) compared to members in the genus Hepacivirus, such as HCV. Our sequence alignment revealed that the BVDV NS5B protein has an extra domain of approximately 130 amino acids at the N terminus. The hydropathy profile comparison (Fig. 1A) between BVDV and HCV NS5Bs indicates that this extra N-terminal domain is rather hydrophilic. Our deletional analysis demonstrated that at least 90 amino acids of the N terminus could be removed without affecting RdRp activities in vitro. For HCV and poliovirus, deletions of 19 and 5 amino acids have been shown to be detrimental for their RdRp activities (16, 23). It has been suggested that the N-terminal domain of poliovirus RdRp may be important for self-interaction to form functional oligomers of the polymerase (11, 21). This is unlikely to be the same for BVDV RdRp, at least not for the first 90 amino acids. A BLAST search of GenBank databases failed to identify any meaningful homologs of the N-terminal domain. More work is needed to elucidate the functions of this unique domain in BVDV.
Similar to that of HCV, BVDV RdRp possesses a hydrophobic domain at the C terminus. Consistent with the results of HCV NS5B, removal of this domain greatly increases the solubility of the BVDV NS5B, suggesting that this domain plays a common and important structural role in RNA replication, possibly as a membrane anchor.
The largely similar effects of mutations on de novo and elongative RNA syntheses confirm that both modes of RNA synthesis are catalyzed by the same active site in the polymerase. It will be interesting to determine which mode of RNA synthesis is more efficient. In poliovirus, oligonucleotide-directed RNA synthesis is far more dominant than protein-primed (de novo initiation) synthesis. However, de novo protein-primed synthesis is the mode of replication acquired by poliovirus (22). We believe that de novo initiation is a plausible mechanism of replication by BVDV and possibly by other members in the Flaviviridae family as well.
ACKNOWLEDGMENTS
We thank Charles Lesburg and Bahige Baroudy for helpful discussions. We are grateful to Ruben Donis for his generosity and for making his valuable reagents available to us.
REFERENCES
- 1.Adkins S, Quadt R, Choi T J, Ahlquist P, German T. An RNA-dependent RNA polymerase activity associated with virions of tomato spotted wilt virus, a plant- and insect-infecting bunyavirus. Virology. 1995;207:308–311. doi: 10.1006/viro.1995.1083. [DOI] [PubMed] [Google Scholar]
- 2.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 3.Chon S K, Perez D R, Donis R O. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhea virus. Virology. 1998;251:370–381. doi: 10.1006/viro.1998.9425. [DOI] [PubMed] [Google Scholar]
- 4.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 5.De Francesco R, Behrens S E, Tomei L, Altamura S, Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996;275:58–67. doi: 10.1016/s0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 6.Elbers K, Tautz N, Becher P, Stoll D, Rumenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov I, McBride M S, Rice C M. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 12.Hong Z, Ferrari E, Wright-Minogue J, Chase R, Risano C, Seelig G, Lee C-G, Kwong A D. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells using the herpes simplex virus amplicon system. J Virol. 1996;70:4261–4268. doi: 10.1128/jvi.70.7.4261-4268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 14.Kao C C, Del Vecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant Flavivirus RNA-dependent RNA polymerase. Virology. 1998;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 15.Lemon S H, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 16.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 18.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 19.Oberste M S, Flanegan J B. Measurement of poliovirus RNA polymerase binding to poliovirion and nonviral RNAs using a filter-binding assay. Nucleic Acids Res. 1988;16:10339–10352. doi: 10.1093/nar/16.21.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Reilly E, Kao C. Analysis of the structure and function of viral RNA-dependent RNA polymerases as guided by computer-assisted structure predictions and the known structures of polymerases. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 21.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 22.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 23.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole T L, Wang C, Popp R A, Potgieter L N, Siddiqui A, Collet M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 26.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 931–960. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 29.Stark R, Meyers G, Rumenapf T, Thiel H J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Adkins S, Faurote G, Kao C. Initiation of (−) strand RNA synthesis catalyzed by the brome mosaic virus RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 31.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a serine proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong W, Gutshall L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]