Abstract
Y-box binding protein YB-1 is a member of a family of DNA and RNA binding proteins which have been shown to affect gene expression at both the transcriptional and translational levels. We have previously shown that YB-1 modulates transcription from the promoters of the ubiquitous human polyomavirus JC virus (JCV). Here we investigate the physical and functional interplay between YB-1 and the viral regulatory protein large T antigen (T-antigen), using JCV as a model system. Results of mobility band shift assays demonstrated that the efficiency of binding of YB-1 to a 23-bp single-stranded viral target sequence was significantly increased when T-antigen was included in the binding reaction mixture. Affinity chromatography and coimmunoprecipitation assays demonstrated that YB-1 and T-antigen physically interact with each other. Additionally, results of transcription studies demonstrated that these two proteins interact functionally on the JCV early and late gene promoters. Whereas ectopic expression of YB-1 and T-antigen results in synergistic transactivation of the viral late promoter, YB-1 alleviates T-antigen-mediated transcriptional suppression of the viral early promoter activity. Furthermore, we have localized, through the use of a series of deletion mutants, the sequences of these proteins which are important for their interaction. The T-antigen-interacting region of YB-1 is located in the cold shock domain of YB-1 and its immediate flanking sequences, and the YB-1-interacting domain of T-antigen maps to the carboxy-terminal half of T-antigen. Results of transient transfection assays with various YB-1 mutants and T-antigen expression constructs confirm the specificity of the functional interaction between YB-1 and T-antigen. Taken together, these data demonstrate that the cellular factor YB-1 and the viral regulatory protein T-antigen interact both physically and functionally and that this interaction modulates transcription from the JCV promoters.
The Y-box binding proteins are the most evolutionarily conserved nucleic acid binding proteins, representing a multigene family identified in a number of organisms ranging from bacteria to higher eukaryotes (52, 53). The first of these proteins from a vertebrate was originally cloned by virtue of its binding to a C/T-rich double-stranded oligonucleotide spanning the DRA X and Y elements found within the promoters of human major histocompatibility complex class I genes (12). The human homologue of Y-box proteins is called YB-1. In general, all vertebrate Y-box binding proteins consist of three general domains: a variable glycine-rich N terminus, a highly conserved central nucleic acid recognition domain, and a hydrophilic C-terminal tail domain. No function has yet been assigned to the glycine-rich N-terminal region. The central nucleic acid recognition domain is also known as the cold shock domain (CSD) and shows 43% homology to the bacterial cold shock protein (CS 7.4) (19). The tail domain contains alternating regions of predominantly basic or acidic amino acids, each about 30 amino acids in length. The basic regions are rich in arginine, glutamine, and proline. The tail domain is thought to stabilize protein-DNA interactions (52). Since Y-box binding proteins exhibit a high degree of primary sequence conservation, particularly in the central DNA binding domain, it has been suggested that they may have essential structural and functional roles in eukaryotic cells. In fact, a growing body of experimental evidence indicates that Y-box binding proteins are involved in a wide variety of biological functions, including regulation of gene expression at both the transcriptional (2, 10, 13, 25, 26, 32, 37, 38) and translational (48) levels, DNA and RNA condensation, and DNA repair (21, 52, 53). In addition, recent reports indicate that the members of the Y-box family of proteins are responsive to many types of stress-related stimuli, including UV irradiation (8, 28), drug treatment (3, 32), DNA damage (24, 28), and interleukin-2 treatment in T cells (45). Our previous results have implicated YB-1 in transcriptional regulation of JC virus (JCV) promoters (26, 46).
JCV, the etiologic agent of the fatal subacute human neurodegenerative disease known as progressive multifocal leukoencephalopathy, lytically infects myelin-producing oligodendrocytes in the central nervous system. Seroepidemiological studies have demonstrated that 70 to 80% of adults are seropositive for JCV (17). Despite this high prevalence of infection, progressive multifocal leukoencephalopathy emerges only in patients with an underlying cellular immunodeficiency, such as AIDS, lymphoma, or chronic lymphocytic leukemia (5, 29).
The major regulatory protein of JCV, large T antigen (T-antigen), is a multifunctional phosphoprotein involved in both viral DNA replication (33–35, 49) and viral gene transcription (27, 30). It exhibits significant sequence homology (72%) to the T-antigens of polyomaviruses simian virus 40 (SV40) and BK virus, with the greatest divergence occurring within the carboxy-terminal region (16). Biochemical and genetic analyses of SV40 T-antigen have defined within the protein discrete subregions with particular biological functions (15). SV40 T-antigen specifically interacts with sequences within the viral origin of DNA replication (ori) and, in the presence of ATP, oligomerizes to form a double hexamer structure capable of unwinding ori sequences (7), thereby creating a replication bubble. Cellular DNA polymerase α-primase recruited to the DNA replication initiation site initiates the elongation process via its helicase-ATPase activity (11, 47). The phosphorylation of T-antigen at selected serine residues, including Ser 120, 123, 677, and 679, appears to diminish its DNA binding and transactivation activities (14). In contrast, phosphorylation at the Thr 124 residue greatly enhances the interaction of T-antigen with ori sequences and, therefore, its replication and transforming activities (14). Moreover, T-antigen is able to transform primary cells in culture, presumably through its ability to transactivate cellular genes and inactivate tumor suppressor proteins, including pRb and p53 (39). JCV T-antigen appears to have functions in viral DNA replication (33–35) and gene transcription similar to those of SV40 T-antigen. JCV T-antigen transactivates JCV late genes but suppresses gene transcription from its early promoter via an autoregulatory loop (17).
We have previously shown that YB-1 regulates JCV gene transcription from the JCV early and late promoters (10, 26, 46). We have also shown that T-antigen stimulates the binding of YB-1 to its target sequences in the pentanucleotide repeat element (PRE), which is found in the regulatory region of JCV (9). Furthermore, a unique sequence element, designated the 23-bp sequence element (23-bpse) (17, 46), found in the regulatory region of some JCV strains, including the JCV archetype strain and some intermediate forms, also contains binding sites for YB-1 (17). Therefore, in the present study, we utilized JCV sequences and promoter reporter constructs as model systems to investigate the stimulatory effect of T-antigen on binding of YB-1 to the 23-bpse as well as physical and functional interaction between YB-1 and T-antigen. In this report, we provide experimental evidence, obtained by several independent assays, that the cellular factor YB-1 physically and functionally interacts with JCV T-antigen.
MATERIALS AND METHODS
Cell lines.
HJC-15b (44) cells were derived from JCV-induced hamster brain tumors (51) and grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum and antibiotics (penicillin-streptomycin, 100 μg/ml). U-87MG, a human glioblastoma cell line, was also grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and the same antibiotics. All cells were maintained at 37°C in a humidified atmosphere with 7% CO2.
Plasmid constructs.
The regulatory region of the JCV archetype strain (JCVCY) was subcloned into the pBLCAT3 vector at the BamHI restriction site in both the early and late orientations, resulting in constructs designated pBLCAT3-CyE and -CyL, respectively. The regulatory region of JCVCY was also subcloned into the pGL3-basic luciferase (Promega, Madison, Wis.) vector in both the early and late orientations at the BglII restriction site, resulting in constructs designated pGL3-basic-CyE and pGL3-basic-CyL, respectively. Luciferase reporter plasmids containing point mutations within the 23-bp region of JCVCY were also constructed and designated pGL3-basic-CyL mut23 (for the late orientation) and pGL3-basic-CyE mut23 (for the early orientation). pGEX2T-YB-1 was a kind gift from Gene MacDonald (36). It was originally cloned into the BamHI and EcoRI sites of the vector. C-terminal deletion mutants of glutathione S-transferase (GST) fusion proteins of YB-1, pGEX2T-YB-1(1–37), pGEX2T-YB-1(1–75), pGEX2T-YB-1(1–125), and pGEX2T-YB-1(1–203), were previously described (46). N-terminal deletion mutants of YB-1, CMV-YB-1 (126–318) and CMV-YB-1(204–318), were previously described (46). Cytomegalovirus (CMV)-JCV T-antigen and pMAL-MBP-YB-1 expression plasmids were described previously (9, 10). pGEX2T-JCV T-antigen and its deletion mutants were created by PCR amplification. A plasmid containing an intronless JCV T-antigen coding region (6) was used as a template in PCR amplification with specific primers. Forward primers were JCT 5′-Eco R1 (5′-CTGAGGAATTCATGGACAAAGTGCTG-3′), FP aa 266 (5′-CTGAGGAATTCGAAGAAACTAAGCAGGTT-3′), FP aa 412: (5′-CTGAGGAATTCCTAAAATGCATTGTATTA-3′), and FP aa 629 (5′-CTGAGGAATTCGACTTTCCTAGAGAGGAA-3′); reverse primers were JCT-3′ Eco R1 (5′-CTGAGGAATTCTTATTTTGGGGGAGG-3′), RP aa 81 (5′-CTGAGGAATTCACTATTCCATGTACCAAA-3′), RP aa 265 (5′-CTGAGGAATTCTGGTTCTTCTGGGTTAAA-3′), RP aa 411 (5′-CTGAGGAATTCAAAGTCATAAATAACAGT-3′), and RP aa 628 (5′-CTGAGGAATTCAAGAATGGGTCTCCCCAT-3′). Resulting PCR fragments were digested with EcoRI and subcloned into the EcoRI site of the pGEX2T vector. The orientations of the subcloned fragments in pGEX2T vector were determined by DNA sequencing. The Epstein-Barr virus (EBV)–His–YB-1 plasmid containing a Rous sarcoma virus (RSV)-driven promoter was previously described (46).
Expression and purification of recombinant proteins.
Expression and purification of a maltose binding protein (MBP)–YB-1 fusion protein was described previously (9). For GST fusion proteins, 50-ml overnight cultures of Escherichia coli DH5α, transformed with either pGEX2T-YB-1 or pGEX2T-JCV T-antigen or the respective deletion mutant plasmid, were diluted 1:10 in fresh Luria-Bertani broth supplemented with ampicillin (100 μg/ml). Cultures were induced with 0.3 M isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 595 nm of 0.5 and incubated for an additional 2 h at 37°C. Cells were collected by centrifugation and resuspended in 10 ml of a lysis buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 and supplemented with 1 mM phenylmethylsulfonyl fluoride, 2 mM pepstatin A, 0.6 mM leupeptin, and 2 mM benzamidine. After sonication, clear cell lysates were prepared by centrifugation at 12,000 × g. Lysates were then each incubated with 150 μl of 50% glutathione-Sepharose beads (Pharmacia, Piscataway, N.J.) overnight at 4°C. GST fusion proteins were purified by three cycles of washing and centrifugation with 5 ml of lysis buffer. Fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
Mobility band shift assays.
For band shift assays, a single-stranded DNA fragment representing the early strand (5′-CAGTTTTAGCCAGCTCCTCCCTA-3′) of the JCVCY 23-bp sequence was 5′ end labeled with [γ-32P]ATP by using polynucleotide kinase and then gel purified. Highly purified bacterially expressed MBP–YB-1 fusion protein and affinity-purified baculovirus-expressed JCV T-antigen were mixed, either alone or in combination with the single-stranded probe (30,000 cpm/reaction), respectively, in a binding buffer containing 0.1 μg of poly(dI-dC)/ml, 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2, and 0.1 mM dithiothreitol and incubated for 30 min at 4°C. Protein concentrations in the binding reactions are indicated under the respective figure legends. The DNA-protein complexes were resolved on 6% polyacrylamide gels in 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 8.0], 89 mM boric acid, and 2 mM EDTA [pH 8.0]). Gels were dried, and complexes were detected by autoradiography at −70°C with intensifying screens.
Coimmunoprecipitation assays.
The pEBV-His-YB-1 expression plasmid was transfected into HJC-15b (a hamster astrocytic cell line that constitutively expresses JCV T-antigen) cells via the calcium phosphate precipitation method (20). At 36 h posttransfection, cells were lysed in a lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.4), and 0.5% Nonidet P-40 and supplemented with a cocktail of proteinase inhibitors consisting of 1 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (1 μg/ml), and 50 mM sodium fluoride. Five hundred micrograms of whole-cell extract (treated with DNase I prior to coimmunoprecipitation assay) in a total volume of 0.5 ml was incubated with either 0.5 μg of an anti-T7 antibody (Novagen, Madison, Wis.) directed against the His-tagged YB-1 or 0.5 μg of preimmune antiserum overnight at 4°C. Immunocomplexes were precipitated by the addition of protein A-Sepharose beads (Pharmacia), with an additional 2-h incubation, and washed extensively with lysis buffer. Immunocomplexes were resolved on an SDS–8% polyacrylamide gel and analyzed by Western blotting. Blots were probed with an anti-T-antigen antibody (Ab-2 416; Oncogene, Uniondale, N.Y.) and developed with an ECL detection kit (Amersham, Arlington Heights, Ill.) in accordance with the recommendations of the manufacturer.
Transient transfection assays.
U-87MG cells were transiently transfected by the calcium phosphate precipitation method (20) with reporter constructs, alone or in combination with expression plasmids. Plasmid concentrations used in transfections are indicated in the respective figure legends, but the total amount of DNA transfected into the cells was normalized by using the respective empty vector. In all transfection studies, 1 μg of RSV β-gal, a plasmid expressing β-galactosidase (β-Gal), was included in the transfection mixture to normalize for transfection efficiency. A glycerol shock procedure was performed at 4 h posttransfection, and the medium was replenished. At 48 h posttransfection, cells were lysed by three freeze-thaw cycles. Cell debris was cleared, and chloramphenicol acetyltransferase (CAT) activity of supernatants was determined and normalized to β-Gal activity. Alternatively, luciferase assays were performed in accordance with the manufacturer’s (Promega) recommendations. Transfections were repeated at least three times with different plasmid preparations.
In vitro transcription and translation assay.
Full-length YB-1 protein and its two N-terminal deletion mutants, YB-1 (126–318) and YB-1 (204–318), were labeled with [35S]methionine by using a TNT coupled in vitro transcription-translation system (Promega) in accordance with the recommendations of the manufacturer.
GST affinity chromatography assays (GST pull-down).
Physical association between JCV T-antigen and His–YB-1 proteins was tested by the GST pull-down assay as described previously (46). Three micrograms of either GST alone or GST–YB-1 immobilized on Sepharose beads was incubated with 0.5 mg of DNase I (2 U/μg of protein)- or ethidium bromide (100 μg/ml)-treated whole-cell extract prepared from HJC cells, which constitutively express JCV T-antigen, for 2 h at 4°C in a lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% Nonidet P-40. The protein-complexed beads were washed extensively with lysis buffer and resolved by SDS–10% PAGE followed by Western blot analysis with an antibody (Ab-2 416) directed against JCV T-antigen. Similarly, either GST or GST-JCV T-antigen immobilized on Sepharose beads was incubated with an extract prepared from HJC cells transfected with the EBV–His–YB-1 expression plasmid. The rest of the procedures for GST pull-down assays were followed as described above. The presence of the His–YB-1 proteins on Western blots was determined by using an anti-T7 antibody directed against His-tagged YB-1.
To map the region(s) within JCV T-antigen responsible for interaction with YB-1, or vice versa, GST pull-down experiments were performed either with 35S-labeled, in vitro-translated YB-1 or with whole-cell extracts prepared from HJC-15b cells, using GST-JCV T-antigen, GST–YB-1, or the respective deletion mutant.
RESULTS
In vitro binding of YB-1 is stimulated by T-antigen.
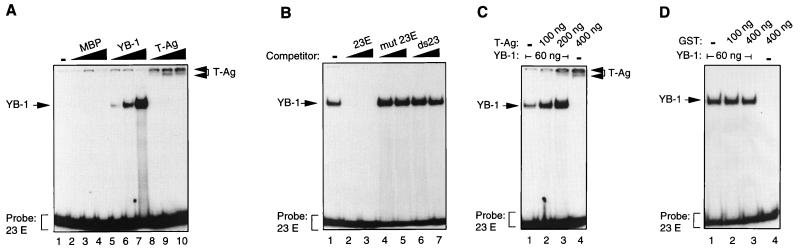
We have previously shown that JCV T-antigen enhances the binding of YB-1 to the PRE early single strand present within the regulatory region of JCV isolates (9). Since the 23-bpse and the PRE exhibit sequence homology in their C/T-rich regions, we wanted to determine whether T-antigen would also stimulate binding of YB-1 to the 23-bp early single-strand sequences. To test this hypothesis, mobility band shift assays were performed with recombinant YB-1 and T-antigen proteins and an end-labeled single-stranded oligonucleotide derived from the early strand of the 23-bpse, designated 23E. As shown in Fig. 1A, MBP–YB-1 interacted with the 23E probe in a dose-dependent manner (lanes 5 to 7), but MBP alone did not (lanes 2 to 4). Interestingly, T-antigen interacted with 23E but formed large complexes with the 23E probe, some of which remained in the wells under native conditions. To examine the specificity of the interaction between YB-1 and the 23E probe, we performed competitive band shift assays. The 23E probe was incubated with YB-1 protein in the absence (lane 1) or presence of 50- and 250-fold molar excesses of unlabeled competitor DNA. As demonstrated in Fig. 1B, wild-type 23E DNA efficiently competes with the labeled 23E probe for YB-1 protein (lanes 2 and 3), whereas neither mutant 23E (lanes 4 and 5) nor the double-stranded 23-bp oligonucleotide (lanes 6 and 7) has an effect on the binding of YB-1 to its target, indicating the specificity of the interaction between YB-1 and the 23E probe.
FIG. 1.
T-antigen induces binding of YB-1 to the 23-bp early strand (23E) in electrophoretic mobility shift assays. (A) Interaction of bacterially expressed MBP, MBP–YB-1 fusion protein, and baculovirus-expressed T-antigen with the 23-bp early single-stranded DNA probe. The 23E probe was incubated with 30, 60, or 240 ng of MBP alone (lanes 2 to 4, respectively) or MBP–YB-1 fusion protein (lanes 5 to 7, respectively) or with 100, 200, or 400 ng of T-antigen (T-Ag) (lanes 8 to 10, respectively). DNA-protein complexes were resolved on a 6% polyacrylamide gel under native conditions. Positions of MBP–YB-1 and T-antigen complexes with the 23E probe are indicated by arrows. (B) Competitive band shift assay. The 23E probe was incubated with MBP–YB-1 protein in the absence (lane 1) or presence of a 50- or 250-fold molar excess of unlabeled competitor DNA as indicated. 23E represents the 23-bp wild-type early single-stranded oligonucleotide (5′-CAGTTTTAGCCAGCTCCTCCCTA-3′) (lanes 2 and 3), mut 23E represents a mutant 23E single-stranded oligonucleotide (5′-AGTATTACACAGATATTTATTAC-3′) (lanes 4 and 5), and ds23 represents the wild-type double-stranded 23-bpse (lanes 6 and 7). (C) T-antigen induces binding of YB-1 to the 23-bp early single strand. The 23E probe was incubated with recombinant YB-1 (MBP–YB-1), either alone (lane 1) or with baculovirus-expressed T-antigen (lanes 2 and 3), or with T-antigen alone (lane 4). Formed DNA-protein complexes were resolved on a 6% polyacrylamide gel under native conditions. Respective protein concentrations used for the binding reactions are shown above the panel. The protein concentration used for all binding reactions was kept constant by addition of MBP. MBP–YB-1:23E and T-Ag:23E complexes are indicated by arrows. (D) Bacterially expressed, purified GST protein does not induce binding of YB-1 to the 23-bp early single strand. The 23-bp probe was incubated with recombinant YB-1, either alone (lane 1) or in combination with GST (lanes 2 and 3), or with GST alone (lane 4). Formed DNA-protein complexes were resolved as described for panel C. Respective protein concentrations used for the binding reactions are shown above the panel. MBP–YB-1:23E complex is indicated by an arrow. In all panels, brackets indicate free probe.
Next, we analyzed whether T-antigen influences the binding of YB-1 to its target sequences on the 23E probe. As shown in Fig. 1C, YB-1 alone exhibited a relatively low affinity for the 23E strand (lane 1) at low protein concentrations; however, simultaneous addition of increasing amounts of T-antigen to the binding reaction resulted in a significant dose-dependent increase in binding of YB-1 to the 23E probe (compare lane 1 with lanes 2 and 3). Addition of T-antigen, however, did not alter the mobility of the YB-1–23E complex. Rather, T-antigen formed a complex with a slower electrophoretic mobility (lane 4), similar to that evident in Fig. 1A. Under no conditions did we observe formation of an additional complex, suggesting that a ternary complex of 23E, YB-1, and T-antigen, if it exists at all, is either transient or unstable under our gel shift conditions. We further examined the specificity of the T-antigen-induced binding of YB-1 to the 23E probe by utilizing an unrelated, bacterially produced GST protein in band shift assays. As shown in Fig. 1D, this GST protein did not stimulate binding of YB-1 to the 23E probe (lanes 2 and 3), indicating that binding of YB-1 to its target sequences is specifically stimulated by T-antigen. GST protein alone does not bind to the 23E probe (lane 4). Taken together, these results demonstrate that YB-1 interacts specifically with the 23E probe and that its interaction is specifically stimulated by T-antigen.
Interaction of YB-1 with T-antigen in the absence of DNA.
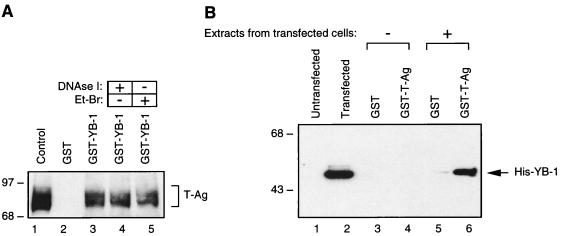
Our findings from mobility band shift assays suggested that YB-1 and T-antigen could physically interact with one another in the absence of DNA. To test this possibility, we performed affinity chromatography (GST pull-down) experiments in which one of the two proteins was expressed as a GST fusion protein and bound to a glutathione resin while the other protein was passed over the resin and analyzed for its ability to be specifically retained by the GST fusion protein. For this purpose, bacterially expressed GST or GST–YB-1 was immobilized on glutathione-Sepharose beads and incubated with whole-cell lysate from HJC-15b cells, which constitutively express T-antigen. Beads were extensively washed, and bound proteins were analyzed by immunoblotting with antibodies directed against T-antigen. As demonstrated in Fig. 2A, T-antigen was retained by GST–YB-1 (lane 3) but not by GST alone (lane 2). Additionally, whole-cell lysate treated with DNase I (lane 4) or ethidium bromide (lane 5) prior to pull-down experiments was also incubated with GST–YB-1 to eliminate the possibility that DNA mediates any interaction between YB-1 and T-antigen. As shown in Fig. 2A, neither DNase I nor ethidium bromide treatment affected the efficiency of the binding observed between GST–YB-1 and T-antigen in GST pull-down assays, indicating that these two proteins physically interact in the absence of DNA. In reciprocal GST pull-down assays, we also demonstrated the physical association of YB-1 and T-antigen proteins. Whole-cell lysates prepared from either untransfected HJC-15b cells or the same cell line transfected with a histidine-tagged YB-1 expression plasmid (pEBV-YB-1) were incubated with either GST or GST–T-antigen columns. The columns were washed, and bound complexes were analyzed by Western blotting with antibodies specific for His-tagged YB-1. As shown in Fig. 2B, His–YB-1 was specifically retained on the Sepharose column containing GST–T-antigen (lane 6) but not on the column containing GST alone (lane 5). The specificity of the association was demonstrated both by the absence of an interaction between expressed His–YB-1 and the GST moiety alone (Fig. 2B, lane 5) and by the absence of such an interaction when protein extract prepared from untransfected cells was used (Fig. 2B, lanes 3 and 4).
FIG. 2.
In vitro interaction between YB-1 and T-antigen. (A) Bacterially expressed GST (lane 2) or GST–YB-1 (lane 3) was immobilized on GST-Sepharose beads and incubated with whole-cell extract prepared from hamster glial cells constitutively expressing T-antigen (HJC-15b). Additionally, whole-cell extracts from HJC-15b cells, treated with DNase I (0.2 U/μg of protein) or ethidium bromide (100 ng/ml) (Et-Br), were also incubated with GST–YB-1 (lanes 4 and 5, respectively). HJC-15b whole-cell extract was loaded as a migration control (lane 1). Nonbinding proteins were removed from the column by extensive washing, and proteins interacting with GST or GST–YB-1 were resolved by SDS–10% PAGE and analyzed by Western blotting with an anti-T-antigen antibody (Ab-2 416). The bracket indicates T-antigen (T-Ag). (B) Whole-cell extracts prepared from untransfected HJC-15b cells (lanes 1, 3, and 4) and from HJC-15b cells transfected with a histidine-tagged YB-1 expression plasmid (pEBV-YB-1) (lanes 2, 5, and 6) were incubated with either GST alone (lanes 3 and 5) or GST–T-antigen (GST-T-Ag) (lanes 4 and 6) as indicated. After being washed, proteins interacting with GST or GST–T-antigen were resolved by SDS–10% PAGE and analyzed by Western blot analysis with anti-T7 antibody for detection of His-tagged YB-1. Whole-cell extracts from HJC-15b cells either untransfected (lane 1) or transfected with pEBV-His-YB-1 expression plasmid (lane 2) were loaded as negative and positive migration controls, respectively. The arrow indicates histidine-tagged YB-1. The positions of molecular mass markers (in kilodaltons) are shown on the left of each panel.
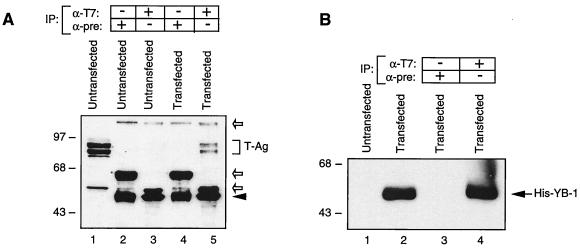
To further examine the physical association of YB-1 and T-antigen, coimmunoprecipitation experiments were performed to determine the interaction between these two proteins in cells. Whole-cell lysates prepared from HJC-15b cells that were either untransfected or transfected with the His–YB-1 expression plasmid were immunoprecipitated with either an anti-T7 antibody that recognizes His-tagged YB-1 or preimmune serum (normal mouse serum). Immunocomplexes were then resolved by SDS–7% PAGE and analyzed by Western blotting for the presence of T-antigen. In extracts from HJC-15b cells transfected with His-tagged YB-1, anti-T7 antibody coimmunoprecipitated T-antigen (Fig. 3A, lane 5). The specificity of this coimmunoprecipitation was investigated by using (i) normal mouse serum, which failed to show any cross-reactivity with proteins prepared from either untransfected cells (lane 2) or cells transfected with His–YB-1 (lane 4), and (ii) the anti-T7 antibody, which did not result in any immunoprecipitation of proteins from untransfected cells (lane 3). Figure 3B demonstrates the expression of the histidine-tagged YB-1 and its immunoprecipitation with anti-T7 antibody (lanes 2 and 4, respectively). Taken together, data from both in vitro affinity chromatography and coimmunoprecipitation experiments demonstrate that T-antigen and YB-1 physically associate with one another in the absence of DNA.
FIG. 3.
T-antigen coimmunoprecipitates with YB-1. (A) Coimmunoprecipitation of T-antigen with His–YB-1. Coimmunoprecipitation experiments were performed as described in Materials and Methods. Antibodies used for the respective lanes are shown at the top. HJC-15b whole-cell extract was loaded as a migration control (lane 1). The bracket indicates T-antigen (T-Ag). An arrowhead indicates the position of immunoglobulin heavy chain detected by the secondary antibody. Open arrows indicate nonspecific bands. IP, immunoprecipitation; α-T7, anti-T7 antibody; α-pre, preimmune serum. (B) Direct immunoprecipitation of His-tagged YB-1 with anti-T7 antibody, demonstrating that this antibody works in immunoprecipitation assays. Whole-cell extract (40 μg) from hamster glial cells (HJC-15b) transfected with the pEBV-YB-1 expression plasmid was immunoprecipitated with preimmune serum or with anti-T7 as indicated. The immunocomplexes were analyzed by Western blotting for His-tagged YB-1, using anti-T7. Whole-cell extracts from HJC-15b cells either untransfected or transfected with His-tagged YB-1 were loaded as negative and positive controls (lanes 1 and 2, respectively). The arrow indicates His-tagged YB-1. The positions of molecular mass markers (in kilodaltons) are shown on the left of each panel.
Functional interaction between T-antigen and YB-1 on the JCVCY promoters.
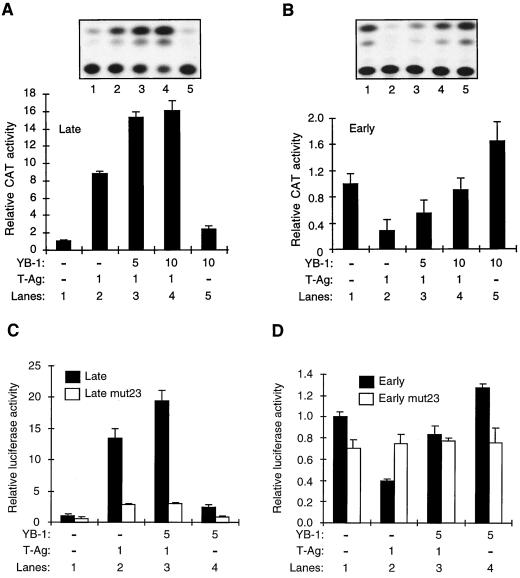
The physical interaction between YB-1 and T-antigen led us to investigate whether any functional interaction between these two proteins occurs. For this purpose, we performed transient transfection experiments with U-87MG cells, using reporter constructs containing the JCV early and late gene promoters. Previous observations have shown that YB-1 transactivates transcription from the JCV early and late promoters (10, 26) whereas T-antigen transactivates transcription only from the JCV late promoter (27) while suppressing its own transcriptional activity from the JCV early promoter (17). In the next series of experiments, we assessed the activity of the viral early and late promoters in the presence of each transactivator, alone or in combination. As shown in Fig. 4A, a CAT reporter construct containing the late promoter was poorly expressed in U-87MG cells in the absence of the transactivators (lane 1). Cotransfection of the reporter construct with a T-antigen expression plasmid resulted in a substantial (eight- to ninefold) increase in the transcriptional activity from the late promoter (compare lanes 1 and 2). However, when a constant amount of T-antigen was cotransfected with increasing amounts of YB-1, we observed a synergistic effect on the transcriptional activity of the late promoter (14- to 18-fold increase) (compare lane 2 to lanes 3 and 4). At the highest YB-1 concentration alone, we observed only a threefold increase in transcription (Fig. 4A, lane 5), demonstrating a synergistic effect of these two transactivators on transcription from the late promoter. This synergistic effect is independent of an increase in DNA replication of the reporter plasmid, which contains the origin of viral replication, because similar results were obtained when transient transfections were performed in the presence of aphidicolin, an inhibitor of DNA replication (data not shown). It is of note that expression of T-antigen or YB-1 remains unaltered upon expression of either transactivator (data not shown), suggesting that the observed transcriptional cooperativity between T-antigen and YB-1 is not due to the effect of one transactivator on the expression of the other.
FIG. 4.
Functional interaction between T-antigen and YB-1. (A) T-antigen and YB-1 synergistically transactivate the JCV late gene promoter in U-87MG cells. A reporter plasmid (7.5 μg) containing the JCV late gene promoter was transfected into U-87MG cells alone or together with YB-1 and T-antigen expression plasmids (T-Ag). Expression plasmid DNA concentrations used in the transfections are indicated at the bottom of the panel (in micrograms per plate). The data are represented as CAT activity relative to basal-level expression of the promoter. The results of a representative CAT assay are shown at the top. (B) YB-1 alleviates T-antigen-mediated transcriptional suppression from the JCV early promoter. Experiments similar to those detailed for panel A were also performed with a reporter plasmid containing the JCV early gene promoter. The results are expressed as CAT activity relative to basal-level expression of the promoter. (C and D) Experiments similar to those detailed for panel A were also performed with luciferase plasmids containing early and late wild-type promoters as well as early and late promoters containing point mutations within the 23-bp region (Late mut23 and Early mut23). Reporter activity of each plasmid is expressed as luciferase activity relative to basal-level expression of the wild-type promoter. Results shown in each panel represent the means of data from three independent experiments. Bars indicate standard deviations.
A similar set of cotransfection experiments was also carried out to evaluate the effect of transcriptional cooperation between YB-1 and T-antigen on the viral early promoter. As shown in Fig. 4B, a CAT reporter construct containing the early promoter showed a notable basal expression level in the absence of the transactivators (lane 1). However, as expected, cotransfection of the reporter construct with the T-antigen expression plasmid resulted in a substantial decrease in transcriptional activity, demonstrating the T-antigen-mediated suppression of transcription from the early promoter (compare lanes 1 and 2). In contrast, when a constant amount of T-antigen was cotransfected with increasing amounts of YB-1, we observed a dose-dependent alleviation of T-antigen-mediated suppression of transcription from the early promoter (compare lane 2 to lanes 3 and 4), again indicating the occurrence of a functional interaction between YB-1 and T-antigen. As expected, YB-1 alone significantly activated transcription from the early promoter (lane 5).
Since YB-1 binds to single-stranded early sequences of the 23-bpse and its binding is stimulated by T-antigen, we assessed the role of the 23-bpse in the transcriptional regulation of JCVCY promoters with respect to YB-1 and T-antigen. For this purpose, we created PCR-based point mutations within the 23-bpse of the viral regulatory region and subcloned them into a luciferase reporter plasmid in both the early and late orientations, resulting in early mut23 and late mut23, respectively. As shown in Fig. 4C, compared to the wild-type promoter activity, the late mut23 promoter showed a relatively low level of transcriptional activity even in the presence of transactivators YB-1 and T-antigen (compare the reporter activity of the late promoter with that of the late mut23 promoter). This indicates that the 23-bp region plays a critical role in the regulation of transcriptional activity from the late promoter by YB-1 and T-antigen. A similar set of transfection studies was also performed to assess the effect of point mutations within the 23-bp region on the transcriptional activity of the early promoter. As demonstrated in Fig. 4D, the presence of a mutant 23-bpse deregulated the effect of T-antigen and YB-1 on the early promoter, reaffirming the importance of the 23-bp region in the transcriptional activity of the early promoter by YB-1 and T-antigen.
Stability of YB-1 in the presence of T-antigen.
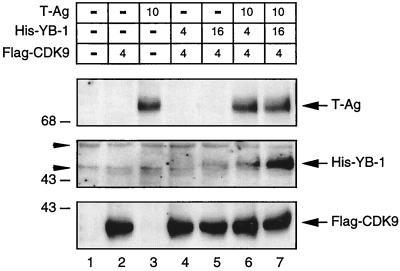
Recent reports have indicated that YB-1 can be induced by a variety of stress-related stimuli, including UV irradiation (28) and anticancer agents (41). It is possible that under stress-related conditions, cellular or viral proteins bind YB-1 and stabilize the protein to prevent its degradation. Since we have demonstrated the occurrence of a physical and functional interaction between T-antigen and YB-1, we further postulated that T-antigen might contribute to YB-1’s stability in cells. To investigate this possibility, we overexpressed YB-1 in U-87MG cells in the presence and absence of T-antigen and analyzed the levels of YB-1 in whole-cell extracts by Western blotting with antibodies specific for T-antigen, His-tagged YB-1, and flag-tagged CDK9 (control). As shown in Fig. 5, in the presence of T-antigen, YB-1 protein was readily detectable (compare lanes 6 and 7 with lanes 4 and 5, respectively). Conversely, we observed a very low level of YB-1 in the absence of T-antigen (lanes 4 and 5), suggesting that the steady-state levels of YB-1 protein were increased in the presence of T-antigen. The observed stability cannot be attributed to the transregulatory effect of T-antigen on the YB-1 expression plasmid (RSV promoter-driven YB-1) because when we coexpressed T-antigen along with an RSV promoter-driven β-Gal reporter plasmid we did not detect any increase in β-Gal activity (data not shown). Further, this stability is not attributable to variations in transfection efficiencies. To account for this variability, we used flag-tagged CDK9 as an internal control in our transfections, and we observed a relatively uniform expression of flag-tagged CDK9 (bottom panel). Hence, our data suggest that the steady-state level of YB-1 is increased in the presence of T-antigen. It is of note that preliminary pulse-chase labeling experiments have also demonstrated that T-antigen stabilizes the YB-1 protein level in U-87MG cells (data not shown).
FIG. 5.
Stability of YB-1 protein is increased in the presence of T-antigen. U-87MG cells were either transfected with CMV–T-antigen alone (lane 3) or cotransfected with pEBV-His-YB-1 and CMV–T-antigen expression plasmids (lanes 6 and 7). Lanes 4 and 5 received only pEBV-His-YB-1. Expression plasmid concentrations used in the transfections are indicated above the panels. The DNA concentrations for the transfection mixtures were normalized by addition of the respective empty expression vector DNA. A flag-tagged CDK9 expression plasmid was also included in the transfection mixture as an internal control for transfection efficiency. Thirty micrograms of whole-cell lysate was resolved by SDS–10% PAGE followed by immunoblotting. The different strips were cut out from the same blot and analyzed for T-antigen (T-Ag), His-tagged YB-1, and flag-tagged CDK9. Whole-cell lysate from untransfected cells (lane 1) was loaded as a negative control. The arrowheads indicate nonspecific bands. The arrows indicate the positions of the respective proteins. The positions of molecular mass markers (in kilodaltons) are shown to the left of the panels.
Localization of T-antigen sequences important for interaction with YB-1.
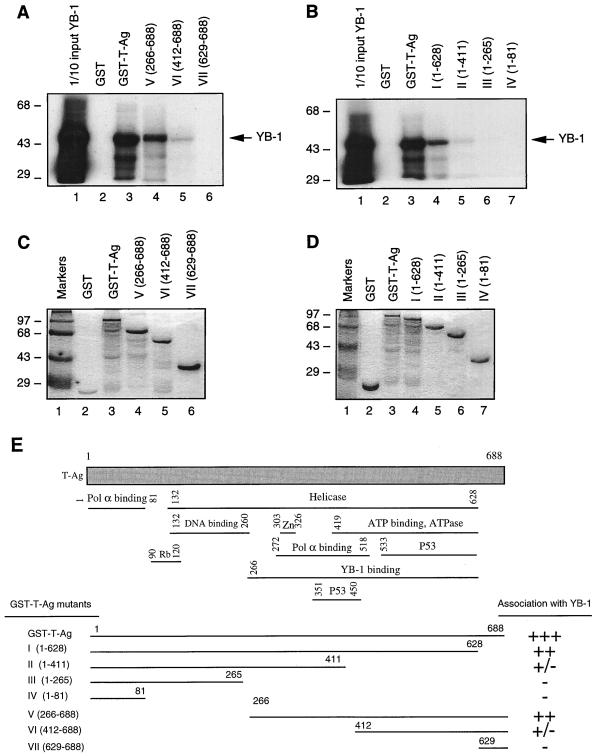
In the next series of studies, we attempted to identify the region(s) of T-antigen necessary for the interaction with YB-1. For this purpose, series of amino-terminal (Fig. 6A) and carboxy-terminal (Fig. 6B) deletion mutants of T-antigen were created as GST fusion proteins and incubated with in vitro-translated 35S-labeled YB-1. The beads were washed extensively, and bound complexes were resolved by SDS–10% PAGE and analyzed by autoradiography. As shown in Fig. 6A, consistent with the data presented above (Fig. 2B), full-length T-antigen fused to GST (GST–T-Ag) efficiently bound to YB-1 (lane 3) but GST alone did not (lane 2). The amino-terminal deletion mutant of T-antigen which retains residues 266 to 688 was able to form a complex with YB-1 (lane 4), although its binding affinity was reduced (compare lanes 3 and 4). The removal of residues 1 to 411 significantly affected the ability of this mutant to interact with YB-1 (lane 5). A further amino-terminal deletion completely abrogated the ability of T-antigen to interact with YB-1 (lane 6). To further define the sequences within T-antigen that are important for its interaction with YB-1, we also created a series of carboxy-terminal deletion mutants of T-antigen and used them in GST pull-down experiments as described for Fig. 6A. As shown in Fig. 6B, a deletion construct of T-antigen lacking the carboxy-terminal 60 amino acids (lane 4) showed a reduced ability to interact with YB-1 compared with full-length T-antigen (compare lanes 3 and 4). A further carboxy-terminal deletion up to amino acid 411 significantly reduced the interaction between these two proteins (lane 5). Furthermore, C-terminal deletion mutants GST–T-antigen (1–265) and GST–T-antigen (1–81) did not interact with YB-1 (lanes 6 and 7). Taken together, these results demonstrate that the minimal region of T-antigen which is important in the interaction with YB-1 resides in the carboxy-terminal half of the protein between amino acids 266 and 628. These results are summarized in Fig. 6E.
FIG. 6.
Localization of the domain of T-antigen that interacts with YB-1. (A and B) GST–T-antigen (GST-T-Ag) and N-terminal (A) and C-terminal (B) T-antigen deletion mutants immobilized on glutathione-Sepharose beads were incubated with in vitro-translated [35S]methionine-labeled YB-1. The Sepharose beads were washed extensively, and bound proteins were resolved by SDS–10% PAGE and analyzed by autoradiography. One-tenth of the input YB-1 used in each reaction was loaded as a migration control (lane 1 in each panel). The arrows indicate the position of in vitro-translated [35S]methionine-labeled YB-1. (C and D) SDS–10% PAGE analysis of GST, GST–T-antigen, and T-antigen N-terminal (C) and C-terminal (D) deletion mutants. (E) Summary of the results obtained from in vitro mapping assays. A schematic representation of T-antigen is shown at the top (not to scale). The abilities of T-antigen and its deletion mutants to interact with YB-1 are shown on the right (+++, specific interaction; ++, reduced interaction; +/−, minimal interaction; and −, no interaction). Pol α binding, polymerase α binding domain. The positions of molecular mass markers (in kilodaltons) are shown to the left of panels A to D.
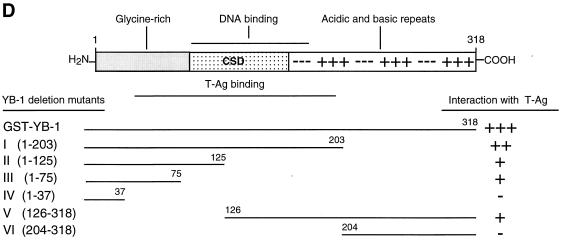
Localization of YB-1 sequences important for interaction with T-antigen.
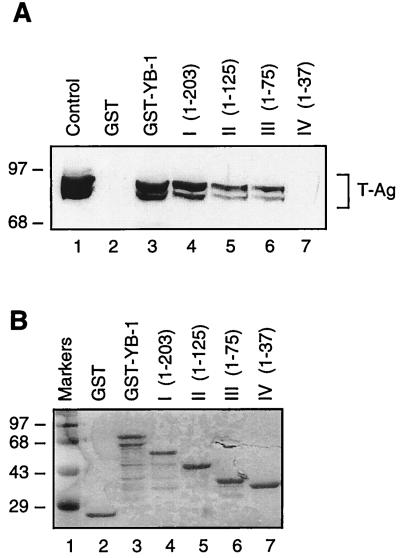
Next, we attempted to map the YB-1 region(s) involved in the interaction with T-antigen. YB-1 C-terminal deletion mutants (Fig. 7A) were created as GST fusion proteins and incubated with whole-cell lysates from HJC-15b cells, which constitutively express T-antigen. As shown in Fig. 7A, substantial removal of alternating acidic and basic clusters in YB-1 had little effect on its interaction with T-antigen (lane 4). Further deletions including two-thirds of the CSD (lane 5) and additional residues from the glycine-rich amino-terminal domain of the protein (lane 6) reduced its affinity for T-antigen by approximately half compared to full-length YB-1 (compare lanes 5 and 6 to lane 3). However, mutant GST–YB-1 (1–37) did not show any detectable association with T-antigen (lane 7).
FIG. 7.
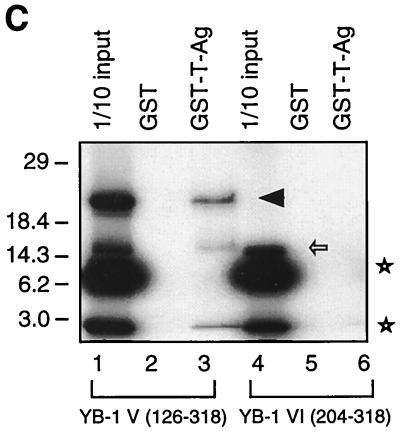
Localization of the domain of YB-1 that interacts with T-antigen. (A) Whole-cell extract from hamster glial cells constitutively expressing T-antigen (HJC-15b) was incubated with either GST alone (lane 2) or C-terminal deletion mutants of GST–YB-1 fusion proteins immobilized on glutathione-Sepharose beads. Bound complexes were washed extensively, resolved by SDS–8% PAGE, and analyzed for T-antigen by Western blotting with an anti-T-antigen antibody (Ab-2 416). HJC-15b whole-cell extract was loaded as a migration control (lane 1). The bracket indicates T-antigen. (B) SDS–10% PAGE analysis of GST and of GST–YB-1 and its C-terminal deletion mutants. (C) Two different 35S-labeled, in vitro-translated N-terminal deletion mutants of YB-1 [YB-1 (126–318) and YB-1 (204–318)] were incubated with GST (lanes 2 and 5) or GST–T-antigen (T-Ag) (lanes 3 and 6). Bound proteins were resolved by SDS-PAGE and analyzed by autoradiography. Lanes 1 and 4 contain 1/10 of the amount used in the pull-down experiments with YB-1 (126–318) and YB-1 (204–318), respectively. An arrowhead and an open arrow designate the positions of the in vitro-translated N-terminal deletion mutants YB-1 (126–318) and YB-1 (204–318), respectively. The asterisks denote nonspecific products of the in vitro transcription-translation reactions. (D) A schematic representation of full-length YB-1 is shown at the top. The abilities of YB-1 and its deletion mutants to interact with T-antigen are depicted on the right (+++, specific interaction; ++ or +, reduced interaction; and −, no interaction). The positions of molecular mass markers (in kilodaltons) are shown to the left of panels A to C.
To further define the amino-terminal boundary of the interaction, in vitro-translated, [35S]methionine-labeled amino-terminal deletion mutants of YB-1, YB-1 V (126–318) and YB-1 VI (204–318), were separately incubated with either bacterially expressed GST or GST–T-antigen in GST pull-down assays. After being washed, bound proteins were resolved by SDS–15% PAGE and analyzed by autoradiography. As shown in Fig. 7B, YB-1 V (126–318) was able to interact with GST–T-antigen (lane 3), indicating that the C-terminal half of the CSD of YB-1, and perhaps residues adjacent to the carboxy terminus of the CSD, contribute to the observed interaction between YB-1 and T-antigen. YB-1 VI (204–318) failed to exhibit a detectable level of binding affinity for T-antigen, suggesting that the alternating basic and acidic clusters near the C terminus of YB-1 do not contribute to the observed interaction. Taken together, the above-described mapping results demonstrate that the minimal region of YB-1 which is important in the interaction with T-antigen lies between amino acids 37 and 204. These results are summarized and shown schematically in Fig. 7D.
Two YB-1 mutants confirm the functional interaction between T-antigen and YB-1.
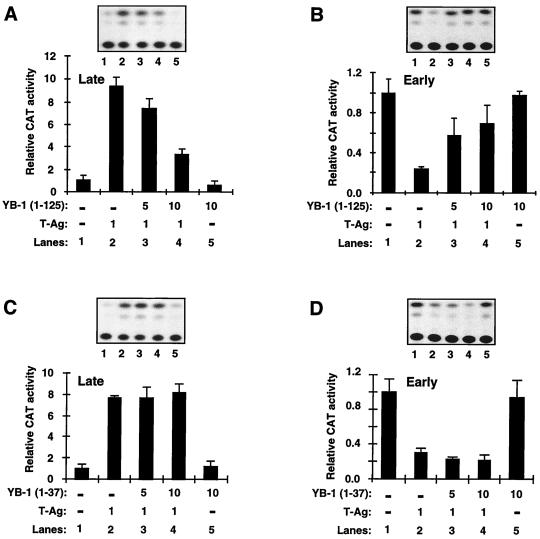
To further assess the interaction between YB-1 and T-antigen, we examined the functional relationship between two YB-1 deletion mutants (46) and T-antigen, using reporter plasmids containing the early and late promoters. One of these mutants, YB-1 (1–125), interacts with T-antigen, whereas the other mutant, YB-1 (1–37), does not (Fig. 7A). Interestingly, YB-1 (1–125), unlike full-length YB-1, did not synergistically activate the late promoter but rather decreased the T-antigen-mediated late-gene transactivation (Fig. 8A; compare lane 2 with lanes 3 and 4). This T-antigen-interacting mutant, however, like full-length YB-1, was able to alleviate T-antigen-mediated suppression of transcription from the early promoter (Fig. 8B; compare lane 2 with lanes 3 and 4). When its transcriptional activity was compared with that of full-length YB-1 (Fig. 4B), this mutant did not show a detectable level of transcription from the early promoter (Fig. 8B, lane 5), suggesting that perhaps the sequences 3′ of the CSD of YB-1 are involved in transcriptional activation. Upon deletion of these sequences, the protein is no longer transcriptionally active, which is consistent with our previous findings (26). We also examined the functional interaction between T-antigen and YB-1 (1–37). As expected, YB-1 (1–37) failed to functionally interact with T-antigen at detectable levels in cotransfection experiments, which is consistent with our findings from in vitro mapping studies (Fig. 7A). As shown in Fig. 8C and D, both T-antigen-mediated transactivation of the late promoter (Fig. 8C) and T-antigen-mediated transcriptional suppression of early-promoter activities were essentially unaltered when YB-1 (1–37) was coexpressed along with T-antigen, demonstrating that YB-1 (1–37) does not functionally interact with T-antigen.
FIG. 8.
Functional interaction of T-antigen with YB-1 deletion mutants. (A and B) Transcriptional activity of deletion mutant YB-1 (1–125), in the presence or absence of T-antigen (T-Ag), on CAT reporter plasmids (7.5 μg) containing the viral late (A) and early (B) promoters. The concentrations of expression plasmids are indicated at the bottom of the respective panels (in micrograms per plate). Representative CAT data are shown above the graphs, and quantitative analysis of results is expressed as CAT activity relative to the basal-level expression of the promoter. (C and D) Transcriptional activity of deletion mutant YB-1 (1–37), in the presence or absence of T-antigen, on CAT reporter plasmids (7.5 μg) containing late (C) and early (D) promoters. The experimental design was virtually identical to that described for panels A and B. The results shown in each panel represent the means of data from three independent experiments. Bars indicate standard deviations.
DISCUSSION
In this study, by means of several independent assays, including coimmunoprecipitation and affinity chromatography experiments, we demonstrated that the Y-box binding protein YB-1 physically associates with the JCV regulatory protein T-antigen. Moreover, we demonstrated that these two proteins interact functionally as well. Coexpression of YB-1 and T-antigen in glial cells synergistically activates transcription from the JCVCY late promoter. In addition, YB-1 abrogates T-antigen-mediated suppression of transcription from the JCVCY early promoter.
Our DNA binding assays using recombinant YB-1 and T-antigen proteins revealed that T-antigen induces binding of YB-1 to its target sequences within the 23E probe (Fig. 1C). Results from these experiments appear to be similar to those found previously for the PRE, in which binding of YB-1 to the PRE early strand was enhanced by T-antigen (9). These data suggest that these two transcription factors may interact with each other in the absence of DNA. It was of interest that the electrophoretic mobility of the YB-1–DNA complex was not altered when T-antigen was included in the binding reaction, suggesting that T-antigen is not part of complexes. Altogether, these results suggest the following possibilities. (i) T-antigen may transiently interact with YB-1, inducing a conformational change in its structure. This may result in an increase in the DNA binding activity of YB-1, and T-antigen is subsequently released from the complex. (ii) T-antigen may function as a stabilizer of the YB-1–DNA complex by forming a ternary complex in solution but may dissociate from this complex under mobility band shift assay conditions and, thus, be undetectable. Nonetheless, the ability of one protein to influence the DNA binding capacity of another is well established. Examples include HMG-I/Y-induced DNA binding of Tst-1 (31) and Phox1-induced DNA binding of the serum response factor (23).
Results from in vitro protein-protein interaction assays have clearly shown a direct association between T-antigen and YB-1 (Fig. 2). It should be noted, however, that this interaction was detected in the absence of their respective DNA binding sites. These results are interesting when compared to our findings from DNA binding assays, in which T-antigen appears to modulate YB-1’s binding to DNA without being involved in a stable ternary complex of DNA, YB-1, and T-antigen. It is of note that T-antigen present in HJC-15b cells exists as several isoforms depending on the phosphorylation state of the protein (18, 44). Interestingly, YB-1 interacts with all of the isoforms of T-antigen (Fig. 2A and 7A). This is similar to the observed interaction between Purα protein and T-antigen, since Purα appears to interact with all T-antigen isoforms (18). Interestingly, the phosphorylation state of T-antigen does not appear to dictate the association between YB-1 and T-antigen, since bacterially expressed T-antigen which does not contain phosphorylated residues is able to interact with YB-1 (Fig. 6A and B). Nonetheless, further experiments will be required to demonstrate the contribution, if any, of the phosphorylation state of T-antigen to the efficiency of formation of the intermolecular complex between YB-1 and T-antigen. In addition, results from in vitro mapping experiments demonstrated that the T-antigen interaction domain of YB-1 is localized to its central DNA binding domain and its immediate flanking sequences. Alternating acidic and basic amino acid clusters of YB-1, localized to the protein’s carboxy terminus, appear to contribute to this interaction but are not essential for it (Fig. 7D). The YB-1 interaction domain of T-antigen maps to the carboxy-terminal half of the protein (Fig. 6E), where it encompasses functionally important domains, including the polymerase α, ATP binding, ATPase, and helicase domains, implying that YB-1 may interfere with those functions by sequestering T-antigen in cells; alternatively, it may cooperate with T-antigen in those functions. Again, further experiments are required to address these questions.
Knowledge of the physical interaction between YB-1 and T-antigen in vitro is complemented by in vivo functional assays. We demonstrated that both transcription factors act in synergy to activate transcription from the JCV late promoter but antagonize each other’s transcriptional activity from the JCV early promoter. Since YB-1 interacts with the 23-bpse early single strand and this interaction is enhanced by T-antigen, we also examined the relevance of this binding in terms of viral gene transcription by creating point mutations within the 23-bp region and performing transient-transfection assays with these mutated constructs. Consistent with our previous findings (46), the 23-bp region appears to play an important role in transcription from the JCV early and late promoters. Point mutations within the 23-bp region significantly reduced the transcriptional activity of JCV promoters compared to the wild-type activity (Fig. 4C and D), suggesting that this region plays a significant role in JCV transcriptional regulation. Additionally, results from transient transfection assays using JCV early promoter reporter constructs suggested that YB-1 may sequester T-antigen in cells and alleviate T-antigen-mediated suppression of transcription from the JCV early promoter. Perhaps the additional accumulation of T-antigen in cells provides an advantage for the virus, facilitating T-antigen-dependent viral DNA replication with subsequent production and assembly of capsid proteins.
The specificity of the functional interaction between YB-1 and T-antigen was tested with specific YB-1 deletion mutants in cotransfection experiments. In contrast to full-length YB-1, YB-1 (1–125), which still interacted with T-antigen in in vitro protein-protein interaction assays, down-regulated T-antigen-mediated activation of transcription from the JCV late promoter. This observation is consistent with previous observations demonstrating that the carboxy terminus of YB-1 contains the transactivation domain of the protein (26). This is further supported by the observation that this mutant does not activate the JCV promoter itself (compare Fig. 4A and 8A) (46). Thus, this mutant, which lacks a transactivation domain but interacts with T-antigen, will be unable to synergistically activate the JCV late promoter. Furthermore, YB-1 (1–125) exerted an antagonistic effect on T-antigen-mediated suppression of transcription from the JCV early promoter, possibly by sequestering T-antigen in cells. Expectedly, YB-1 (1–37), which does not interact with T-antigen in vitro, showed no detectable increase or decrease in transcription from the JCV promoters in the presence of JCV T-antigen.
What is the functional significance of the observed interaction between T-antigen and YB-1 in terms of viral biology? Since T-antigen interacts not only with YB-1 but also with other cellular proteins, including TATA-binding protein (22), it is possible that T-antigen mediates contact between the transcriptional activator YB-1 and the basal transcriptional machinery, potentiating transcriptional cooperativity between T-antigen and YB-1 on the JCV late promoter. In previous studies (46), we demonstrated that YB-1 interacts with another single-stranded cellular DNA binding protein, Purα. These two proteins, YB-1 and Purα, which recognize C/T- and GC/GA-rich sequences, respectively, directly interact with the 23-bpse and can modulate the association of each other with this DNA element. Moreover, ectopic expression of these proteins in cells results in synergistic transactivation through the 23-bpse. In this study, we demonstrated that YB-1 interacts with the viral protein T-antigen and modulates T-antigen-mediated transactivation from the JCV early and late gene promoters. Future studies detailing the interplay between YB-1, Purα, and T-antigen will further elucidate the mechanisms underlying transcriptional control by these proteins.
Recent reports indicate that the members of the Y-box family of proteins are responsive to a wide spectrum of stress-related stimuli, including UV irradiation (8, 28), drug treatment (3, 32), DNA damage-inducing antineoplastic agents (24, 40), and interleukin-2 treatment in T cells (45). Our present and previous (10, 26, 46) data demonstrate that YB-1 is involved in transcriptional regulation of the JCV promoters. Since viral infection certainly causes cellular stress, YB-1 may be a candidate for an inducible protein secondary to stress and inflammation by viruses. Such an assumption leads to the following reasoning with respect to the viral life cycle: it is likely that in the initial phase of infection, YB-1 levels are up-regulated due to viral infection, which in turn activates transcription from the early promoter, leading to T-antigen production. As the lytic infection progresses, T-antigen stabilizes the steady-state levels of YB-1 by physical interaction and prevents YB-1 from rapid degradation. Simultaneously, T-antigen promotes viral DNA replication (33–35) and transcription from the viral late promoter (27, 30). Although T-antigen suppresses its own gene transcription via an autoregulatory loop (17), this suppression appears to be alleviated by YB-1, resulting in further accumulation of T-antigen in infected cells. T-antigen then, by stimulating the binding of YB-1 to the 23-bp region, may potentiate the transcriptional activity of viral late genes and thereby the production of the viral capsid proteins, with cell lysis eventually occurring.
A growing body of experimental evidence suggests that the regulation of gene expression is not mediated solely by the presence or absence of a particular transcription factor. The interactions among transcription factors or interactions between host and viral proteins may be an important determinant in gene regulation. Here we have presented evidence of specific physical and functional interactions between a cellular transcription factor, YB-1, and the JCV regulatory protein T-antigen. YB-1 has also been shown to interact with other cellular and viral transcriptional factors and to modulate transcription of various genes, including the MDR1 gene (2), the chicken α-2 collagen gene (4), the grp78 gene (32), the matrix metalloproteinase 2 gene (38), the major histocompatibility complex class II HLA-DR-α gene (40), the thyrotropin receptor gene (42), the gamma-interferon gene (50), the myosin light-chain 2 gene (54), and the human immunodeficiency virus type 1 long terminal repeat (1). Mechanisms involved in regulation of transcription of these genes may be related to the ability of YB-1 to functionally interact with both basal promoter and enhancer binding proteins, such as Purα, AP2, and NF-κB (37, 43, 46). In addition, interactions between YB-1 and several viral proteins, including human immunodeficiency virus type 1 tat (1) and T-antigen, also modulate gene transcription. The interactions of YB-1 with both cellular and viral transcription factors reflect its diverse functions in gene regulation.
ACKNOWLEDGMENTS
We thank G. MacDonald for providing the full-length YB-1 DNA and A. Rice for providing CDK9 DNA. Additionally, we also thank the past and present members of the Center for NeuroVirology and NeuroOncology for sharing ideas and for insightful discussions. We thank Cynthia Schriver for editorial assistance.
This work was made possible by grants awarded by the National Institutes of Health to K.K.
REFERENCES
- 1.Ansari S A, Safak M, Gallia G L, Sawaya B E, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type and TAR RNA modulates viral promoter activity. J Gen Virol. 1999;80:229–236. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 2.Asakuno K, Kohno K, Uchiumi T, Kubo T, Sato S, Isono M, Kuwano M. Involvement of a DNA binding protein, MDR-NF1/YB-1, in human MDR1 gene expression by actinomycin D. Biochem Biophys Res Commun. 1994;199:1428–1435. doi: 10.1006/bbrc.1994.1390. [DOI] [PubMed] [Google Scholar]
- 3.Bargou R C, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara M Y, Winzer K J, Dietel M, Dorken B, Royer H D. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 4.Bayarsaihan D, Enkhmandakh B, Lukens L N. Y-box proteins interact with the S1 nuclease-sensitive site in the chicken alpha 2(1) collagen gene promoter. Biochem J. 1996;319:203–207. doi: 10.1042/bj3190203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger J R, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 6.Bollag B, Mackeen P C, Frisque R J. Purified JC virus T antigen derived from insect cells preferentially interacts with binding site II of the viral core origin under replication conditions. Virology. 1996;218:81–93. doi: 10.1006/viro.1996.0168. [DOI] [PubMed] [Google Scholar]
- 7.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 8.Boulikas T. DNA lesion-recognizing proteins and the p53 connection. Anticancer Res. 1996;16:225–242. [PubMed] [Google Scholar]
- 9.Chen N N, Chang C F, Gallia G L, Kerr D A, Johnson E M, Krachmarov C P, Barr S M, Frisque R J, Bollag B, Khalili K. Cooperative action of cellular proteins YB-1 and Pur alpha with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc Natl Acad Sci USA. 1995;92:1087–1091. doi: 10.1073/pnas.92.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N N, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur α in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePamphilis M L, Breadley M K. Replication of SV40 and polyomavirus chromosomes. In: Salzman E, editor. The polyomaviruses. 1. The Papovaviridae. New York, N.Y: Plenum Press; 1986. pp. 99–346. [Google Scholar]
- 12.Didier D K, Schiffenbauer J, Woulfe S L, Zacheis M, Schwartz B D. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci USA. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duh J L, Zhu H, Shertzer H G, Nebert D W, Puga A. The Y-box motif mediates redox-dependent transcriptional activation in mouse cells. J Biol Chem. 1995;270:30499–30507. doi: 10.1074/jbc.270.51.30499. [DOI] [PubMed] [Google Scholar]
- 14.Fanning E, Westphal K H, Brauer D, Corlin D. Subclasses of simian virus 40 large T antigen: differential binding of two subclasses of T antigen from productively infected cells to viral and cellular DNA. EMBO J. 1982;1:1023–1028. doi: 10.1002/j.1460-2075.1982.tb01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 16.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisque R J, White F A. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. In: Roos R, editor. Molecular neurovirology: pathogenesis of viral CNS infections. Clifton, N.J: Humana Press; 1992. pp. 25–158. [Google Scholar]
- 18.Gallia G L, Safak M, Khalili K. Interaction of the single-stranded DNA-binding protein Purα with the human polyomavirus JC virus early protein T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein J, Pollitt N S, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Grant C E, Deeley R G. Cloning and characterization of chicken YB-1: regulation of expression in the liver. Mol Cell Biol. 1993;13:4186–4196. doi: 10.1128/mcb.13.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruda M C, Zabolotny J M, Xiao J H, Davidson I, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grueneberg D A, Natesan S, Alexandre C, Gilman M Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 24.Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamota T, Ohga T, Uchiumi T, Kuwano M, Kohno K. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–346. [PubMed] [Google Scholar]
- 25.Kashanchi F, Duvall J F, Dittmer J, Mireskandari A, Reid R L, Gitlin S D, Brady J N. Involvement of transcription factor YB-1 in human T-cell lymphotropic virus type I basal gene expression. J Virol. 1994;68:561–565. doi: 10.1128/jvi.68.1.561-565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr D, Chang C-F, Chen N, Gallia G, Raj G, Schwartz B, Khalili K. Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J Virol. 1994;68:7637–7643. doi: 10.1128/jvi.68.11.7637-7643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili K, Feigenbaum L, Khoury G. Evidence for a shift in 5′-termini of early viral RNA during the lytic cycle of JC virus. Virology. 1987;158:469–472. doi: 10.1016/0042-6822(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 28.Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kuwano M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–394. doi: 10.1016/s0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 29.Kure K, Llena J F, Lyman W D, Soeiro R, Weidenheim K M, Hirano A, Dickson D W. Human immunodeficiency virus-1 infection of the nervous system: an autopsy study of 268 adult, pediatric, and fetal brains. Hum Pathol. 1991;22:700–710. doi: 10.1016/0046-8177(91)90293-x. [DOI] [PubMed] [Google Scholar]
- 30.Lashgari M S, Tada H, Amini S, Khalili K. Regulation of JCVL promoter function: transactivation of JCVL promoter by JCV and SV40 early proteins. Virology. 1989;170:292–295. doi: 10.1016/0042-6822(89)90381-4. [DOI] [PubMed] [Google Scholar]
- 31.Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W W, Hsiung Y, Wong V, Galvin K, Zhou Y, Shi Y, Lee A S. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol Cell Biol. 1997;17:61–68. doi: 10.1128/mcb.17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch K J, Frisque R J. Factors contributing to the restricted DNA replicating activity of JC virus. Virology. 1991;180:306–317. doi: 10.1016/0042-6822(91)90035-a. [DOI] [PubMed] [Google Scholar]
- 34.Lynch K J, Frisque R J. Identification of critical elements within the JC virus DNA replication origin. J Virol. 1990;64:5812–5822. doi: 10.1128/jvi.64.12.5812-5822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch K J, Haggerty S, Frisque R J. DNA replication of chimeric JC virus-simian virus 40 genomes. Virology. 1994;204:819–822. doi: 10.1006/viro.1994.1600. [DOI] [PubMed] [Google Scholar]
- 36.MacDonald G H, Itoh-Lindstrom Y, Ting J P. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J Biol Chem. 1995;270:3527–3533. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- 37.Mertens P R, Alejandra Alfonso-Jaume M, Steinmann K, Lovett D H. A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J Biol Chem. 1998;273:32957–32965. doi: 10.1074/jbc.273.49.32957. [DOI] [PubMed] [Google Scholar]
- 38.Mertens P R, Harendza S, Pollock A S, Lovett D H. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272:22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- 39.Monier R. Transformation by SV40 and polyomaviruses. In: Salzman N P, editor. The Papovaviridae. 1. The polyomaviruses. New York, N.Y: Plenum Press; 1986. pp. 247–294. [Google Scholar]
- 40.Montani V, Taniguchi S I, Shong M, Suzuki K, Ohmori M, Giuliani C, Napolitano G, Saji M, Fiorentino B, Reimold A M, Ting J P, Kohn L D, Singer D S. Major histocompatibility class II HLA-DR alpha gene expression in thyrocytes: counter regulation by the class II transactivator and the thyroid Y box protein. Endocrinology. 1998;139:280–289. doi: 10.1210/endo.139.1.5673. [DOI] [PubMed] [Google Scholar]
- 41.Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, Kuwano M, Kohno K. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- 42.Ohmori M, Shimura H, Shimura Y, Kohn L D. A Y-box protein is a suppressor factor that decreases thyrotropin receptor gene expression. Mol Endocrinol. 1996;10:76–89. doi: 10.1210/mend.10.1.8838147. [DOI] [PubMed] [Google Scholar]
- 43.Raj G V, Safak M, MacDonald G H, Khalili K. Transcriptional regulation of human polyomavirus JC: evidence for a functional interaction between RelA (p65) and the Y-box-binding protein, YB-1. J Virol. 1996;70:5944–5953. doi: 10.1128/jvi.70.9.5944-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj G V, Gordon J, Logan T J, Hall D J, Deluca A, Giordano A, Khalili K. Characterization of glioma cells derived from polyomavirus-induced brain tumors in hamsters. Int J Oncol. 1995;7:801–808. doi: 10.3892/ijo.7.4.801. [DOI] [PubMed] [Google Scholar]
- 45.Sabath D E, Podolin P L, Comber P G, Prystowsky M B. cDNA cloning and characterization of interleukin 2-induced genes in a cloned T helper lymphocyte. J Biol Chem. 1990;265:12671–12678. [PubMed] [Google Scholar]
- 46.Safak M, Gallia G L, Khalili K. Reciprocal interaction between two cellular proteins, Purα and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol Cell Biol. 1999;19:2712–2723. doi: 10.1128/mcb.19.4.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stillman B. Smart machines at the replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 48.Tafuri S R, Wolffe A P. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;268:24255–24261. [PubMed] [Google Scholar]
- 49.Tavis J E, Frisque R J. Altered DNA binding and replication activities of JC virus T-antigen mutants. Virology. 1991;183:239–250. doi: 10.1016/0042-6822(91)90136-y. [DOI] [PubMed] [Google Scholar]
- 50.Ting J P, Painter A, Zeleznik-Le N J, MacDonald G, Moore T M, Brown A, Schwartz B D. YB-1 DNA-binding protein represses interferon gamma activation of class II major histocompatibility complex genes. J Exp Med. 1994;179:1605–1611. doi: 10.1084/jem.179.5.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker D L, Padgett B L, ZuRhein G M, Albert A E, Marsh R F. Human papovavirus (JC): induction of brain tumors in hamsters. Science. 1973;181:674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- 52.Wolffe A P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 53.Wolffe A P, Tafuri S, Ranjan M, Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992;4:290–298. [PubMed] [Google Scholar]
- 54.Zou Y, Chien K R. EFIA/YB-1 is a component of cardiac HF-1A binding activity and positively regulates transcription of the myosin light-chain 2v gene. Mol Cell Biol. 1995;15:2972–2982. doi: 10.1128/mcb.15.6.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]