Abstract
The nucleocapsid protein (NP) (56 kDa) of human influenza A viruses is cleaved in infected cells into a 53-kDa form. Likewise, influenza B virus NP (64 kDa) is cleaved into a 55-kDa protein with a 62-kDa intermediate (O. P. Zhirnov and A. G. Bukrinskaya, Virology 109:174–179, 1981). We show now that an antibody specific for the N terminus of influenza A virus NP reacted with the uncleaved 56-kDa form but not with the truncated NP53 form, indicating the removal of a 3-kDa peptide from the N terminus. Amino acid sequencing revealed the cleavage sites ETD16*G for A/Aichi/68 NP and sites DID7*G and EAD61*V for B/Hong Kong/72 NP. With D at position −1, acidic amino acids at position −3, and aliphatic ones at positions −2 and +1, the NP cleavage sites show a recognition motif typical for caspases, key enzymes of apoptosis. These caspase cleavage sites demonstrated evolutionary stability and were retained in NPs of all human influenza A and B viruses. NP of avian influenza viruses, which is not cleaved in infected cells, contains G instead of D at position 16. Oligopeptide DEVD derivatives, specific caspase inhibitors, were shown to prevent the intracellular cleavage of NP. All three events, the NP cleavage, the increase of caspase activity, and the development of apoptosis, coincide in cells infected with human influenza A and B viruses. The data suggest that intracellular cleavage of NP is exerted by host caspases and is associated with the development of apoptosis at the late stages of infection.
Influenza viruses are enveloped viruses (52) containing segmented negative-strand RNA as their genome (38). RNA segments interact with four viral proteins to form ribonucleoprotein (RNP) segments (7, 16, 19, 48). Depending on the RNA length, each RNP segment contains from 30 to 100 molecules of the major nucleocapsid protein (NP) (19, 45) and several molecules of three high-molecular-mass (∼90 kDa) polymerase proteins: PB1, PB2, and PA (10, 40, 50). The viral RNP structures mediate transcription and replication of the viral genome (13, 25, 30) and participate in the morphogenesis and assembly process of virus particles (37, 54, 60) in infected cells. NP plays significant roles in these events by regulating intracellular transport of viral RNPs (5, 35, 55) and metabolic processes of transcription and replication (6, 9, 28, 53). To exert these functions NP has RNA-binding sites (1, 32), a cytoskeleton-binding domain (5), and a nuclear localization signal (17, 42, 55, 59).
NP was found to be phosphorylated (2, 4, 31, 47) and to be cleaved by proteases (62, 63) in infected cells. The influenza A virus NP (56 kDa) (NP56) is converted proteolytically into a 53-kDa form (NP53), and the influenza B virus NP (64 kDa) is cleaved at two sites into a 62- and a 55-kDa form (NP62 and NP55) (63). Both phosphorylation and proteolytic cleavage of NP are known to be host-dependent events and vary in different cells (31, 35, 62). The regulatory roles of these modifications for NP functions are poorly understood. However, NP cleavage appears to prevent incorporation of viral RNP into virus, since only uncleaved NP56 was found to be assembled into virions (62). Phosphorylation of NP was shown to be necessary for influenza virus replication by a yet-unknown mechanism (4, 31).
There is evidence that the NP gene is a determinant of the host tropism of influenza A viruses. (8, 51, 58). At least two main classes of NPs can be discriminated; each one is typical for either nonhuman or human strains (12, 22, 23). These data suggest that NP determines host tropism by interacting with species-specific host factors. Compatible with this concept is our previous observation that cleavability of NP in infected cells correlated with the host origin of the virus strain. NP of human influenza viruses was shown to be sensitive to host proteases and was cleaved in infected cells, whereas NP of animal influenza viruses was resistant to intracellular proteolytic cleavage and failed to be cleaved (63, 65). The mechanism responsible for these differences in cleavage remained unclear.
In order to find out which host factors may be involved in viral NP cleavage, we examined the primary structure of NP proteolytic sites and characterized host proteases responsible for this cleavage. NPs of human influenza viruses A and B were found to be cleaved at the amino acid sequences EXD/X and DXD/X characteristic for caspase proteases (3), which play a key role in apoptosis (15, 41). With human influenza viruses, cleavage of NP coincided with the activation of host caspases at the late stage of infection and was sensitive to suppression by specific caspase inhibitors. NP of animal influenza viruses failed to contain such proteolytic sites and therefore was resistant to intracellular cleavage. The data imply that NP cleavage of human influenza viruses is accomplished by host caspases and associated with the development of apoptosis in infected cells at the late stage of infection.
MATERIALS AND METHODS
Viruses.
Influenza viruses A/WSN/33 (H1N1), A/Aichi/68 (H3N2), and B/Hong Kong/72 (HK/72) were propagated in embryonated chicken eggs as described previously (64). NP gene sequences of the following viruses were used: A/NT/68 (H3N2), A/New Jersey/8/76 (H1N1) (ANJ/76), A/Hong Kong/156/97 (H5N1) (AHK/97), A/Swine/Hong Kong/76 (H3N2) (ASWHK/76), A/equi/Miami/63 (H3N8) (AEQMI/63), B/Lee/40 (BLE/40), B/Ann Arbor/1/66 (BAA/66), B/Singapore/222/79 (BSN/79), B/Ann Arbor/1/86 (BAA/86), B/Yamagata/16/88 (BYM/88), B/Texas/37/88 (BTX/88), and B/Panama/45/90 (BPM/90).
Cells.
The porcine embryo kidney cell line SPEV and the canine kidney cell line MDCK2 (21) were grown as monolayers in Dulbecco minimal essential medium (DMEM) supplemented with 10% fetal calf serum (GIBCO-BRL, Karlsruhe, Germany). For infection, 2-day-old confluent cell monolayers were incubated with allantoic fluid containing influenza A or B virus (multiplicity of infection = 1 PFU/cell) for 1 h at 37°C. After infection, cells were washed and incubated with DMEM without serum. At 9 (influenza A viruses) and 12 (influenza B virus) hours postinfection (hpi), either one of the caspase inhibitors Z-DEVD-chloromethylketone (CMK) or biotinyl-DEVD-aldehyde (pseudoacid) (Bachem, Heidelberg, Germany) was added to culture medium at a final concentration of 200 μM, and the cells were incubated at 37°C for different periods of time. Cells were then prepared either for protein gel electrophoresis, examination of caspase activity, or DNA fragmentation analysis.
Western blot (WB) analysis.
After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) the polypeptides were transferred from the gel to polyvinylidene difluoride (PVDF) 0.45-μm-pore-size membranes (Millipore, Eschborn, Germany) by semidry electroblotting with a Tris-HCl–ɛ-aminocaproic acid buffer system (pH, ∼9.8) (33). Membranes were washed with 150 mM phosphate-buffered saline (PBS) and incubated overnight at 4°C in 10% dried milk in PBS. After a washing with PBS, membranes were incubated for 2 h at room temperature in PBS containing 0.5% bovine serum albumin, 0.01% Tween 20, and either anti-NP monoclonal antibodies or anti-pNNP (anti-N-terminal peptide 1-34 of NP) guinea pig antibodies. After that, membranes were exposed to horseradish peroxidase (HRP)-conjugated secondary anti-species antibodies (Dako, Glostrup, Denmark), followed by visualization of positive bands with the Pierce (Rockford, Ill.) enhanced chemiluminescence (ECL) procedure by using Kodak BioMax film.
PAGE.
Polypeptides were electrophoresed in 12% polyacrylamide gels containing SDS, as described earlier (66, 67). For protein gel staining with Coomassie Blue R-350, the protocol recommended by Pharmacia (Freiburg, Germany) was applied. An Amersham (Freiburg, Germany) marker polypeptide kit was used containing myosin (200 kDa), phosphorylase b (97.4 kDa), bovine serum albumin (69 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.3 kDa).
Antibodies.
Oligopeptide NH2-MASQGTKRSYEQMETDGERQNATEIRASVGKMIDGC-COOH (pNNP) containing the 34 N-terminal amino acids of influenza A NP (23) was covalently conjugated through a C-terminal cysteine with activated keyhole lympet hemocyanin (KLH) protein (Pierce, Rockford, Ill.). The conjugate (1 mg of pNNP) was mixed with complete Freund adjuvant and injected subcutaneously into guinea pigs. Immunization was repeated two times at 3-week intervals using 0.5 mg of KLH-pNNP conjugate emulsified with incomplete Freund adjuvant. On day 5 after the third immunization guinea pig blood was sampled and serum was prepared by a standard clotting technique. The obtained serum was used in WB experiments.
DNA fragmentation analysis.
MDCK-2 cells (106 cells) infected with influenza viruses were lysed in 0.5 ml of PBS containing 0.4% of the nonionic detergent NP-40 and centrifuged at 12,000 × g for 10 min to remove high-molecular-weight chromatin structures. The supernatant was treated sequentially with RNase T (200 U/ml) and proteinase K (0.5 mg/ml) for 30 min at 37°C and then mixed with 0.6% SDS (final concentration). DNA was extracted once with buffered phenol and once with buffered chloroform and then ethanol precipitated in the presence of 0.3 M NaCl. DNA was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.6), electrophoresed in 1.8% agarose-TBE buffer, and stained with ethidium bromide. Bacteriophage lambda DNA cleaved with PstI was used as molecular weight marker.
Edman microsequencing analysis of truncated NP forms.
Truncated forms of NP of influenza A and B viruses were obtained from viral RNP accumulated in infected cells at 20 to 24 hpi. SPEV cells were used because the cleavage of NP in this cell line was reported to be processed with high efficiency (62). Cells were infected with either A/Aichi/68 or B/HK/72 virus at a multiplicity of 1 PFU per cell and were incubated for 20 and 24 hpi, respectively. Infected cells were disrupted with a Dounce homogenizer and clarifed at 12,000 × g for 20 min. Supernatants were loaded onto 55 to 20% (wt/wt) sucrose gradients in PBS and centrifuged in an SW41 rotor (Beckman, Munich, Germany) at 34,000 rpm for 16 h at 4°C. The sucrose gradients were fractionated and fraction aliquots were analyzed by PAGE. Viral RNP containing fractions were 10-fold diluted with PBS and pelleted at 38,000 rpm for 5 h at 4°C in an SW41 rotor. RNP pellets were then dissolved in 1% SDS containing 5 mM dithiothreitol and electrophoresed in 10% polyacrylamide gel; this was followed by WB transfer onto a 0.45-μm-pore-size PVDF membrane (Millipore). The membrane was stained with 0.1% Coomassie blue R-250, destained with an ethanol (46%)-acetic acid (8%)-water (47%) mixture, and washed with deionized water. Membrane pieces with ANP53, BNP62, and BNP55 bands were excised and processed for N-terminal amino acid analysis by the Edman microsequencing technique on a 477A pulsed liquid-phase sequencer with an online 120A phenylthiohydantoin-derivative analyzer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.) (49). Sequencing reagents were purchased from Perkin-Elmer/Applied Biosystems.
Caspase activity assay.
MDCK infected cells (2 × 106 cells) were dissolved in 0.2 ml of PBS, containing 20 mM Tris-HCl (pH 7.6) and 4 mM dithiothreitol, and then sonicated to prepare cell homogenates. Then, 100-μl aliquotes normalized by protein concentration (∼60 μg per sample) were mixed with 12 μl (40 μg of substrate peptide) of caspase substrate (DEVD-pNA; Bachem, Babendorf, Switzerland) solution. The reaction mixtures were incubated at 37°C for 4 h and centrifuged at 12,000 × g for 5 min. Caspase activity was measured with a Titertak Multiscan type 310C reader (Eflab Oy, Helsinki, Finland) at 405 nm as the intensity of the color of free para-nitroanilide (pNA) appearing in the supernatants.
RESULTS
It was previously shown that intracellular cleavage removed two acidic peptides from NP of human influenza A virus (62). Inspection of the primary structure of NP had shown that these acidic peptides were localized in the N-terminal part of NP, and it was suggested that the N terminus of NP was removed in infected cells (29, 61). To prove this assumption, we tested the presence of the excised peptide in truncated and noncleaved NP molecules by WB analysis with antibodies (anti-pNNP) directed to the N terminus of uncleaved NP56 protein.
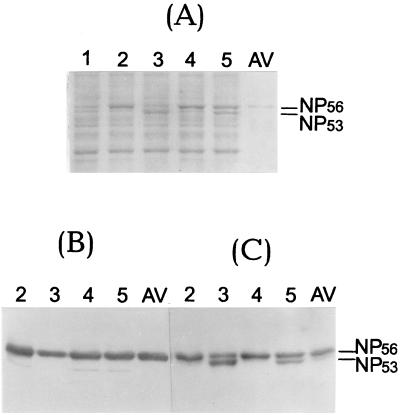
The main question was whether truncated NP53 reacted with these antibodies. The results are shown in Fig. 1. As indicated by a Coomassie blue-stained gel, noncleaved NP prevailed early (9 h) after infection, whereas NP53 was seen predominantly at late (18 h) stages of infection. WB analysis of this gel showed that the anti-pNNP antibody specifically reacted only with noncleaved NP56 and did not recognize truncated NP53 of both virus strains (Fig. 1B). As a control, antibodies specific to the central part of NP (mouse monoclonal antibody clone A1 obtained from the Centers for Disease Control, Atlanta, Ga.) were shown to recognize both NP56 and NP53 (Fig. 1C). Taken together, these observations clearly showed that intracellular cleavage of NP of human influenza A viruses resulted in the loss of the N-terminal 3-kDa peptide.
FIG. 1.
WB analysis of NP with an antiserum specific for the N terminus. MDCK cells were infected with strains A/Aichi/68 (lanes 2 and 3) and A/WSN/33 (lanes 4 and 5) and incubated for 9 h (lanes 2 and 4) and 18 h (lanes 3 and 5). After PAGE, proteins were analyzed either directly by Coomassie brilliant blue staining (A) or by WB with an antiserum specific for the N terminus (anti-pNNP) (B) or an antiserum specific for the central part of NP (anti-NP/A1) (C). Antibody specific bands were developed by the ECL procedure. Lane 1, uninfected MDCK cells; lane AV, purified A/Aichi/68 virus.
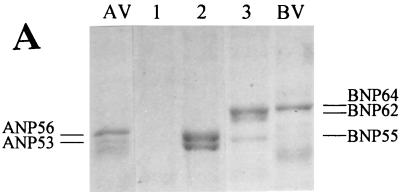
Next we identified the primary structures of the cleavage sites of A/Aichi/68 (H3N2) and B/HK/72 NP. For this, the N-terminal amino acid sequences in the truncated forms ANP53, BNP62, and BNP55 isolated from intracellular viral RNPs (Fig. 2A) were determined. To localize the cleavage sites in the NP molecules, the obtained sequences were then aligned with known NP sequences of influenza A and B viruses. Because there were no NP sequences of A/Aichi/68 and B/HK/72 viruses available, NPs of the related A/NT/68 and B/Lee/40 viruses were used for comparison. The alignment data are shown in Fig. 2B. It can be seen that cleavage took place at Asp16 in ANP56 and Asp7 and Asp61 in BNP64. In each case the cleavage motifs are EXD*X or DXD*X, where X is an uncharged amino acid. Such motifs were found to be typical for cleavage sites recognized by caspases (3), known to be key enzymes in the development of programmed cell death, apoptosis (15, 41).
FIG. 2.
Amino acid alignments of truncated and uncleaved NPs of influenza A and B viruses. Truncated NP forms of A/Aichi/68 (ANP53) and B/HK/72 (BNP62 and BNP55) viruses were purified from viral RNPs accumulated in infected cells at 20 hpi. (A) PAGE analysis of RNP fractions from mock-infected (lane 1), influenza A virus-infected (lane 2), and influenza B virus-infected (lane 3) cells followed by Coomassie brilliant blue staining. Lane AV, purified A/Aichi/68 virus; lane BV, purified B/HK/72 virus. (B) Either six or seven N-terminal amino acids of truncated NP forms were identified by an Edman microsequencing procedure. One-letter abbreviations of amino acids were used. Z, unidentified amino acid. Position 1 of BNP62 can be either S or G. The primary structures of the NP genes of influenza A and B viruses have been published before (11, 14, 18, 23, 24, 36, 49, 56). The EMBL and GenBank accession numbers for the NP genes were as follows: M63754 (ANJ/76), AF028710 (AHK/97), K01395 (BLE/40), M20173 (BAA/66), K01139 (BSN/79), X14217 (BAA/86), L49385 (BYM/88), L49384 (BTX/88), and AF005739 (BPM/90). Identical amino acids (|) and conservative substitutions (:) in NPs of influenza A and B viruses are indicated.
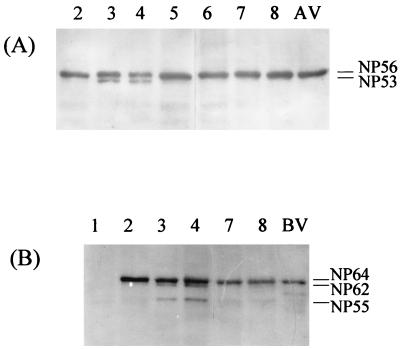
It was now of interest to find out if NP cleavage was indeed accomplished by caspases. To this end, the influence of the specific caspase inhibitors Z-Asp-Glu-Val-Asp-CMK and biotinyl-Asp-Glu-Val-Asp-aldehyde (pseudoacid) (3) on NP cleavage were investigated. The presence of NP56 and NP53 in infected cells incubated with these protease inhibitors was analyzed. As can be seen in Fig. 3, infected cells incubated without inhibitor (lanes 3 to 4) displayed both NP56 and NP53, indicating cleavage. In contrast, after inhibitor treatment (lanes 5 to 8) only uncleaved NP56 was found. It is important to mention that DEVD derivatives containing either aldehyde (pseudoacid) or CMK groups had a similar effect on intracellular NP cleavage. This finding excluded the possibility that NP cleavage was suppressed nonspecifically by DEVD-CMK due to the alkylation effect of the CMK group. Similarly, inhibition of BNP64 cleavage precluding the formation of the BNP62 and BNP55 products was observed in cells infected with influenza B virus after treatment with DEVD-CMK (Fig. 3B). Taken together, these results show that cleavage of influenza virus NP (i) is caused by caspase-type proteases and (ii) is specifically prevented by inhibitors of caspases.
FIG. 3.
Suppression of NP cleavage in influenza virus-infected cells by caspase inhibitors. MDCK cells were infected with either A/Aichi/68 (A) or B/HK/72 (B) viruses and incubated for 9 or 12 h (lanes 2), respectively. Thereafter, the caspase inhibitors DEVD-aldehyde (lanes 5 and 6) and DEVD-CMK (lanes 7 and 8) were added to the culture fluids, and cells were incubated for an additional 3.5 h (lanes 3, 5, and 7) or 7 h (lanes 4, 6, and 8). Cell homogenates were electrophoresed by 12% PAGE, and NP proteins were identified by the WB-ECL technique with monoclonal antibodies against influenza A NP (clone A1) and influenza B NP (clone B017; Serotec, Oxford, England) with secondary anti-mouse HRP conjugates. Lane AV, purified A/Aichi/68 virus; lane BV, purified B/HK/72 virus.
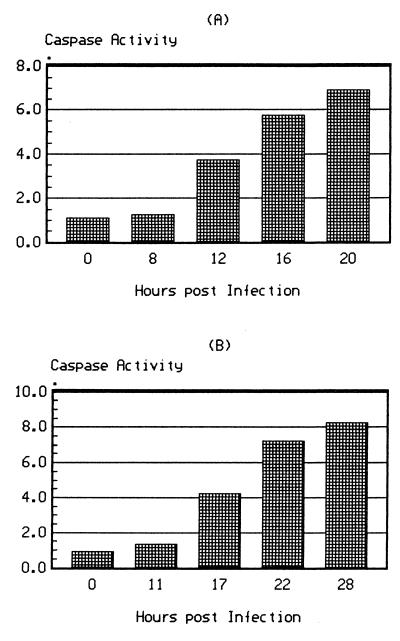
Caspase proteases are known to play a key role in programmed cell death (15, 41). We analyzed, therefore, whether influenza infection induced caspase activity in infected cells, and whether enzyme induction and NP cleavage coincided with the development of apoptosis. Caspase activity in cell homogenates prepared at different times after infection was assayed by in vitro hydrolysis of the specific caspase substrate Asp-Glu-Val-Asp-pNA. As shown in Fig. 4A, caspase activity increased markedly in influenza A virus-infected cells after 12 hpi. In influenza B virus-infected cells the kinetics were similar but developed more slowly (Fig. 4B). The kinetics of caspase increase clearly coincided with the appearance of truncated NP at 12 to 14 hpi in influenza A virus-infected cells and at 17 to 19 hpi in influenza B virus-infected cells.
FIG. 4.
Kinetics of caspase activity in cells infected with influenza A and B viruses. Cells were infected with A/Aichi/68 (A) or B/HK/72 (B) virus and incubated for different times after infection. Caspase activity in cell homogenates was determined by an in vitro hydrolysis test by using DEVD-pNA as a caspase-specific color substrate. The ordinate gives the caspase activity evaluated as the optical density at 405 nm per milligram of cell protein.
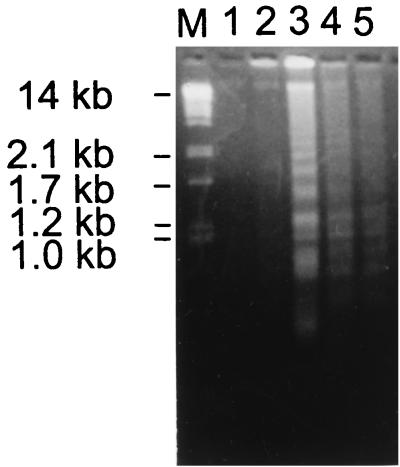
To monitor apoptosis, fragmentation of chromatin DNA has been analyzed in infected MDCK cells (34, 43). Low-molecular-weight host cell DNA was isolated at different times after infection and analyzed by agarose gel electrophoresis. As shown in Fig. 5, the DNA of MDCK cells infected with influenza B virus began to be fragmented into the typical 50- to 0.2-kb ladder by 17 hpi and was clearly evident at 22 and 28 hpi, indicating the high level of apoptosis at this period of infection. No fragmentation was observed in uninfected cells or in infected cells during the initial 11 hpi. In cells infected with influenza A virus, the initial DNA fragmentation ladder was detected at 12 hpi, and marked fragmentation was seen at 16 and 20 hpi (not shown). From these data it follows that apoptotic DNA fragmentation developed by 12 and 17 hpi in cells infected with ca. 1 PFU/cell of influenza A and B viruses, respectively. Interestingly, the temporal appearance of caspase activity, DNA fragmentation, and NP cleavage in infected cells was observed to depend on the multiplicity of virus infection. An increase in the dosage of input virus accelerated the development of apoptosis and NP cleavage (not shown). Taken together, these results suggest that in cells infected with human influenza A and B viruses NP cleavage, increase of caspase activity, and development of apoptosis are tightly linked events.
FIG. 5.
DNA fragmentation analysis of cells infected with influenza virus. The low-molecular-weight DNA fraction was isolated from MDCK cells infected with B/HK/72 virus at 0 (lane 1), 11 (lane 2), 17 (lane 3), 22 (lane 4), and 28 (lane 5) hpi. DNA samples were analyzed by electrophoresis in 1.8% agarose gels, followed by ethidium bromide staining. Lane M, DNA markers obtained from PstI-digested lambda phage DNA.
DISCUSSION
It was previously shown that the NP protein of human influenza A and B viruses is cleaved by host proteases at the late stage of infection (62). Influenza A virus NP (56 kDa) gives rise to one major cleavage product of 53 kDa, whereas influenza B virus NP (64 kDa) yields 62- and 55-kDa proteins, suggesting two cleavage sites (63). The present study was undertaken to identify the cleavage sites and the proteases responsible for cleavage. We found that cleavage occurs in the amino-terminal part of NP at the amino acid sequences EXD*X and DXD*X, where the X’s are hydrophobic amino acids. Such sequences are typical recognition motifs for caspases (3). We also show that caspase inhibitors prevent NP cleavage. These observations indicate that host caspases are responsible for the NP cleavage in infected cells. As shown previously, cleavage was more extensive in chicken fibroblasts and porcine (SPEV) cells than in canine (MDCK) cells and was greatly reduced in a human (HeLa) cell line (62). It therefore appears that the extent of caspase induction by influenza virus infection varies significantly with different cells.
Using pulse-chase experiments with labeled amino acids, we have shown that the cleavage of NP takes place predominantly in the assembled viral RNP (unpublished data) and that cleaved NPs accumulate in RNP (Fig. 2A). In the light of these data, the finding that the N terminus of NP is accessible to proteases in infected cells suggests that it is localized at the surface of the viral RNP and that it is therefore not involved in NP-NP interactions, which have been proposed to promote the formation of the protein backbone in viral RNP structures (50). It has to be pointed out, however, that the N terminus of influenza A virus NP contains phosphorylation (4, 31), RNA-binding (1, 32), and nuclear localization (42, 55, 59) sites that are presumably of great functional importance. It has also been reported that the N terminus of NP may have a role in the specific incorporation of viral RNPs into virus, since only uncleaved NP was found in virions (62). It therefore appears that the N terminus of NP extruding externally may provide interactions with other viral proteins and host cell factors in assembly and migration events in infected cells. Removal of the N-terminal fragment containing these signals by caspase should therefore render nucleocapsids no longer available for virus assembly. On the other hand, released fragments may compete with intact NP in the assembly process. By either mechanism caspase action would interfere with virus replication.
Cleavage of influenza A NP takes place at Asp16, and it has to be pointed out that the respective consensus sequence required for caspase cleavage is a specific trait of human strains. In contrast, NP of nonhuman influenza A viruses has Gly in this position (22, 23, 24). The elimination of the caspase recognition motif resulting from this substitution explains the cleavage resistance of NP of nonhuman strains in infected cells. The concept that caspase susceptibility is linked to the human origin is also supported by influenza B viruses, which all contain the consensus sequences at Asp7 and Asp61 (11, 18, 36, 49). The evolutionary stability of the caspase sensitivity of NP implies that it is a host-specific trait of influenza viruses and that it may play a role in host tropism. On the other hand, it is interesting to see that influenza A/Swine/Hong Kong/6/76 (H3N2) virus that has been transmitted from humans to pigs retained NP cleavability (63, 65) and the caspase cleavage site ETD16*G (23), whereas the NP of A/New Jersey/76 (H1N1) and A/Hong Kong/97 (H5N1) viruses transmitted from animals to humans retained Gly16 (14, 24, 56) and resistance to intracellular cleavage (65). These observations suggest that acquisition or loss of caspase sensitivity results from a longer adaptation process but is not necessary for the initial crossing of the species barrier.
Several laboratories have reported that apoptosis is an important mechanism for the induction of cell death by influenza viruses (20, 26, 27, 39, 46, 57). In compliance with this conclusion is the observation that cells transfected with the proto-oncogene bcl-2, an apoptosis inhibitor, did not undergo DNA fragmentation after virus infection (27, 44). The data presented here indicate that cleavage of NP reflects the induction of caspases which play a central role in apoptosis (15, 41). It remains to be seen, however, whether NP cleavage is merely a marker for the onset of apoptosis in infected cells, or whether it has a specific function in virus-host interaction. It has also to be pointed out that our studies have been performed in cell cultures, and it will be interesting to see whether caspase-dependent cleavage occurs as well in cells involved in the natural infection process, such as epithelial cells of the respiratory tract or cells of the immune system. It is conceivable, for instance, that NP cleavage may contribute to the pathogenetic properties of the virus. Alternatively, apoptosis may exert its protective effects on the host not only by elimination of infected cells, but also by NP cleavage-mediated downregulation of virus replication.
ACKNOWLEDGMENTS
We thank Ralph Wagner, Thorsten Wolff, and Anke Feldmann for helpful discussions and Astrid Herwig and Svetlana Zaitseva for excellent technical assistance.
This work was supported by The Howard Hughes Medical Institute (grant 75195-546302), the Deutsche Forschungsgemeinschaft (SFB 286), and The Russian Foundation of Basic Research (grants 98/04-48901 and 96/04-00124). O.P.Z. is very grateful to I. O. and A. O. Zhirnov and to K. O. Zhirnova for helpful assistance with the manuscript.
REFERENCES
- 1.Albo C A, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond J W, Felsenreich V. Phosphorylation of the nucleoprotein of an avian influenza virus. J Gen Virol. 1982;60:295–305. doi: 10.1099/0022-1317-60-2-295. [DOI] [PubMed] [Google Scholar]
- 3.Alnemeri E S, Livingston D J, Nicholson D W, Salsesen G, Thomberry N A, Wong W W, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171–172. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Arrese M, Portela A. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza A/Victoria/3/75. J Virol. 1996;70:3385–3391. doi: 10.1128/jvi.70.6.3385-3391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avalos R T, Yu Z, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcena J, Ochoa M, de la Luna S, Melero J A, Nieto A, Ortin J, Portela A. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin F, Bach C, Cusack S, Ruigrok R W H. Structure of influenza RNP. 1. Influenza virus ribonucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bean W J. Correlation of influenza A virus nucleoprotein genes with host species. Virology. 1984;133:438–442. doi: 10.1016/0042-6822(84)90410-0. [DOI] [PubMed] [Google Scholar]
- 9.Beaton A R, Krug R M. Transcription antitermination during influenza virus template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braam J, Ulmanen I, Krug R M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983;34:609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- 11.Briedis D J, Tobin M B. Influenza B virus genome: complete nucleotide sequence of the influenza B/Lee/40 virus genome RNA segment 5 encoding the nucleoprotein and comparison with the B/Singapore/222/79 nucleoprotein. Virology. 1984;133:448–455. doi: 10.1016/0042-6822(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 12.Buckler-White A J, Murphy B R. Nucleotide sequence analysis of the nucleoprotein gene of an avian and a human influenza virus strain identifies two classes of nucleoproteins. Virology. 1986;155:345–355. doi: 10.1016/0042-6822(86)90198-4. [DOI] [PubMed] [Google Scholar]
- 13.Bukrinskaya A G, Vorkunova G K, Vorkunova N K. Cytoplasmic and nuclear input virus RNPs in influenza virus-infected cells. J Gen Virol. 1979;45:557–567. doi: 10.1099/0022-1317-45-3-557. [DOI] [PubMed] [Google Scholar]
- 14.Claas E C J, Osterhaus A D M E, vab Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A/H5N1 virus related to a highly pathogenic avian virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 15.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;15:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compans R W, Content J, Duesberg P H. Structure of ribonucleoprotein of influenza virus. J Virol. 1972;10:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey J, Dimmock N J, Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985;40:667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- 18.DeBorde D C, Donabedian A M, Herlocher M L, Naeve C W, Maassab H F. Sequence comparison of wild-type and cold-adapted B/Ann Arbor/1/66 influenza virus genes. Virology. 1988;163:429–443. doi: 10.1016/0042-6822(88)90284-x. [DOI] [PubMed] [Google Scholar]
- 19.Duesberg P. Distinct subunits of the ribonucleoprotein of influenza virus. J Mol Biol. 1969;42:485–499. doi: 10.1016/0022-2836(69)90237-x. [DOI] [PubMed] [Google Scholar]
- 20.Fesq H, Bacher M, Nain M, Gemsa D. Programmed cell death (apoptosis) in human monocytes infected with influenza A virus. Immunobiology. 1994;190:175–182. doi: 10.1016/S0171-2985(11)80292-5. [DOI] [PubMed] [Google Scholar]
- 21.Fuller S, von Bonsdorf C H, Simons K. Vesicular stomatitis virus infects and matures only through the basolateral surface of the polarized epithelial cell line, MDCK. Cell. 1984;38:65–77. doi: 10.1016/0092-8674(84)90527-0. [DOI] [PubMed] [Google Scholar]
- 22.Gammelin M, Mandler J, Scholtissek C. Two subtypes of nucleoproteins (NP) of influenza A viruses. Virology. 1989;170:71–80. doi: 10.1016/0042-6822(89)90353-x. [DOI] [PubMed] [Google Scholar]
- 23.Gorman O T, Bean W J, Kawaoka Y, Webster R G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990;64:1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorman O T, Bean W J, Kawaoka Y, Donatelli I, Guo Y J, Webster R G. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jennings P A, Finch J T, Winter G, Robertson J S. Does the higher structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1987;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 26.Hechtfischer A, Marschall M, Helten A, Boswald C, Meier-Ewert H. A highly cytopathogenic influenza C variant induces apoptosis in cell culture. J Gen Virol. 1997;78:1327–1330. doi: 10.1099/0022-1317-78-6-1327. [DOI] [PubMed] [Google Scholar]
- 27.Hinshaw V G, Olsen C W, Dybdahi-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 29.Huddleston J A, Brownlee G G. The sequence of the nucleoprotein gene of human influenza A virus strain, A/NT/60/78. Nucleic Acids Res. 1982;10:1029–1037. doi: 10.1093/nar/10.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson J B, Flawith J, Dimmock N J. Early events in influenza virus infection. III. The formation of a nucleoplasmic ribonucleoprotein complex from the input virion. Virology. 1978;86:167–176. doi: 10.1016/0042-6822(78)90017-x. [DOI] [PubMed] [Google Scholar]
- 31.Kistner O, Muller K, Scholtissek C. Differential phosphorylation of the nucleoprotein of influenza A viruses. J Gen Virol. 1989;70:2421–2431. doi: 10.1099/0022-1317-70-9-2421. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi M, Toyoda T, Adyshev D M, Azuma Y, Ashihama A. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol. 1994;68:8433–8436. doi: 10.1128/jvi.68.12.8433-8436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyhse-Anderson J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 34.Lagarkova M A, Iarovaia O V, Razin S V. Large-scale fragmentation of mammalian DNA in the course of apoptosis proceeds via excision of chromosomal DNA loops and their oligomers. J Biol Chem. 1995;270:20239–20341. doi: 10.1074/jbc.270.35.20239. [DOI] [PubMed] [Google Scholar]
- 35.Leavitt J. Tumorigenic potential of human fibroblasts as a function of ability to express a novel form of influenza A nucleocapsid protein. Carcinogenesis. 1983;4:1229–1237. doi: 10.1093/carcin/4.10.1229. [DOI] [PubMed] [Google Scholar]
- 36.Londo D R, Davis A R, Nayak D P. Complete nucleotide sequence of the nucleoprotein gene of influenza B virus J. Virol. 1983;47:642–648. doi: 10.1128/jvi.47.3.642-648.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCauley J, Mahy B W J. Structure and function of influenza virus genome. Biochem J. 1985;211:281–294. doi: 10.1042/bj2110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori I, Komatsu T, Takeuchi K, Nakakuki K, Sudo M, Kimura Y. In vivo induction of apoptosis by influenza virus. J Gen Virol. 1995;76:2869–2873. doi: 10.1099/0022-1317-76-11-2869. [DOI] [PubMed] [Google Scholar]
- 40.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases on influenza viral ribonucleoproteins by immunogold labelling. Virology. 1988;164:562–565. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 41.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 42.Neumann G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oberhammer F, Wilson J W, Dive C, Morris I D, Hickman J A, Wakeling A E, Walker P R, Sikorska M. Apoptotic death in epithelials cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;9:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen C W, Kehren J C, Dybdahl-Sissoko R, Hinshaw V S. Bcl-z alters influenza virus yield, spread, and hemagglutinin glycosylation. J Virol. 1996;70:663–666. doi: 10.1128/jvi.70.1.663-666.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pons M W, Schulze I T, Hirst G K. Isolation and characterisation of the ribonucleoprotein of influenza virus. Virology. 1969;39:250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- 46.Price G E, Smeeth H, Sweet C. Differential induction of cytotoxicity and apoptosis by influenza virus strains of different virulence. J Gen Virol. 1997;78:2821–2829. doi: 10.1099/0022-1317-78-11-2821. [DOI] [PubMed] [Google Scholar]
- 47.Privalsky M L, Penhoet E E. Phosphorylated protein component present in influenza virions. J Virol. 1975;24:401–405. doi: 10.1128/jvi.24.1.401-405.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rees P J, Dimmock N J. Electrophoretic separation of influenza virus ribonucleoproteins. J Gen Virol. 1991;53:125–130. doi: 10.1099/0022-1317-53-1-125. [DOI] [PubMed] [Google Scholar]
- 49.Rickers A, Brockstedt E, Mapara M Y, Otto A, Dorken B, Bommert K. Inhibition of CPP32 blocks surface IgM-mediated apoptosis and D4-GDI cleavage in human BL60 Burkitt lymphoma cells. Eur J Immunol. 1998;28:296–304. doi: 10.1002/(SICI)1521-4141(199801)28:01<296::AID-IMMU296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 50.Ruigrok R W, Baudin F. Structure of influenza virus ribonucleoprotein particles. 2. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishible from the intact influenza virus ribonucleoprotein particles. J Gen Virol. 1995;76:1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
- 51.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 52.Schulze I T. The structure of influenza virus. 2. A model based on the morphology and composition of subviral particles. Virology. 1972;47:181–196. doi: 10.1016/0042-6822(72)90251-6. [DOI] [PubMed] [Google Scholar]
- 53.Shapiro G L, Krug R M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sklyanskaya E I, Varich N L, Amvrosieva T V, Kaverin N V. Virions and intracellular nucleocapsids produced during mixed heterotypic influenza infection of MDCK cells. J Virol. 1985;53:679–683. doi: 10.1128/jvi.53.2.679-683.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens M P, Barclay W S. The N-terminal extention of the influenza B virus nucleoprotein is not required for nuclear accumulation or the expression and replication of a model RNA. J Virol. 1998;72:5307–5312. doi: 10.1128/jvi.72.6.5307-5312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 57.Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- 58.Tian S-F, Buckler-White A J, London W T, Reck L J, Chanock R M, Murphy B R. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/NY/78 virus and its reassortants in squirrel monkey respiratory tract. J Virol. 1985;53:771–775. doi: 10.1128/jvi.53.3.771-775.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P, Palese P, O’Neil R. The NPI-1/NPI-3 (karyopherin-alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter G, Fields S. The structure of the gene encoding the nucleoprotein of human influenza virus A/PR/8/34. Virology. 1981;114:423–428. doi: 10.1016/0042-6822(81)90223-3. [DOI] [PubMed] [Google Scholar]
- 62.Zhirnov O P, Bukrinskaya A G. Two forms of influenza virus nucleoprotein in infected cells and virions. Virology. 1981;109:174–179. doi: 10.1016/0042-6822(81)90482-7. [DOI] [PubMed] [Google Scholar]
- 63.Zhirnov O P, Bukrinskaya A G. Nucleoproteins of animal influenza viruses, in contrast to those of human strains, are not cleaved in infected cells. J Gen Virol. 1984;65:1127–1134. doi: 10.1099/0022-1317-65-6-1127. [DOI] [PubMed] [Google Scholar]
- 64.Zhirnov O P, Ovcharenko A V, Bukrinskaya A G. Myxovirus replication in chicken embryos can be suppressed by aprotinin due to the blockage of viral glycoprotein cleavage. J Gen Virol. 1985;66:1633–1638. doi: 10.1099/0022-1317-66-7-1633. [DOI] [PubMed] [Google Scholar]
- 65.Zhirnov O P. The host origin on influenza viruses can be assessed by the intracellular cleavage of the viral nucleocapsid protein. Arch Virol. 1988;99:277–284. doi: 10.1007/BF01311077. [DOI] [PubMed] [Google Scholar]
- 66.Zhirnov O P. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 67.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]