Graphical Abstract
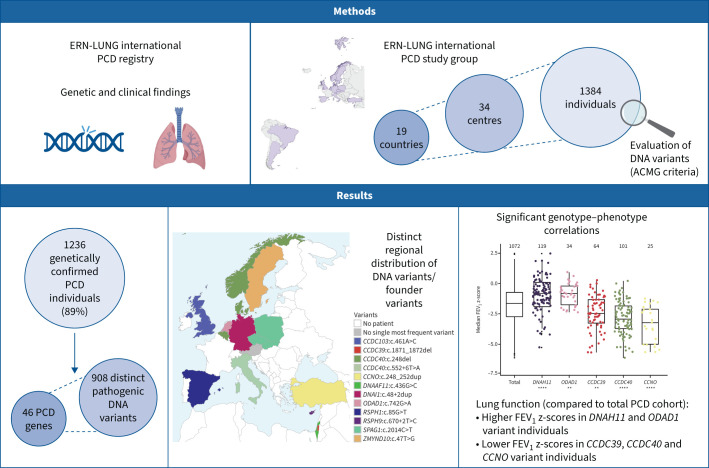
Outline of the study. ERN: European Reference Network; PCD: primary ciliary dyskinesia; ACMG: American College of Medical Genetics and Genomics; FEV1: forced expiratory volume in 1 s. **: p≤0.01; ****: p≤0.0001.
Abstract
Background
Primary ciliary dyskinesia (PCD) represents a group of rare hereditary disorders characterised by deficient ciliary airway clearance that can be associated with laterality defects. We aimed to describe the underlying gene defects, geographical differences in genotypes and their relationship to diagnostic findings and clinical phenotypes.
Methods
Genetic variants and clinical findings (age, sex, body mass index, laterality defects, forced expiratory volume in 1 s (FEV1)) were collected from 19 countries using the European Reference Network's ERN-LUNG international PCD Registry. Genetic data were evaluated according to American College of Medical Genetics and Genomics guidelines. We assessed regional distribution of implicated genes and genetic variants as well as genotype correlations with laterality defects and FEV1.
Results
The study included 1236 individuals carrying 908 distinct pathogenic DNA variants in 46 PCD genes. We found considerable variation in the distribution of PCD genotypes across countries due to the presence of distinct founder variants. The prevalence of PCD genotypes associated with pathognomonic ultrastructural defects (mean 72%, range 47–100%) and laterality defects (mean 42%, range 28–69%) varied widely among countries. The prevalence of laterality defects was significantly lower in PCD individuals without pathognomonic ciliary ultrastructure defects (18%). The PCD cohort had a reduced median FEV1 z-score (−1.66). Median FEV1 z-scores were significantly lower in CCNO (−3.26), CCDC39 (−2.49) and CCDC40 (−2.96) variant groups, while the FEV1 z-score reductions were significantly milder in DNAH11 (−0.83) and ODAD1 (−0.85) variant groups compared to the whole PCD cohort.
Conclusion
This unprecedented multinational dataset of DNA variants and information on their distribution across countries facilitates interpretation of the genetic epidemiology of PCD and indicates that the genetic variant can predict diagnostic and phenotypic features such as the course of lung function.
Shareable abstract
The distribution of affected PCD genes and pathogenic gene variants differs markedly within Europe and beyond due to several founder variants. The PCD genotype can predict diagnostic and phenotypic features such as the course of lung function. https://bit.ly/44AbHTY
Introduction
Primary ciliary dyskinesia (PCD) (Mendelian Inheritance in Man (MIM) 244400) represents a group of rare genetic disorders characterised by impaired function, structure or generation of multiple motile cilia on epithelial cells lining the airways. Impaired mucociliary clearance leads to chronic mucopurulent airway disease that progresses to irreversible lung damage. Dysfunctional motile cilia present in other tissues can result in non-respiratory disease manifestations such as infertility, laterality defects or, less commonly, hydrocephalus [1]. PCD demonstrates a considerable phenotypic and genetic heterogeneity that often hampers diagnosis. The estimated prevalence ranges from one in 4000 to one in 20 000 and, so far, more than 50 genes have been described to be involved in PCD [1–6]. Notably, data on the regional prevalence of PCD genotypes across Europe are limited to a handful of country-specific studies [4, 6–10]. Therefore, we analysed the regional prevalence of PCD genotypes across countries using data compiled by the European Reference Networks’ ERN-LUNG network (https://ern-lung.eu). The ERN-LUNG international PCD Registry systematically collects data from PCD individuals such as diagnostic results, natural history, incidence, clinical presentation, treatment and course of disease [11–13]. We assembled data for 19 different countries across Europe, Asia and South America to explore the global impact of genotypes on clinical aspects of the disease, including lung function. In this largest multinational cohort to date of genetically diagnosed PCD individuals, we revealed marked regional differences of PCD genotypes and identified substantial genotype–phenotype correlations.
Methods
Summary of applied methods
Please see the supplementary material for a detailed method section.
The study used data from previously genotyped individuals of the ERN-LUNG international PCD Registry. Genetic variants were evaluated according to American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) guidelines [14]. Only pathogenic variants were included for further analyses. Several clinical parameters, such as age, sex, body mass index (BMI), laterality status and forced expiratory volume in 1 s (FEV1), were evaluated. Groups were categorised according to genotypes associated with different ciliary ultrastructural phenotypes. Statistical analysis was performed using R (www.r-project.org), with adjustments for multiple comparisons.
Results
Study population
In this study, 34 centres from 19 countries participated: 15 from Europe, two from Asia (Israel, Palestine) and two from South America (Brazil, Argentina). The number of included PCD individuals differed between centres, ranging from three to 190 (median 25, interquartile range (IQR) 10–38.5), and between countries, ranging from three to 321 (median 32, IQR 17.5–92). Following independent evaluation at the coordinating centre, 148 among 1384 individuals submitted to the study (11%, 0–36% per country) did not have a confirmed genetic diagnosis according to ACMG/AMP guidelines [14]. The most frequent reasons for genetically unconfirmed diagnoses were 1) a single variant in a PCD gene without a second variant identified (in cases of autosomal-recessive inheritance); 2) two heterozygous variants in two different PCD genes (in cases of autosomal-recessive inheritance); 3) genetic variants in candidate but not in known PCD genes; 4) bi-allelic yet not reported genetic variants of unknown significance (class 3) without a consistent clinical phenotype or further confirmatory diagnostic findings such as transmission electron microscopy (TEM), immunofluorescence microscopy analyses or high-speed videomicroscopy; or 5) benign/likely benign (class 1/class 2) variants in known PCD genes. The remaining 1236 individuals with confirmed genetic diagnoses were included in further analyses. The median age of the study population was 21.6 years (IQR 15.4–32.2 years, as of January 2023), 428 individuals (35%) were <18 years old and 808 (65%) were >18 years old. Data on age at diagnosis were available for 947 individuals, showing a median of 10 years for age at diagnosis (IQR 4.4−17 years, range 0–77.7 years). The median age at diagnosis for participants with laterality defects was 8 years (IQR 1.08–16.3 years) compared to 11 years for participants without laterality defects (IQR 6–17.9 years) (p<0.0001).
A total of 615 individuals were male (50%) and 621 were female (50%) (supplementary figure E1). The median BMI of the cohort was 20.3 kg·m−2 (IQR 17.4–23.8 kg·m−2) with a median BMI z-score for individuals <19 years old of 0.00 (IQR −0.7–0.07, n=652). The median BMI for individuals >19 years old was 22.7 kg·m−2 (IQR 20.4–25.6 kg·m−2, n=441). There were no significant differences in median BMI and age between the gene groups (supplementary table E1).
Genotypes in PCD individuals
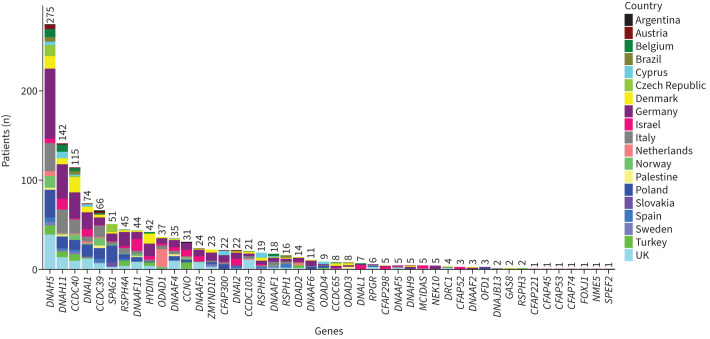
Overall, 908 distinct disease-causing variants in 46 PCD-associated genes were detected in the group of 1236 PCD individuals (supplementary table E2), of whom 687 (56%) had homozygous and 528 (43%) had compound heterozygous DNA variants. Only 20 individuals (2%) had hemizygous, X-linked variants (OFD1, DNAAF6 and RPGR), while one individual carried an autosomal dominant variant (FOXJ1). The majority of allele frequencies (99.7%) for the genetic variants of the study were <0.005, according to the Genome Aggregation Database (gnomAD) for European (non-Finnish) ancestry (supplementary table E2). The most frequently affected genes, in individuals with bi-allelic pathogenic variants, were DNAH5 (n=275, 22%), DNAH11 (n=142, 11%), CCDC40 (n=115, 9%), DNAI1 (n=74, 6%), CCDC39 (n=66, 5%) and SPAG1 (n=51, 4%) (figure 1).
FIGURE 1.
The regional distribution and number of individuals with confirmed pathogenic variants in primary ciliary dyskinesia-associated genes among 34 centres from 19 different countries.
Regional distribution of the mutated PCD genes and pathogenic variants
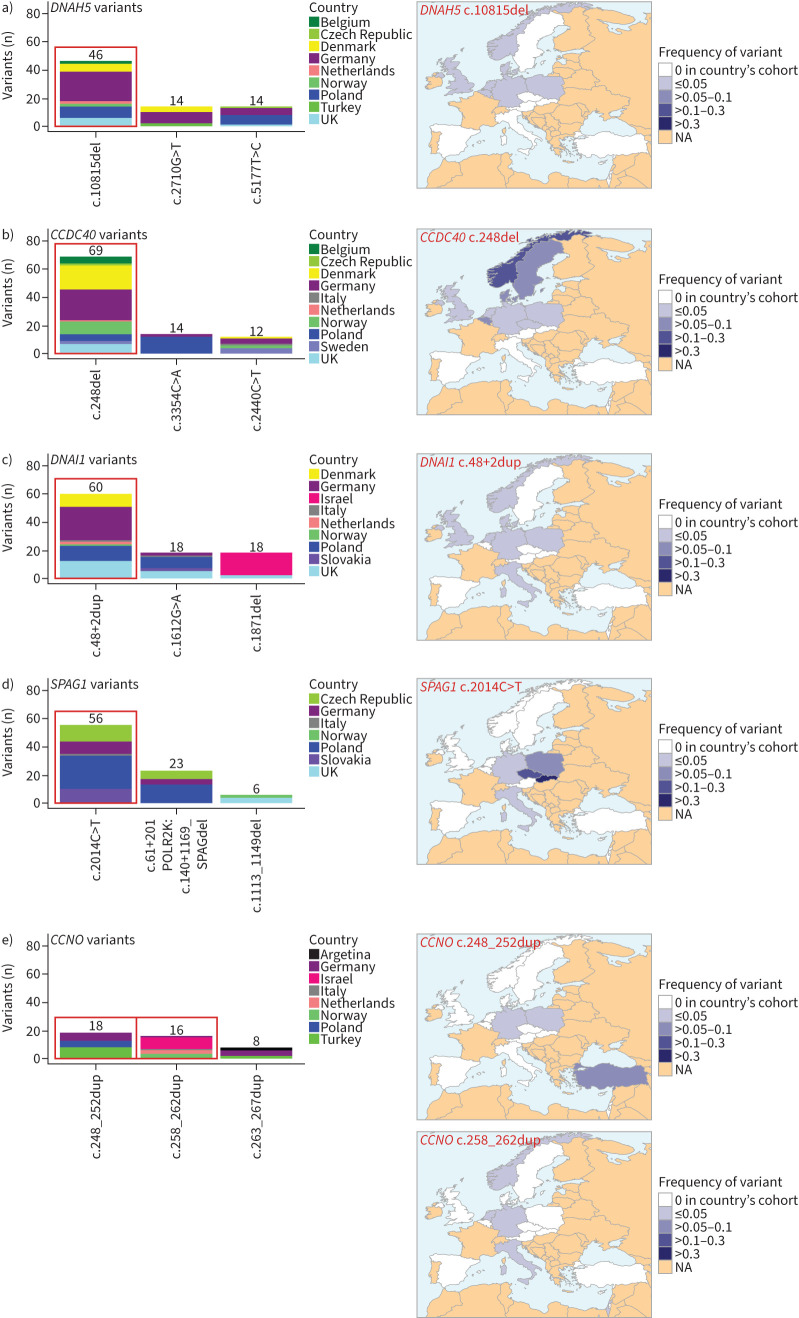
The spectrum of mutated PCD-associated genes differed markedly across the 19 countries. DNAH5 was overall the most frequently affected gene, both in the whole study cohort (figure 1) and in 12 of 19 participating countries (63%). The most common DNAH5 variant, c.10815del, was primarily observed in Northern and Central Europe (×46; figure 2a). The regional distribution of the most frequent pathogenic variants in other selected PCD genes (CCDC40, DNAI1, SPAG1, CCNO) is illustrated in figure 2b–e. The CCDC40 variant c.248del (×69) was frequently observed in the northern and central parts of Europe. The DNAI1 variant c.48+2dup (×60) was prevalent in Northern and Central Europe. Interestingly, the SPAG1 variant c.2014C>T (×56) showed a high frequency in the Slavic region including Poland, Czech Republic and Slovakia. The CCNO variant c.248_252dup (×18) was mainly present in Turkey, whereas the CCNO variant c.258_262dup (×16) was mainly present in Israel. Analysis of the most frequent variants per country provided another perspective for PCD-associated genetic diversity. Interestingly, in spite of the overall high involvement of DNAH5, none of its variants was reported as the most frequent in any of the analysed countries. The most frequent variants per country were found in 11 different PCD genes (figure 3). More data on the distribution of frequent variants in PCD-associated genes per country can be found in supplementary figure E2.
FIGURE 2.
The most frequent pathogenic genetic variants in selected primary ciliary dyskinesia genes and their regional distribution. The most common variants in DNAH5, CCDC40, DNAI1, SPAG1 and CCNO show regional clusters. a) The most common DNAH5 variant c.10815del (×46) is prevalent in northern Europe. b) The most common CCDC40 variant c.248del (×69) is also frequently reported in the northern parts of Europe. c) The most common DNAI1 variant c.48+2dup (×60) predominantly occurs in northern Europe and neighbouring countries. d) The most common SPAG1 variant c.2014C>T (×56) shows a dominant regional distribution in the Slavic countries Poland, Czech Republic and Slovakia. e) In CCNO, there are two frequent genetic variants: c.248_252dup (×18) mainly occurs in Turkey, whereas c.258_262dup (×16) is mainly reported in Israel. NA: not available.
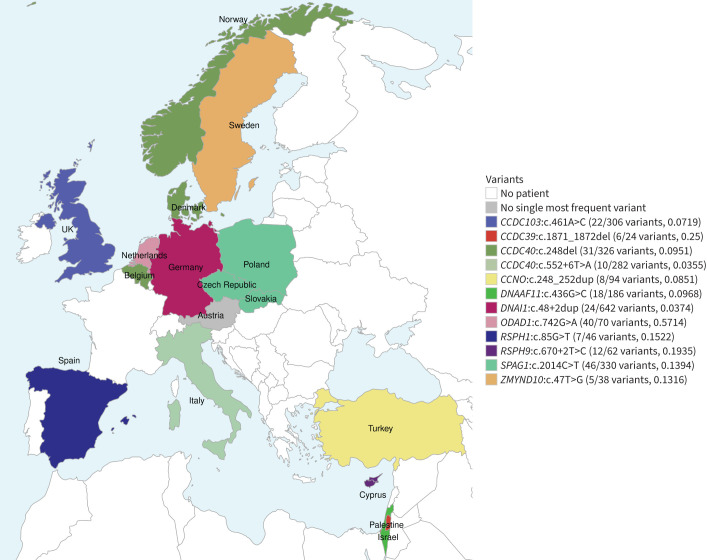
FIGURE 3.
The most frequent pathogenic gene variants associated with primary ciliary dyskinesia per country. There are clear regional differences between countries. The total absolute and relative frequency of the most frequent variant per country is shown in brackets. The variant c.2014C>T in SPAG1 (mint green) is the most frequently reported variant in Poland (24 out of 268 variants, 0.0896), the Czech Republic (12 out of 46 variants, 0.2609) and Slovakia (10 out of 16 variants, 0.625). The variant c.742G>A in ODAD1 (pale pink) prevails in the Netherlands. The variant c.248_252dup in CCNO (yellow) is the most frequently detected variant in Turkey and c.248del in CCDC40 (dark green) is the most frequently detected variant in Denmark (17 out of 182 variants, 0.0934), Norway (9 out of 80 variants, 0.1125) and Belgium (5 out of 64 variants, 0.0781). Despite the overall high involvement of DNAH5 (figure 1), none of its variants was identified as the most frequent in any of the countries. Argentina and Brazil are not shown (no most frequent genetic variant).
Genotype–phenotype correlations
Distribution of predicted ultrastructural ciliary phenotypes
Based on the genotypes, we assessed the proportion of patients who could have been successfully diagnosed by TEM [5, 15]. In total, 894 individuals (72%) had DNA variants associated with pathognomonic ciliary ultrastructure defects detectable by TEM (class I defects). The remaining 342 individuals (28%) had DNA variants not associated with hallmark pathognomonic ciliary ultrastructure defects. The proportion of PCD individuals with genetic variants associated with hallmark pathognomonic ciliary ultrastructure defects differed significantly among countries, ranging from 47% to 100% (supplementary figure E3).
Laterality defects
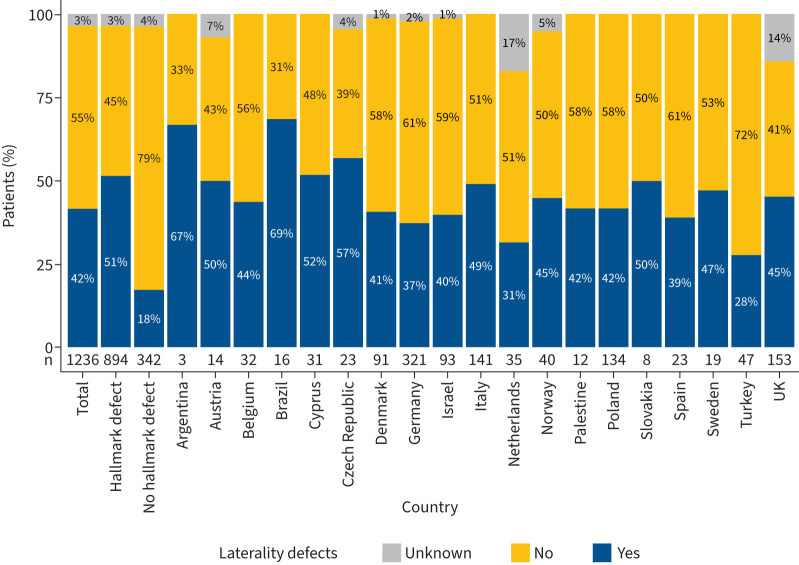
Information on the laterality status was available for 1195 of 1236 individuals (97%). 676 PCD individuals (55%) had normal body composition (situs solitus). 519 individuals were reported to have laterality defects (42%) (figure 4), of whom 482 had situs inversus totalis (39%) and 37 had situs ambiguous (3%). Laterality defects were present in individuals with DNA variants in both the genes associated with hallmark pathognomonic ciliary ultrastructure defects (CCDC103, ODAD1, ODAD2, ODAD3, ODAD4, DNAH5, DNAH9, DNAI1, DNAI2, DNAL1, DNAAF1, DNAAF2, DNAAF3, DNAAF4, DNAAF5, DNAAF6, DNAAF11, CFAP298, CFAP300, SPAG1, ZMYND10, CCDC39 and CCDC40) and in the genes DNAH11, FOXJ1, CFAP45, CFAP52, CFAP53 and OFD1, which are not associated with pathognomonic ciliary ultrastructure defects. No laterality defects were present in individuals with DNA variants in the genes HYDIN, SPEF2, CFAP221, CFAP74, RSPH1, RSPH3, RSPH4A, RSPH9, DNAJB13, NME5, GAS8, DRC1, CCDC65, RPGR, CCNO, MCIDAS and NEK10. The overall prevalence of laterality defects was significantly higher in the group of PCD individuals with genetic variants associated with hallmark pathognomonic ultrastructure defects than in the rest of the cohort (51% versus 18%; p<0.0001) (figure 4). The regional distribution of laterality defects varied widely among the participating countries with the lowest prevalence in Turkey (28%), the Netherlands (31%), Germany (37%), Spain (39%) and Israel (40%) (figure 4).
FIGURE 4.
Prevalence of laterality defects per predicted ciliary ultrastructure and country. The prevalence of laterality defects is 42% in the total study cohort (n=519 individuals with laterality defects). There is a significant difference between the groups stratified according to the predicted effect of genetic variants on ciliary ultrastructure. In the group of 894 individuals with genetic variants associated with pathognomonic ciliary ultrastructure defects detectable by transmission electron microscopy, 51% of individuals (n=457) have laterality defects. In contrast, in the group of individuals with genetic variants not associated with defective ciliary ultrastructure hallmark, only 18% of individuals (n=55) have laterality defects (p<0.0001). The prevalence of laterality defects varies widely among the participating countries and ranges from 28% to 69%. It is lowest in Turkey (28%), the Netherlands (31%), Germany (37%), Spain (39%) and Israel (40%).
Lung function
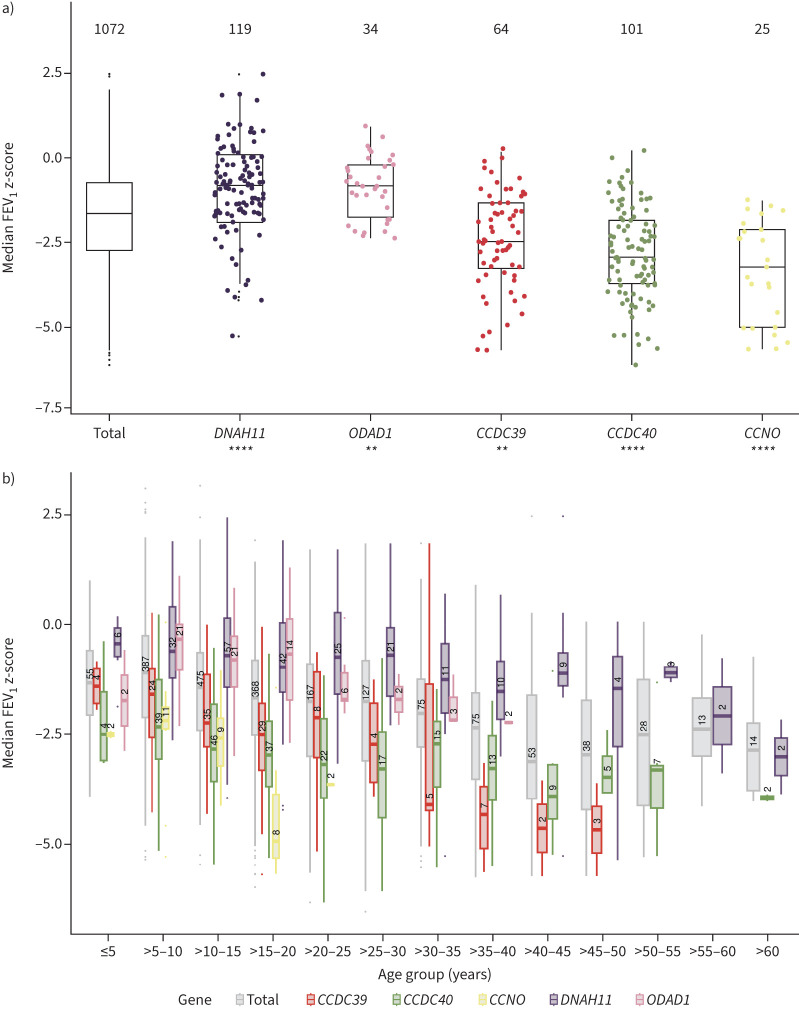
Lung function data were available for 1072 genotyped individuals, with 10 022 FEV1 values in total. The median number of FEV1 z-scores of the participants was 4 (IQR 2–8, range 1–268). 948 individuals had more than one FEV1 measurement; 833 individuals had measurements in a period of more than 1 year, with a median time period of 3.8 years (IQR 2.2–7.9 years, range 1–40.9 years). The median FEV1 z-score was −1.66 (IQR −2.75– −0.752) for the whole study cohort (figure 5a), with progressively lower FEV1 z-scores in the groups of older individuals (figure 5b). 528 PCD subjects had FEV1 data represented in more than one age bin. Individuals with laterality defects had a median FEV1 z-score of −1.65 (IQR −2.69– −0.753) compared to the median FEV1 z-score of −1.67 for individuals without laterality defects (IQR −2.81– −0.779) (nonsignificant, p>0.05). However, we found that distinct gene defects were associated with either a more severe or a more subtle loss of lung function (figure 5, supplementary figure E4). The group of individuals with CCNO variants (n=25) showed the poorest median FEV1 z-score, which was significantly lower than the rest of the cohort (−3.26, IQR −5.04– −2.13, p<0.0001; figure 5a, b; supplementary figure E4). Genetic defects in CCDC39 (n=64) and CCDC40 (n=101) also resulted in significantly lower median FEV1 z-scores compared to the rest of the cohort (CCDC39: −2.49, IQR −3.28– −1.37, p<0.01; CCDC40: −2.96, IQR −3.77– 1.86, p<0.00001), and showed lower values over the entire age range (figure 5a, b; supplementary figure E4). In contrast, the subgroups of individuals with variants in DNAH11 (n=119) and in ODAD1 (n=34) had significantly higher median FEV1 z-scores compared to the rest of the cohort (DNAH11: −0.83, IQR −1.57– −0.098, p<0.0001; ODAD1: −0.85, IQR −1.80– −0.15, p<0.01; figure 5a, b). Detailed information regarding median FEV1 z-scores and FEV1 % predicted for all gene groups is provided in supplementary table E1.
FIGURE 5.
Median forced expiratory volume in 1 s (FEV1) z-scores of the whole primary ciliary dyskinesia (PCD) cohort and distinct PCD groups. a) The median FEV1 z-score of the overall PCD cohort is −1.66 (interquartile range (IQR) −2.75– −0.752). Individuals with CCNO variants (n=25) show a significantly lower median FEV1 z-score (−3.26, IQR −5.04– −2.13, p<0.0001) compared to the rest of the cohort. Individuals with DNA variants in CCDC39 (n=64) and CCDC40 (n=101) associated with microtubular disorganisation and inner dynein arm defects exhibit median FEV1 z-scores significantly lower than the rest of the cohort (CCDC39: −2.49, IQR −3.28– −1.37, p<0.01; CCDC40: −2.96, IQR −3.77– −1.86, p<0.00001). The group of individuals with DNAH11 (n=119) and ODAD1 variants (n=34) show significantly higher median FEV1 z-scores compared to the rest of the cohort (DNAH11: −0.831, IQR −1.57– −0.0984, p<0.0001; ODAD1 −0.850, IQR −1.57– −0.0984, p<0.01). Significant differences between distinct gene groups and the rest of the cohort are marked with asterisks. p≤0.05 was considered significant. **: p≤0.01; ****: p≤0.0001. b) The study cohort was divided into consecutive 5-year age groups to analyse age-dependence of FEV1 z-scores. 528 PCD individuals have FEV1 data represented in more than one age bin. Groups of older PCD individuals have increasingly lower FEV1 z-scores (grey bars). The groups of individuals with CCNO, CCDC39 and CCDC40 variants have lower FEV1 z-scores, while individuals with DNAH11 and ODAD1 variants have higher FEV1 z-scores in most age bins compared to the total cohort. However, the median FEV1 z-scores of the DNAH11 and ODAD1 variant group of individuals aged >60 or >30–35 years, respectively, show similarly low values as the total PCD cohort.
Discussion
In this multinational study, the genetic diagnosis was confirmed in 1236 individuals (89%), who harboured 908 different disease-causing genetic variants in 46 different PCD genes, confirming the high degree of genetic heterogeneity in PCD (figure 1). In the whole study cohort, DNAH5 was the most frequently implicated gene, consistent with previous reports [8, 16, 17]. Our study revealed marked regional differences in this distribution within and beyond Europe (figures 1–3), suggesting the presence of several different founder variants. It is known that the presence of founder variants results in a highly variable prevalence of monogenic diseases in Europe and other parts of the world, e.g. F508del in the CFTR gene responsible for cystic fibrosis [18, 19]. This is also true for PCD, but much more complex because of the high degree of genetic heterogeneity. Our findings are consistent with previous studies that have reported recurrent gene variants, including DNAH5:c.10815del [17], DNAI1:c.48+2dup [20], CCDC40:c.248del [21], ODAD1:c.742G>A [22, 23], SPAG1:c.2014C>T [24, 25], CCDC39:c.1871_1872del [10], CCDC103:c.461A>C [26, 27], HYDIN:c.922A>T [28], CFAP300:c.198_200del [29, 30], CCNO:c.258_262dup [31], MCIDAS:c.1142G>A [32, 33], DNAL1:c.449A>G [34], RSPH9:c.670+2T>C [9], CFAP300:c.98_106del [35], CCDC40:c.552+6T>A [36], RSPH4A:c.1391G>A [37, 38] and ZMYND10:c.47T>G [39]. In our study, the most pronounced regional cluster was seen for the founder variant c.2014C>T in SPAG1, which prevailed in the Slavic countries Poland, Czech Republic and Slovakia. We also found regional clustering of other founder variants (supplementary figure E2).
Recently, Hannah et al. [16] estimated the global prevalence of PCD and predicted the most frequent pathogenic genetic variants and genes associated with PCD for different ethnicities, using the Hardy–Weinberg calculation of the prevalence of bi-allelic variants based on publicly available allele frequencies in large genome sequence databases. In our study, the most frequently affected genes were DNAH5, DNAH11, CCDC40, DNAI1 and CCDC39, consistent with the prediction. However, the order of the affected genes and detection of certain alleles differed slightly. For example, variants predicted to be present in the Ashkenazi population were detected in our Israeli PCD group (CFAP298:c.735C>G; CCNO:c.638T>C; DNAI1:c.1490G>A), but other frequent alleles were not detected at all (e.g. DNAAF1:c.1698+1G>A; ZMYND10:c.599+1G>A). Therefore, predictions based on available allele frequencies from large sequence databases are helpful, but real patient data are important to understand the genetic spectrum in defined geographical regions, because publicly available genome information only contains a limited number of genomes and does not reflect all population ancestries. In addition, Hannah et al. [16] reported that PCD is more common than previously assumed, especially in individuals of African ancestry who appear to be under-recognised for PCD.
Next, we investigated whether regional differences in the distribution of PCD genotypes might influence the outcome of non-genetic tests used to diagnose PCD, such as TEM, which is recommended by current guidelines [40, 41]. Overall, 72% of study participants had a genotype associated with hallmark pathognomonic ciliary ultrastructure defects detectable by TEM, consistent with previous reports [3, 42]. Interestingly, we found that this proportion varied from country to country depending on the regional prevalence of distinct PCD gene defects (supplementary figures E2 and E3). For example, in Turkey, where variants in CCNO and DNAH11 (not associated with hallmark ciliary ultrastructure defect) are frequently involved in PCD, TEM failed to diagnose more than half of PCD individuals (53%). Thus, knowledge of the regional distribution of distinct PCD gene variants is important to estimate the sensitivity of a test (e.g. TEM) used to diagnose PCD. Previous studies in the USA and UK have reported a higher proportion of hallmark ciliary ultrastructure defects in PCD (from 83% to 86%), which might reflect regional differences in the prevalence of affected PCD genes associated with class I ciliary defects [5, 43]. In some countries, the use of TEM as the first-line diagnostic test, prior to selecting individuals for genetic testing, might result in failure to identify PCD individuals without pathognomonic ciliary ultrastructure defects. This may also explain the discrepancy in the relative frequency of pathogenic variants detected in similarly sized DNAH11 and DNAH5. The highest prevalence of pathogenic DNAH11 variants was reported in a study based on variant frequencies available in public databases [16], while in the present cohort, where patients were selected based on regional diagnostic schemes often relying on TEM, the frequency of pathogenic variants in DNAH11 (×142) was much lower than DNAH5 (×275).
This registry study included participating centres from many countries with different diagnostic resources (e.g. TEM) and expertise. Accordingly, the proportion of PCD individuals with genetic variants associated with hallmark pathognomonic ciliary ultrastructure defects differed considerably among countries (47–100%), likely reflecting distinct regional prevalence of genetic variants. In addition, strategies and techniques used to establish the genetic PCD diagnosis differed among these centres and ranged from targeted Sanger sequencing of individual high-prevalence genes to comprehensive next-generation sequencing. This is a limitation, because it leads to bias in terms of the genes and genetic variants reported. However, our study only reported PCD individuals with a confirmed genetic diagnosis, rendering the diagnostic accuracy very high when compared to other studies where probable PCD diagnoses had been included [44, 45]. Correct interpretation of genetic reports is a common problem [46]. It is even more difficult in rare, genetically and phenotypically heterogeneous diseases such as PCD. The fact that 148 individuals (11% of our cohort) did not meet the ACMG/AMP criteria [14] confirms that genetic PCD diagnosis is very complex, and indicates the need to train specialised respiratory physicians in the interpretation of genetic reports. Genetic diagnosis is increasingly important for patient-centred management in PCD. In other respiratory diseases, such as cystic fibrosis, genetic testing has been instrumental for diagnosis and the development of successful genotype-specific therapies that require recognition of specific pathogenic CFTR variants [18, 19, 47–49]. Personalised therapies for PCD, such as gene-specific mRNA replacement, are currently under investigation [50], and the inclusion of PCD individuals in randomised clinical trials will require genetic confirmation of the diagnosis.
The large size of the genotyped PCD cohort in this registry study enabled detailed genotype–phenotype studies. We here investigated the distribution of laterality defects and FEV1 z-scores in genetically confirmed PCD individuals. The overall proportion of individuals with laterality defects in our cohort was 42%. Laterality defects were only present in individuals with genetic defects known to be associated with laterality defects, confirming the good quality of the genetic diagnosis [1]. Interestingly, several studies reporting situs information in PCD populations showed higher rates of laterality defects [51–53]. Those studies mainly diagnosed PCD by TEM and therefore had a bias to identify more PCD individuals with laterality defects (49–54%), including situs ambiguous (6–12%) [51–53]. Here, we chose genetics for diagnosis and included many PCD types that are not associated with laterality defects. This might explain why we report a higher proportion of PCD individuals with situs solitus (55%) and lower proportions of laterality defects (42%: 39% situs inversus totalis; 3% situs ambiguous). The prevalence of laterality defects differed considerably among countries, reflecting regional distribution of the relevant genotypes. Laterality defects were significantly more frequent in PCD individuals with hallmark pathognomonic ciliary ultrastructure defects than in individuals without hallmark defects, consistent with previous findings in a smaller PCD cohort [3]. It is known that the absence of laterality defects and the lack of pathognomonic ultrastructural ciliary defects make the PCD diagnosis very difficult. [3, 54]. Moreover, PCD individuals without ciliary ultrastructure defects appear to have higher nasal nitric oxide production rates, further hampering PCD diagnosis [3, 6]. Our study confirmed that diagnosis of PCD by standard (non-genetic) tests may be less efficient in populations characterised by a low prevalence of genetic variants causing laterality defects and/or leading to hallmark ciliary ultrastructure defects (such as Turkey in our cohort).
Studies of genotype–phenotype correlations concerning the decline of lung function in PCD have been so far reported only in small genotyped PCD cohorts [4, 5, 10, 55–59]. Analysis of the large cohort in our study demonstrated substantial correlations, indicating that PCD lung function outcomes are related to individual genotypes (figure 5). We showed that the lowest FEV1 z-scores in the whole PCD cohort were associated with pathogenic variants in CCNO (n=25), followed by CCDC39 (n=64) and CCDC40 (n=101). Significant genotype–phenotype correlations for CCNO and CCDC40 have not been reported so far. A smaller study has shown a significant reduction of FEV1 z-score only in individuals with CCDC39 variants (n=35) [4]. However, the same study only recruited 25 CCDC40-variant individuals without significant reduction of FEV1 scores, possibly due to small sample size. Consistent with our findings, several reports lacking genetic test results have shown a severe reduction of FEV1 in individuals with microtubular disorganisation and inner dynein arm defects revealed by TEM that are frequently associated with either CCDC39 or CCDC40 variants [4, 5, 41, 55, 56, 58, 60]. Interestingly, smaller studies including individuals with microtubular disorganisation and inner dynein arm defects showed heterogeneous results concerning BMI: a study in 41 PCD individuals in the USA showed a reduction of BMI [56] whereas a study in Italy with 31 individuals showed a normal BMI [36]. In our large cohort comprising 181 PCD individuals, we did not see a significant reduction in BMI.
We observed that the reduction of FEV1 z-scores was milder in individuals with DNAH11 variants than in the whole PCD cohort, consistent with findings in a smaller DNAH11 cohort [4]. FEV1 z-scores were also significantly higher in individuals with ODAD1 variants than in the whole cohort, which has not been reported previously. FEV1 z-scores associated with DNAH11 or ODAD1 variants differed among the age groups: their reduction was milder in younger individuals (<60 years old (DNAH11) and <30–35 years old (ODAD1)), whereas median FEV1 z-scores were similarly low in older individuals (>60 years old (DNAH11) and >30–35 years old (ODAD1)) as in the total PCD cohort (figure 5b). Thus, PCD individuals with genotypes associated with a mild reduction of FEV1 z-scores should be closely monitored. A limitation of this study is the limited or lack of data from older individuals (>50 years old) for most of the gene groups. Larger longitudinal studies investigating the age-dependency of lung function in the different PCD gene groups are needed.
We observed further interesting correlations between genotypes and FEV1 z-score-associated pulmonary phenotypes, but the respective gene groups were too small for statistical analyses (supplementary table E1). For example, a strong reduction of FEV1 z-scores was seen in the group of individuals with MCIDAS variants (n=5), consistent with a severe ciliogenesis defect, resembling findings in individuals with CCNO variants [31–33]. Interestingly, the group of individuals with RSPH1 variants (n=15) showed a lower median FEV1 z-score than the total PCD cohort, suggesting that the respiratory disease course in these individuals might not be as subtle as previously reported [61]. This is also consistent with the FEV1 z-scores in individuals with pathogenic variants in genes encoding other radial spoke head proteins (RSPH4A, RSPH9, RSPH3; supplementary table E1).
In conclusion, we demonstrated that a high proportion of PCD individuals are difficult to diagnose due to the absence of pathognomonic defects of the ciliary ultrastructure and absence of laterality defects, confirming the importance of genetic testing for PCD diagnosis. The unprecedented use of a multinational dataset of DNA variants and clinical characteristics in PCD individuals allowed us to reveal substantial correlations of genotypes with FEV1 z-scores, suggesting that genetic diagnosis might help to predict the clinical prognosis in affected individuals. Individuals with pathogenic variants in certain genes (CCNO, CCDC39 and CCDC40) may require more rigorous and intensive clinical management.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-01769-2023.Supplement (1.1MB, pdf)
Supplementary table E2: Genetic variants of the study cohort including allele frequencies ERJ-01769-2023.Table_E2 (50KB, xlsx)
Shareable PDF
Acknowledgements
The authors thank the individuals with PCD and their families for participating in the studies and especially acknowledge the national patient support groups. Several participating centres are healthcare providers in the European Reference Network ERN-LUNG and/or are members of BEAT-PCD clinical research collaboration supported by the European Respiratory Society. For excellent organisational assistance, the authors thank the study nurses S. Helms and M. Tekaat (Department of General Pediatrics, University Hospital Muenster, Muenster, Germany). For their excellent technical assistance, the authors thank M. Herting and L. Schwiddessen (Department of General Pediatrics, University Hospital Muenster, Muenster, Germany), C. Westermann (Gerhard Domagk Institute of Pathology, University Hospital Muenster, Muenster, Germany) and K. Wohlgemuth, A. Borgscheiper and S. Sivalingam (Department of General Pediatrics, University Hospital Muenster, Muenster, Germany). We thank G.W. Dougherty for editing the manuscript and C. Rieck for assistance with the design of the graphical abstract (both Department of General Pediatrics, University Hospital Muenster, Muenster, Germany). The authors thank the following persons for substantial support in this study: N.T. Loges, S. Freischem, L. Biebach, J. Wallmeier, A. Schramm and R.D. Tenardi (Department of General Pediatrics, University Hospital Muenster, Muenster, Germany), A. Moreno-Galdó (Paediatric Pulmonology Section, Department of Paediatrics, Vall d'Hebron Hospital Universitari, Vall d'Hebron Barcelona Hospital Campus, Universitat Autònoma de Barcelona, Spain; and Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, Madrid, Spain), J. Korfmacher (Department of Pediatrics, Faculty of Medicine and University Hospital, University of Cologne, Cologne, Germany), S. Scigliano (Respiratory Center, Ricardo Gutiérrez Children's Hospital, Buenos Aires, Argentina), F. Santamaria (Department of Translational Medical Sciences, Pediatric Pulmonology, Federico II University, Naples, Italy), R. Cutrera and A. Allegorico (Pneumology and Cystic Fibrosis Unit, Academic Department of Pediatrics, Bambino Gesù Children's Hospital, Rome, Italy), P. Pohunek and L. Borek-Dohalska (Department of Paediatrics, Second Faculty of Medicine, Charles University and Motol University Hospital, Prague, Czech Republic), A. Holubova (Department of Biology and Medical Genetics, Second Faculty of Medicine, Charles University and Motol University Hospital, Prague, Czech Republic), B. Karadag (Department of Pediatric Pulmonology, Marmara University School of Medicine, Istanbul, Turkey), M. Dotan (Pulmonary Institute, Schneider Children's Medical Center of Israel, Petach-Tikva, Israel; and Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel), D. Prais (Pulmonary Institute, Schneider Children's Medical Center of Israel, Petach-Tikva, Israel; and Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel), E. Rietschel (Department of Pediatrics, Faculty of Medicine and University Hospital, University of Cologne, Cologne), F. Brinkmann (University Children's Hospital, Ruhr University Bochum, Katholisches Klinikum Bochum, Bochum, Germany) and D.J. Morris-Rosendahl (Clinical Genetics and Genomics, Royal Brompton Hospital, Guy's and St Thomas' NHS Foundation Trust and NHLI, Imperial College London, London, UK). The graphical abstract was created with BioRender.com. The authors would like to thank J. Strub and the varSEAK Team as well as the groups that provided their variant data.
Footnotes
Ethics statement: The study was conducted in accordance with the Declaration of Helsinki and was approved by the local ethics committee of the Westphalian Wilhelms–University of Muenster (Muenster, Germany; AZ 2011–270-f-S). Ethical approval was obtained from the institutional ethics and/or national research committees of each centre involved in the study. Each participant or his or her legal guardian gave written informed consent prior to participation.
The study is registered on ClinicalTrials.gov (NCT:04717115).
This article has an editorial commentary: https://doi.org/10.1183/13993003.01026-2024
Conflict of interest: The authors have no potential conflicts of interest to disclose.
Support statement: This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG OM6/7, OM6/8, OM6/10, OM6/14, CRU 326 (to H. Omran, J. Raidt); OL 450/1 (H. Olbrich); CRC 1449 (project 431232613; sub-project Z02 to M.A. Mall)), the Interdisziplinaeres Zentrum für Klinische Forschung Muenster (Om2/009/12, Om2/015/16 and Om2/010/20), Registry Warehouse (Horizon2020 GA 777295) and the German Federal Ministry of Education and Research (82DZL009B1 to M.A. Mall). J.F. Roehmel is a participant of the Case Analysis and Decision Support (CADS) programme funded by the Berlin Institute of Health. The authors thank the “Børnelungefonden” and “Rigshospitalet's research fund” (K.G. Nielsen). A. Shoemark is supported by Asthma and Lung UK. The National UK PCD service is supported by NHS England. Some study authors and data contributors participate in the BEAT-PCD clinical research collaboration supported by the European Respiratory Society. This study was supported and promoted as part of work package 3 during the 2020-2023 BEAT-PCD funding period. H.M. Mitchison is supported by the NIHR Biomedical Research Centre at Great Ormond Street Hospital. S. Rovira-Amigo was funded by a grant from Instituto de Salud Carlos III (ISCIII) through the project “PI20/01419” and co-funded by the EU and a grant of the Spanish Society of Paediatrics (Invest-AEP 2021). V. Martinu was supported by Ministry of Health of the Czech Republic, grant number NV19-07-00210 and supported by Motol University Hospital, Prague, Czech Republic, 00064203 (conceptual development of research). E. Ziętkiewicz was supported by the Polish National Science Centre, grant 2018/31/B/NZ2/03248. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Wallmeier J, Nielsen KG, Kuehni CE, et al. Motile ciliopathies. Nat Rev Dis Primers 2020; 6: 77. doi: 10.1038/s41572-020-0209-6 [DOI] [PubMed] [Google Scholar]
- 2.Pennekamp P, Raidt J, Wohlgemuth K, et al. Primary ciliary dyskinesia. In: Wagner TOF, Humbert M, Wijsenbeek M, et al., eds. Rare Diseases of the Respiratory System. Sheffield, European Respiratory Society, 2023; pp. 118–134. [Google Scholar]
- 3.Raidt J, Krenz H, Tebbe J, et al. Limitations of nasal nitric oxide measurement for diagnosis of primary ciliary dyskinesia with normal ultrastructure. Ann Am Thorac Soc 2022; 19: 1275–1284. doi: 10.1513/AnnalsATS.202106-728OC [DOI] [PubMed] [Google Scholar]
- 4.Shoemark A, Rubbo B, Legendre M, et al. Topological data analysis reveals genotype–phenotype relationships in primary ciliary dyskinesia. Eur Respir J 2021; 58: 2002359. doi: 10.1183/13993003.02359-2020 [DOI] [PubMed] [Google Scholar]
- 5.Kinghorn B, Rosenfeld M, Sullivan E, et al. Airway disease in children with primary ciliary dyskinesia: impact of ciliary ultrastructure defect and genotype. Ann Am Thorac Soc 2023; 20: 539–547. doi: 10.1513/AnnalsATS.202206-524OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legendre M, Thouvenin G, Taytard J, et al. High nasal nitric oxide, cilia analyses, and genotypes in a retrospective cohort of children with primary ciliary dyskinesia. Ann Am Thorac Soc 2022; 19: 1704–1712. doi: 10.1513/AnnalsATS.202110-1175OC [DOI] [PubMed] [Google Scholar]
- 7.Emiralioğlu N, Taşkıran EZ, Koşukcu C, et al. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol 2020; 55: 383–393. doi: 10.1002/ppul.24583 [DOI] [PubMed] [Google Scholar]
- 8.Fassad MR, Patel MP, Shoemark A, et al. Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet 2020; 57: 322–330. doi: 10.1136/jmedgenet-2019-106501 [DOI] [PubMed] [Google Scholar]
- 9.Yiallouros PK, Kouis P, Kyriacou K, et al. Implementation of multigene panel NGS diagnosis in the national primary ciliary dyskinesia cohort of Cyprus: an island with a high disease prevalence. Hum Mutat 2021; 42: e62–e77. doi: 10.1002/humu.24196 [DOI] [PubMed] [Google Scholar]
- 10.Rumman N, Fassad MR, Driessens C, et al. The Palestinian primary ciliary dyskinesia population: first results of the diagnostic and genetic spectrum. ERJ Open Res 2023; 9: 00714-2022. doi: 10.1183/23120541.00714-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner C, Lablans M, Ataian M, et al. An international registry for primary ciliary dyskinesia. Eur Respir J 2016; 47: 849–859. doi: 10.1183/13993003.00776-2015 [DOI] [PubMed] [Google Scholar]
- 12.Ardura-Garcia C, Goutaki M, Carr SB, et al. Registries and collaborative studies for primary ciliary dyskinesia in Europe. ERJ Open Res 2020; 6: 00005-2020. doi: 10.1183/23120541.00005-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raidt J, Maitre B, Pennekamp P, et al. The disease-specific clinical trial network for primary ciliary dyskinesia: PCD-CTN. ERJ Open Res 2022; 8: 00139-2022. doi: 10.1183/23120541.00139-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemark A, Boon M, Brochhausen C, et al. International consensus guideline for reporting transmission electron microscopy results in the diagnosis of primary ciliary dyskinesia (BEAT PCD TEM Criteria). Eur Respir J 2020; 55: 1900725. doi: 10.1183/13993003.00725-2019 [DOI] [PubMed] [Google Scholar]
- 16.Hannah WB, Seifert BA, Truty R, et al. The global prevalence and ethnic heterogeneity of primary ciliary dyskinesia gene variants: a genetic database analysis. Lancet Respir Med 2022; 10: 459–468. doi: 10.1016/S2213-2600(21)00453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornef N, Olbrich H, Horvath J, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med 2006; 174: 120–126. doi: 10.1164/rccm.200601-084OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong T, Ramsey BW. Cystic fibrosis: a review. JAMA 2023; 329: 1859–1871. doi: 10.1001/jama.2023.8120 [DOI] [PubMed] [Google Scholar]
- 19.Bell SC, Mall MA, Gutierrez H, et al. The future of cystic fibrosis care: a global perspective. Lancet Respir Med 2020; 8: 65–124. doi: 10.1016/S2213-2600(19)30337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zariwala MA, Leigh MW, Ceppa F, et al. Mutations of DNAI1 in primary ciliary dyskinesia: evidence of founder effect in a common mutation. Am J Respir Crit Care Med 2006; 174: 858–866. doi: 10.1164/rccm.200603-370OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antony D, Becker-Heck A, Zariwala MA, et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat 2013; 34: 462–472. doi: 10.1002/humu.22261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onoufriadis A, Paff T, Antony D, et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet 2013; 92: 88–98. doi: 10.1016/j.ajhg.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kos R, Israëls J, van Gogh CDL, et al. Primary ciliary dyskinesia in Volendam: diagnostic and phenotypic features in patients with a CCDC114 mutation. Am J Med Genet C Semin Med Genet 2022; 190: 89–101. doi: 10.1002/ajmg.c.31968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djakow J, Kramná L, Dušátková L, et al. An effective combination of Sanger and next-generation sequencing in diagnostics of primary ciliary dyskinesia. Pediatr Pulmonol 2016; 51: 498–509. doi: 10.1002/ppul.23261 [DOI] [PubMed] [Google Scholar]
- 25.Knowles MR, Leigh MW, Ostrowski LE, et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am J Hum Genet 2013; 92: 99–106. doi: 10.1016/j.ajhg.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panizzi JR, Becker-Heck A, Castleman VH, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet 2012; 44: 714–719. doi: 10.1038/ng.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemark A, Moya E, Hirst RA, et al. High prevalence of CCDC103 p.His154Pro mutation causing primary ciliary dyskinesia disrupts protein oligomerisation and is associated with normal diagnostic investigations. Thorax 2018; 73: 157–166. doi: 10.1136/thoraxjnl-2017-209999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olbrich H, Schmidts M, Werner C, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left–right body asymmetry. Am J Hum Genet 2012; 91: 672–684. doi: 10.1016/j.ajhg.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zietkiewicz E, Bukowy-Bieryllo Z, Rabiasz A, et al. CFAP300: mutations in Slavic patients with primary ciliary dyskinesia and a role in ciliary dynein arms trafficking. Am J Respir Cell Mol Biol 2019; 61: 440–449. doi: 10.1165/rcmb.2018-0260OC [DOI] [PubMed] [Google Scholar]
- 30.Höben IM, Hjeij R, Olbrich H, et al. Mutations in C11orf70 cause primary ciliary dyskinesia with randomization of left/right body asymmetry due to defects of outer and inner dynein arms. Am J Hum Genet 2018; 102: 973–984. doi: 10.1016/j.ajhg.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallmeier J, Al-Mutairi DA, Chen CT, et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet 2014; 46: 646–651. doi: 10.1038/ng.2961 [DOI] [PubMed] [Google Scholar]
- 32.Amirav I, Wallmeier J, Loges NT, et al. Systematic analysis of CCNO variants in a defined population: implications for clinical phenotype and differential diagnosis. Hum Mutat 2016; 37: 396–405. doi: 10.1002/humu.22957 [DOI] [PubMed] [Google Scholar]
- 33.Boon M, Wallmeier J, Ma L, et al. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun 2014; 5: 4418. doi: 10.1038/ncomms5418 [DOI] [PubMed] [Google Scholar]
- 34.Mazor M, Alkrinawi S, Chalifa-Caspi V, et al. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet 2011; 88: 599–607. doi: 10.1016/j.ajhg.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yiallouros PK, Kouis P, Pirpa P, et al. Wide phenotypic variability in RSPH9-associated primary ciliary dyskinesia: review of a case-series from Cyprus. J Thorac Dis 2019; 11: 2067–2075. doi: 10.21037/jtd.2019.04.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pifferi M, Bush A, Mulé G, et al. Longitudinal lung volume changes by ultrastructure and genotype in primary ciliary dyskinesia. Ann Am Thorac Soc 2021; 18: 963–970. doi: 10.1513/AnnalsATS.202007-816OC [DOI] [PubMed] [Google Scholar]
- 37.Frommer A, Hjeij R, Loges NT, et al. Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am J Respir Cell Mol Biol 2015; 53: 563–573. doi: 10.1165/rcmb.2014-0483OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boaretto F, Snijders D, Salvoro C, et al. Diagnosis of primary ciliary dyskinesia by a targeted next-generation sequencing panel: molecular and clinical findings in Italian patients. J Mol Diagn 2016; 18: 912–922. doi: 10.1016/j.jmoldx.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 39.Moore DJ, Onoufriadis A, Shoemark A, et al. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am J Hum Genet 2013; 93: 346–356. doi: 10.1016/j.ajhg.2013.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas JS, Barbato A, Collins SA, et al. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J 2017; 49: 1601090. doi: 10.1183/13993003.01090-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro AJ, Davis SD, Polineni D, et al. Diagnosis of primary ciliary dyskinesia. an official American Thoracic Society clinical practice guideline. Am J Respir Crit Care Med 2018; 197: e24–e39. doi: 10.1164/rccm.201805-0819ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouis P, Yiallouros PK, Middleton N, et al. Prevalence of primary ciliary dyskinesia in consecutive referrals of suspect cases and the transmission electron microscopy detection rate: a systematic review and meta-analysis. Pediatr Res 2017; 81: 398–405. doi: 10.1038/pr.2016.263 [DOI] [PubMed] [Google Scholar]
- 43.Shah A, Shoemark A, MacNeill SJ, et al. A longitudinal study characterising a large adult primary ciliary dyskinesia population. Eur Respir J 2016; 48: 441–450. doi: 10.1183/13993003.00209-2016 [DOI] [PubMed] [Google Scholar]
- 44.Goutaki M, Pedersen ESL. Phenotype–genotype associations in primary ciliary dyskinesia: where do we stand? Eur Respir J 2021; 58: 2100392. doi: 10.1183/13993003.00392-2021 [DOI] [PubMed] [Google Scholar]
- 45.Halbeisen FS, Jose A, de Jong C, et al. Spirometric indices in primary ciliary dyskinesia: systematic review and meta-analysis. ERJ Open Res 2019; 5: 00231-2018. doi: 10.1183/23120541.00231-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayitoğlu M. Clinical interpretation of genomic variations. Turk J Haematol 2016; 33: 172–179. doi: 10.4274/tjh.2016.0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mall MA, Mayer-Hamblett N, Rowe SM. Cystic fibrosis: emergence of highly effective targeted therapeutics and potential clinical implications. Am J Respir Crit Care Med 2020; 201: 1193–1208. doi: 10.1164/rccm.201910-1943SO [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barry PJ, Mall MA, Álvarez A, et al. Triple therapy for cystic fibrosis Phe508del-gating and -residual function genotypes. N Engl J Med 2021; 385: 815–825. doi: 10.1056/NEJMoa2100665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Middleton PG, Mall MA, Dřevínek P, et al. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single Phe508del allele. N Engl J Med 2019; 381: 1809–1819. doi: 10.1056/NEJMoa1908639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paff T, Omran H, Nielsen KG, et al. Current and future treatments in primary ciliary dyskinesia. Int J Mol Sci 2021; 22: 9834. doi: 10.3390/ijms22189834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy MP, Omran H, Leigh MW, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 2007; 115: 2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038 [DOI] [PubMed] [Google Scholar]
- 52.Shapiro AJ, Davis SD, Ferkol T, et al. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest 2014; 146: 1176–1186. doi: 10.1378/chest.13-1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barber AT, Shapiro AJ, Davis SD, et al. Laterality defects in primary ciliary dyskinesia: relationship to ultrastructural defect or genotype. Ann Am Thorac Soc 2023; 20: 397–405. doi: 10.1513/AnnalsATS.202206-487OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuehni CE, Frischer T, Strippoli MP, et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J 2010; 36: 1248–1258. doi: 10.1183/09031936.00001010 [DOI] [PubMed] [Google Scholar]
- 55.Pifferi M, Bush A, Mariani F, et al. Lung function longitudinal study by phenotype and genotype in primary ciliary dyskinesia. Chest 2020; 158: 117–120. doi: 10.1016/j.chest.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 56.Davis SD, Rosenfeld M, Lee HS, et al. Primary ciliary dyskinesia: longitudinal study of lung disease by ultrastructure defect and genotype. Am J Respir Crit Care Med 2019; 199: 190–198. doi: 10.1164/rccm.201803-0548OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frija-Masson J, Bassinet L, Honoré I, et al. Clinical characteristics, functional respiratory decline and follow-up in adult patients with primary ciliary dyskinesia. Thorax 2017; 72: 154–160. doi: 10.1136/thoraxjnl-2015-207891 [DOI] [PubMed] [Google Scholar]
- 58.Davis SD, Ferkol TW, Rosenfeld M, et al. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am J Respir Crit Care Med 2015; 191: 316–324. doi: 10.1164/rccm.201409-1672OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roehmel JF, Doerfler FJ, Koerner-Rettberg C, et al. Comparison of the lung clearance index in preschool children with primary ciliary dyskinesia and cystic fibrosis. Chest 2022; 162: 534–542. doi: 10.1016/j.chest.2022.02.052 [DOI] [PubMed] [Google Scholar]
- 60.Irving S, Dixon M, Fassad MR, et al. Primary ciliary dyskinesia due to microtubular defects is associated with worse lung clearance index. Lung 2018; 196: 231–238. doi: 10.1007/s00408-018-0086-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knowles MR, Ostrowski LE, Leigh MW, et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med 2014; 189: 707–717. doi: 10.1164/rccm.201311-2047OC [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-01769-2023.Supplement (1.1MB, pdf)
Supplementary table E2: Genetic variants of the study cohort including allele frequencies ERJ-01769-2023.Table_E2 (50KB, xlsx)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01769-2023.Shareable (2.2MB, pdf)