Abstract
In the receptor for ecotropic murine leukemia viruses, tyrosine 235 contributes a critical hydrophobic side chain to the virus-receptor interaction (14). Here we report that tryptophan 142 in ecotropic Moloney murine leukemia virus envelope protein is essential to virus binding and infection. Replacement of tryptophan 142 by alanine or serine resulted in misfolding. However, replacement by methionine (W142M) allowed correct folding of the majority of glycoprotein molecules. W142M virus showed a marked reduction in virus binding and was almost noninfectious, suggesting that tryptophan 142 is involved in receptor binding. In contrast, W142Y virus containing a replacement of tryptophan 142 with an aromatic residue (tyrosine) was as efficient as wild-type virus in infection and binding of cells expressing the wild-type receptor. However, W142Y virus was 100-fold less efficient than wild-type virus in infection of cells expressing a mutant receptor containing tryptophan instead of the critical tyrosine. These results strongly support tryptophan 142 being an essential residue on the virus envelope protein that interacts directly with the critical hydrophobic residue at position 235 of the ecotropic receptor. Tryptophan 142 forms one side of a shallow hydrophobic pocket on the surface of the envelope protein, suggesting that it might comprise the complete putative binding site for tyrosine 235. We discuss the implications of our findings with respect to two models of the envelope protein trimer. Interestingly, both models place tryptophan 142 at the interface between adjacent subunits of the trimer.
Four critical residues have been identified in the receptor for ecotropic murine leukemia viruses (MLV). Asparagine 232, valine 233, tyrosine 235, and glutamic acid 237 are critical for efficient virus infection (1, 18) and binding of purified MLV envelope protein (1). Although each of these residues is critical, none is essential since replacement of any one of them alone does not cause a significant change in virus infection (1). To observe a decrease, at least two of them must be replaced. For example, when tyrosine 235 is changed to proline and glutamic acid 237 is changed to valine, envelope protein binding is abolished and infection is decreased 10,000-fold, even though a single tyrosine-to-proline or glutamic acid-to-valine change does not reduce infection (1). A receptor that contained the double mutations of glutamic acid 237 to valine and tyrosine 235 to the hydrophobic residue phenylalanine, methionine, tryptophan, or histidine was as efficient at mediating virus infection as was the wild-type receptor (14). In contrast, replacing residue 235 with serine, threonine, or alanine in combination with the glutamic acid 237-to-valine change almost completely abolished receptor function, indicating that the side chain at position 235 is involved in a hydrophobic interaction with envelope protein (14).
The specific residues on the ecotropic MLV envelope glycoprotein that interact with the critical receptor positions are not known. At least the first 230 residues in SU are essential for recognition of the receptor (3). This amino-terminal domain contains three regions, called variable regions A, B, and C, that are highly conserved in ecotropic MLV but are divergent in retroviruses that use different receptors, suggesting that they contain the sequences specifying receptor binding (3, 7). Only a few residues involved in receptor recognition have been identified. In particular, aspartate 84 and arginines 83 and 95 in variable region A of ecotropic Moloney MLV (Mo MLV) have been shown to be critical for virus binding, but the specific receptor residues with which they interact have not been determined (2, 13).
In this study, we attempted to identify the residue(s) on the virus envelope protein that interacts with tyrosine 235 on the receptor. Since this receptor residue provides a hydrophobic side chain, we hypothesized that its binding partner on SU is a hydrophobic residue. We selected for analysis a segment extending from residues 134 to 142 in the SU domain of Mo MLV that contains four hydrophobic residues. This segment had not been analyzed before, and no information as to a possible role in envelope function was available. Although it is not part of variable region A, B, or C, two residues within it (aspartate 135 and phenylalanine 137) are conserved among ecotropic MLV but vary among retroviruses that use different receptors. In addition, this segment lies between two disulfide bridges formed by cysteines 131 and 144 and cysteines 139 and 119, a constraint that suggested that an important spatial relationship must be maintained between these and other residues of SU (12).
Here we report evidence identifying tryptophan 142 within this segment as an essential residue for virus binding and infection. We also report genetic evidence supporting its direct interaction with the critical tyrosine 235 on the receptor. Tryptophan 142 is on the surface of the folded SU, forming one side of a shallow hydrophobic pocket or patch that also includes tyrosine 151, tryptophan 152, and tyrosine 210. We propose a model in which this shallow hydrophobic pocket is the binding site for tyrosine 235 on the receptor. Surprisingly, this patch is over 30 Å from aspartate 84 in the folded molecule (7), raising the question of what their relative positions are in the envelope trimers on virions. We discuss the implications of our findings with respect to two trimer models, the model proposed by Fass and coworkers (6, 7) and an alternative model that we propose in which the hydrophobic pocket on one subunit of the trimer is about 11 Å from aspartate 84 on the adjacent subunit. Interestingly, both models place the shallow hydrophobic pocket at the interface between adjacent subunits of the trimer.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T3 fibroblasts and nonpermissive human 293 fetal kidney cells were cultured in Dulbecco’s modified Eagle’s medium and 8% donor calf serum. The 293 cell-derived stable transfectants expressing the receptor and mutant receptor and human cDNAs have been described elsewhere (14). These cells and virus producer H1-BAG cells (20) were maintained in Dulbecco’s modified Eagle’s medium with 8% fetal bovine serum and 250 μg of G418 (Sigma) per ml. Using oligonucleotide-directed mutagenesis, we replaced the codons of each of the selected residues with alanine codons or with glutamic acid, serine, tyrosine, or methionine codons. The absence of unscheduled substitutions and the presence of desired mutations were confirmed by DNA sequence analysis. Virus particles transducing lacZ were produced by transient transfection of pcDNA Mo MLV containing wild-type or mutated env genes into H1-BAG cells (human 293 cells stably expressing the recombinant, replication-defective, Mo MLV-derived BAG genome) as previously described (20). An aliquot of cell-free virus stock from each transfection was used to determine the 50% tissue culture infectious dose (TCID50) to quantitate virus entry. The remaining 7 ml of stock was immediately pelleted through a cushion of 25% sucrose in a Beckman SW41 rotor (30,000 rpm, 2 h, 4°C) and subjected to immunoblot analysis to assess incorporation of the altered envelope proteins into virions relative to the incorporation of the structural capsid (CA) protein (19). Envelope proteins (SU and precursor) were detected with goat anti-Rauscher gp70 antiserum (1:100) (identification no. 80S000018; Quality Biotech Inc.), structural CA protein was detected with goat anti-Rauscher p30 antiserum (1:10,000) (identification no. 81S000263; Quality Biotech Inc.), and TM protein was detected with rabbit anti-p15E antiserum (1:250) (gift of Alan Rein). We also assessed envelope protein processing in virus-producing cells and quantified virus binding as previously described (19, 20).
Virus binding assays.
Binding assays were performed essentially as previously described (4, 11, 20). Briefly, virus-containing supernatants were concentrated 10- to 15-fold on Centricon-100 concentrators (Amicon). The volume of each virus stock was adjusted to achieve comparable particle concentrations based on reverse transcriptase activity. 293 cells (106) expressing wild-type virus receptor or parental 293 cells were incubated with 107 infectious units of Mo MLV or with comparable amounts of virions from mutant virus stocks for 1 h at 4°C. Propidium iodide (Sigma) was added to the binding reaction mixture 5 min prior to analysis to identify live cells (negative for propidium iodide) for quantification of bound virus by flow cytometry with goat anti-gp70 antiserum (Epics Profile Analyzer; Coulter Cytometry). Background fluorescence intensity was determined by incubating cells lacking the receptor with virus particles and antibodies, as well as by incubating cells expressing the virus receptor with antibodies in the absence of virus. Experiments were repeated two times.
Molecular models.
The diagram of the structure of residues 9 to 234 of the wild-type Mo MLV SU was generated with the crystal structure coordinates of the closely related ecotropic Friend MLV surface protein (7) in the RasMol 2.6 visualization program (17), based on the assumption that the structure is conserved between Moloney and Friend SU domains because they have 89% amino acid identity and 97% conservation in this region. Molecular distances were measured with the distance tool of the Swiss PdbViewer 3.5β3 program (8, 9). The diagram of the Fass trimer model was made from the unpublished coordinates of the Fass trimer model (6, 7) (with permission from P. Kim) with RasMol 2.6. The alternative trimer model was generated manually with Swiss PdbViewer 3.5β3 according to the following criteria: (i) carbohydrates must face toward the solvent; (ii) variable regions A, B, and C are exposed to the surface; and (iii) steric clashes are not allowed. Adherence to the first two criteria was confirmed by visual inspection. The absence of steric clashes was initially assessed by visual inspection with the slab mode and then confirmed with the “select for amino acids that clash” function with the trimer coordinates created from merging the three monomer layers.
RESULTS
We reported elsewhere that replacement of aspartate 135 with alanine had no effect on virus infection or on envelope protein folding (19). We report here the results of replacing the adjacent conserved proline 134 (which might provide critical structure to this region) and the four aromatic residues phenylalanine 137, tyrosines 141 and 138, and tryptophan 142 (which might be candidates for the binding partner of tyrosine 235) with alanine. As the work progressed, the tryptophan at position 142 was also replaced with tyrosine, with a nonaromatic but hydrophobic residue (methionine), or with a polar residue (serine).
Effect of virus entry and glycoprotein processing of replacement by alanine.
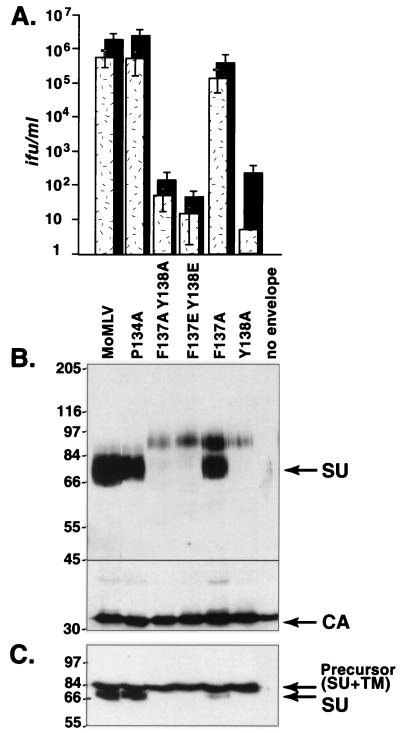
A proline 134-to-alanine, phenylalanine 137-to-alanine, or tyrosine 141-to-alanine change did not affect virus infectiousness, as infectious titers were comparable to titers of wild-type Mo MLV on susceptible mouse NIH 3T3 or human 293 cells stably expressing the receptor cDNA (Fig. 1 and 2). Western blot analysis of the proteins present on purified virions showed that processing and incorporation of P134A or Y141A mutant envelope protein were indistinguishable from those of wild-type envelope protein, suggesting that these positions are not crucial for virus entry or envelope protein folding. However, F137A virions contained an 85-kDa envelope species in addition to the 70-kDa species found in wild-type virions (Fig. 1B).
FIG. 1.
Substitution of phenylalanine 137 did not affect infection, whereas substitution of tyrosine 138 markedly reduced infection. (A) Infectious titers on mouse NIH 3T3 cells bearing the endogenous ecotropic MLV receptor (stippled bars) and human 293 cells expressing the exogenous receptor (black bars). Titers were calculated from the endpoint dilutions (n = 4) after exposure to virions pseudotyped with envelope proteins containing the indicated substitutions. Each value is the average of results from five independent experiments. MoMLV, wild-type ecotropic Mo MLV; no envelope, supernatant or lysate of cells transfected with a virus genome that lacks an env gene; ifu, infectious units. (B) Western blot analysis of virions containing mutant envelope proteins. Samples were denatured by boiling them in the presence of 100 mM dithiothreitol, chilled on ice, and separated on sodium dodecyl sulfate (SDS)–8% polyacrylamide gels. Then proteins were transferred onto nitrocellulose membranes (Protran; Schleicher & Schuell). The membrane was cut into two parts at the position indicated by the black line, roughly that of the 45-kDa molecular mass standard. Envelope proteins (SU and precursor) were detected on the top portion by incubation with goat anti-Rauscher gp70 antiserum and structural CA protein was detected on the bottom portion by incubation with goat anti-Rauscher p30 antiserum. Subsequent incubation with mouse anti-goat or mouse anti-rabbit antiserum conjugated to horseradish peroxidase (Sigma) was performed, and immunoblots were developed with Renaissance chemiluminescence substrate (NEN). (C) Western blot analysis of virus producer cell lysates. Proteins were separated on SDS–8% polyacrylamide gels and then blotted to anti-SU antiserum. Numbers to the left of panels B and C show molecular masses in kilodaltons.
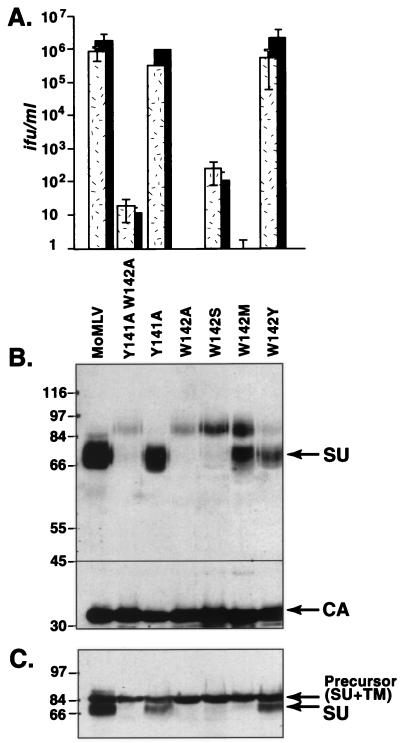
FIG. 2.
Replacement of tryptophan 142 with alanine, serine, or methionine, but not with tyrosine, profoundly reduced infection. Substitution of tyrosine for tryptophan 142 did not affect envelope protein processing or virus infectivity, but replacement of that residue with an alanine, serine, or methionine residue greatly reduced virus entry and interfered with envelope protein processing. (A) Virus titers on NIH 3T3 cells (stippled bars) and human 293 cells expressing the exogenous wild-type receptor (black bars). ifu, infectious units. (B) Western blot analysis of virions containing mutant env genes. (C) Western blot analysis of virus producer cell lysates. Molecular masses (in kilodaltons) are noted at the left of the blots.
Infection was reduced at least 10,000-fold by changes of tyrosine 138 to alanine (Y138A), tyrosine 138 plus phenylalanine 137 to alanine (F137A Y138A) or to glutamate (F137E Y138E), or tryptophan 142 to alanine (W142A) (Fig. 1A and 2A). However, all these mutant viruses lacked the 70-kDa species of mature SU, instead containing sparse amounts of an 85-kDa protein species (Fig. 1B). Moreover, the producer cells also lacked the mature SU species found in wild-type virus producers (Fig. 1C). These results suggested that these substitutions caused glycoprotein misfolding, in light of which the cause of their effect on entry could not be determined.
Conservative replacement of tryptophan 142 rescued envelope protein processing and revealed a role for this residue in virus entry.
It was at this point in our study that the crystal structure of residues 9 through 236 of the homologous ecotropic MLV Friend 57 virus was published (7). In the structure, the residue corresponding to tyrosine 138 was folded within the molecule, suggesting that this residue was more likely to be involved in glycoprotein folding than in receptor recognition. The residue corresponding to tryptophan 142 was on the surface of the fragment, keeping open the possibility that this residue interacts with the receptor. We hypothesized that a conservative substitution of tryptophan 142 with a hydrophobic residue rescues proper protein folding, which would allow us to determine if a change in this residue altered infection.
For this purpose we replaced tryptophan 142 with an aromatic residue (tyrosine, W142Y) and a hydrophobic but nonaromatic amino acid (methionine, W142M). As a control, we also replaced it with a polar residue (serine, W142S). W142S virions were poorly infectious. They contained modest amounts of the 85-kDa envelope protein; no mature SU was detectable (Fig. 2A). Lysates of cells producing this mutant also lacked mature SU, indicating that the W142S change did not rescue glycoprotein folding or virus entry. In contrast, the predominant envelope species on W142M virions was mature SU yet the W142M change reduced infection even more markedly than the W142S change. This result suggested that tryptophan 142 plays a role in virus entry as well as in glycoprotein folding. Replacement with tyrosine did not reduce infection or affect envelope processing and incorporation of mature SU.
Characterization of the minor 85-kDa species on F137A and W142M virions.
Although the majority of envelope protein on W142M virions was mature SU, some 85-kDa protein was consistently present and the level of mature SU in W142M producer cells was below detection (Fig. 2C). These results agreed with our previous finding that mature envelope proteins appear to be preferred substrates for virion assembly (19). Two possibilities might account for the minor 85-kDa species. Both possibilities imply subtle misfolding. First, the cleavage site between SU and TM might be sequestered, resulting in incorporation of the 85-kDa precursor protein. Alternatively, a greater-than-normal amount of carbohydrate might be added at any or all of the glycosylation sites of SU. To determine which was the case, we analyzed the glycosidase F digestion products of W142M and F137A virions by Western blotting to anti-SU antiserum. In addition, we immunoblotted purified virion proteins to anti-TM antiserum.
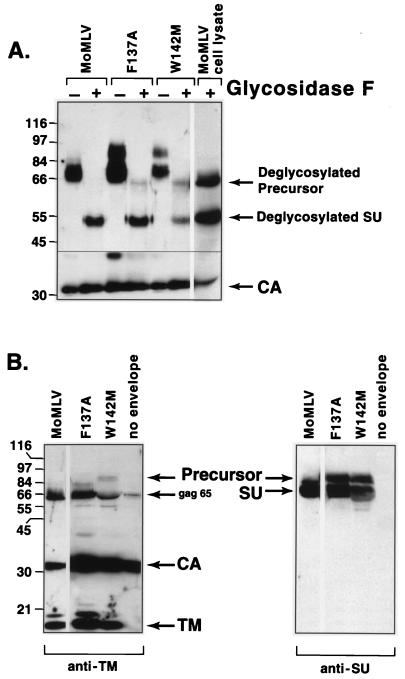
If the 85-kDa species is a hyperglycosylated SU, then removal of carbohydrates would yield a 52-kDa protein. If it is uncleaved precursor, then deglycosylation would yield a 68-kDa species. Lysates of wild-type virus producer cells were used to obtain precise size standards for deglycosylated precursor and SU, as well as to determine if digestion had proceeded to completion. After overnight digestion with an excess of glycosidase F, anti-SU antiserum recognized the expected 68- and 52-kDa proteins in Mo MLV producer cell lysate and a single 52-kDa species in purified Mo MLV virions (Fig. 3A). After digestion, 90% of SU protein in F137A virions was 52 kDa and 10% was 68 kDa, even though approximately 40% was 85 kDa and 60% was 70 kDa prior to digestion (estimated by densitometry of a blot subjected to a shorter exposure than that of the blot shown in Fig. 3). These data suggested that the F137A substitution results in hyperglycosylation of SU. We consistently observed very small amounts of the 68-kDa protein after digestion, suggesting that the change might slightly decrease the efficiency of envelope precursor cleavage in addition to resulting in hyperglycosylation. In contrast, 25% of SU in W142M virions was 85 kDa and 75% was 70 kDa prior to digestion while 40% had a molecular mass of 68 kDa and 60% was 52 kDa after digestion (estimated by densitometry of blots subjected to shorter exposures). These results suggested that the minor species in W142M virions is not hyperglycosylated SU but the envelope precursor.
FIG. 3.
The 85-kDa envelope protein species represents a hyperglycosylated form of SU in F137A virions but represents uncleaved precursor protein in W142M virions. (A) Digestion with glycosidase F to remove N-linked carbohydrates. Sucrose gradient-purified wild-type and mutant virions were incubated overnight in the absence (−) or presence (+) of excess glycosidase F, and then proteins were separated on SDS–8% polyacrylamide gels and immunoblotted with anti-SU (top portion) or anti-CA (bottom portion) antiserum. To determine if glycosidase digestion had been completed, lysates of wild-type Mo MLV producer cells were also analyzed. (B) Immunoblot analysis of virions with anti-TM antiserum. Virion proteins separated on SDS–6 to 15% polyacrylamide gradient gels were immunoblotted with rabbit anti-p15E antiserum (1:250; gift of Alan Rein) with Immobilon membranes to determine if the 85-kDa species contained TM sequences. After detection of the anti-TM antiserum-reactive proteins (left blot), membranes were washed and incubated with anti-SU antiserum (right blot). Molecular mass markers (in kilodaltons) are noted at the left.
In addition, we asked if the 85-kDa species contained sequences derived from TM. Wild-type, F137A, and W142M virions contained comparable amounts of 15-kDa mature TM, indicating that substantial precursor cleavage of both mutants occurred. Anti-TM antiserum reacted only with the protein in the W142M sample (Fig. 3B, left blot) yet SU antiserum showed that the F137A virions contained as much of the 85-kDa protein as did the W142M virions when the same blot was reprobed (Fig. 3B, right blot). These results are in agreement with those of glycosidase F digestion, suggesting that about one-fourth of the W142M molecules misfolded sufficiently to suppress precursor cleavage.
It was possible that the results in Fig. 3B were artifacts resulting from cross-reactivity of the anti-TM antiserum with SU or CA sequences. However, we ruled out that possibility based on the following points. First, we showed previously that this antiserum did not cross-react with SU sequences (19). Gag precursor is normally a 65-kDa cytosolic protein, but use of alternative upstream translation start sites can add a signal peptide sequence to the amino terminus of Gag, which results in an 80-kDa species of the glycosylated Gag precursor (19). Although the anti-TM antiserum cross-reacts with CA sequences in the Gag precursor, the 85-kDa protein species we observed cannot be a contamination of Gag resulting from that cross-reactivity because the constructs we used to provide the gag, pol, and env genes for production of recombinant viruses contain a deletion of the encapsidation signal that also deletes the sequences containing the alternative translation start sites from which the 80-kDa glycosylated Gag protein arises.
Virus binding.
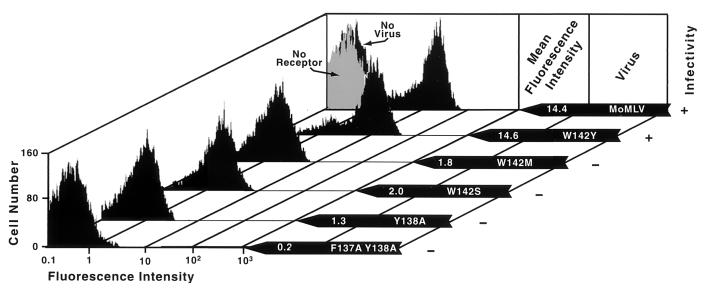
To determine if the W142M change affected the ability of virus to bind the cellular receptor, we performed equilibrium virus binding assays. We typically observed a 50-fold increase in mean fluorescence intensity in host cells incubated with wild-type particles over the background fluorescence intensity. W142Y virions bound cells expressing the receptor as efficiently as did wild-type virions, giving a 49-fold increase in mean fluorescence intensity (Fig. 4). In contrast, W142M virions showed greatly reduced receptor binding even though they contained greater amounts of mature SU than were found in W142Y virions, suggesting that W142 is involved in attachment of virus to the receptor.
FIG. 4.
Virus binding. Binding assays were performed essentially as described previously (4, 11) with modifications as previously reported (20). No virus, nonspecific binding of antiserum in the absence of virus to 293 cells stably expressing receptor; No receptor, binding of wild-type Mo MLV to parental 293 cells lacking ecotropic receptor. The mean fluorescence intensities of the no-receptor and no-virus peaks were 0.2 and 0.3, respectively. Infectivity was summarized from the data in Fig. 1 and 2. +, titer within 10-fold of that of wild-type Mo MLV; −, titer 10,000-fold less than that of wild-type Mo MLV.
F137A Y138A, Y138A, and W142S virions also showed an absence of or <10-fold increases in fluorescence intensity (Fig. 4). This marked reduction in virus binding and infection might have suggested that the altered residues are involved in receptor binding. However, since all molecules of envelope proteins on these virions showed evidence of misfolding and all these virions contained much less envelope protein than wild-type virions, it is more likely that the loss of function resulted from a scarcity of envelope protein and/or its misfolding. The structure of the envelope protein fragment supports the conclusion that residues 134, 137, and 138 are not involved in receptor binding since their corresponding positions lie buried within the folded molecule (7).
Genetic evidence that tryptophan 142 interacts directly with tyrosine 235 on the virus receptor.
We previously showed that tyrosine 235 is critical but not essential for virus entry or binding because its replacement alone by a nonhydrophobic residue does not affect virus entry or envelope protein binding. It must be replaced in combination with at least one other critical residue, such as glutamic acid 237 or asparagine 232 plus valine 233, to abolish binding and reduce entry (1). The human version of the mouse receptor cannot serve as an ecotropic virus receptor because of sequence variation, specifically the lack of the critical tyrosine and the glutamic acid, as well as the asparagine and valine residues (1, 18). However, it acquires receptor function when any two of these residues are introduced into the corresponding positions (1, 18). More importantly, in the human protein, where multiple critical residues are lacking, receptor function is comparable to that of the mouse receptor only when tyrosine or phenylalanine is present in addition to glutamic acid (14). It was reduced 100-fold when tryptophan and glutamic acid were present and 20,000-fold when histidine and glutamic acid were present (14).
These results had indicated that when multiple residues of the virus binding site on the receptor are altered, the virus-receptor interaction is more sensitive to the identity of the critical hydrophobic residue at position 235 than it is in the context of a complete binding site. We reasoned that under these conditions the interaction should also be sensitive to the identity of the side chain on the envelope residue that interacts directly with tyrosine 235. If so, then any difference between infection of W142Y virus and that of wild-type virus should correlate specifically with a change at residue 235 in the receptor or at the corresponding position of the human protein (position 242). For example, the W142Y virus might use a human mutant receptor with tryptophan and glutamic acid or with histidine and glutamic acid more efficiently than the wild-type virus or it might fail to use a mutant receptor with a change that wild-type virus can use. It might also be possible to identify a receptor with a change at position 235 that the W142M virus can use efficiently.
Figure 5 shows the results obtained after exposing human 293 cells stably expressing comparable amounts of mutant receptors and mutant human proteins on their surfaces (14) to serial dilutions of wild-type Mo MLV and W142Y and W142M viruses. W142M virus consistently failed to use any of the mutant receptors or human proteins efficiently. W142Y virus used mutant receptors containing tryptophan, histidine, or methionine instead of tyrosine at position 235 in combination with a valine substitution for glutamic acid 237 as efficiently as did wild-type Mo MLV (Fig. 5A). Similarly, W142Y virus used the human protein containing tyrosine and glutamic acid substitutions as effectively as did wild-type virus (Fig. 5B). In contrast, the TCID50 of W142Y virus on cells expressing a human protein containing tryptophan and glutamic acid was 100-fold less than the TCID50 of wild-type virus on parallel cultures of these cells. These results provide genetic evidence supporting a direct interaction between tryptophan 142 and the critical residue at position 235 in the receptor.
FIG. 5.
A marked loss of infection resulting from a tryptophan 142-to-tyrosine change in the envelope protein correlates with a change from tyrosine to tryptophan in the critical position corresponding to tyrosine 235 in the receptor. Gray bars, wild-type Mo MLV; black bars, W142Y virus stock; white bars, W142M virus stock. Human cells expressing the altered receptor and human cDNAs have been previously described (14). (A) TCID50s for human cells expressing the wild-type or mutant receptor cDNAs. Receptor mutants contained the following changes: a single E237V change (M-65) (Ygv), Y235W and E237V changes (M-38W) (Wgv), Y237H and E237V changes (M-38H) (Hgv), and Y235M and E237V changes (M-38M) (Mgv). (B) TCID50s for human cells expressing cDNAs encoding mutant proteins of the homologous human protein. In two independent titrations, wild-type Mo MLV showed a TCID50 of 2 × 103 infectious units (ifu) per ml on cells expressing the human protein containing proline 242-to-tryptophan and valine 244-to-glutamic acid changes (Wge), whereas W142Y virus showed a TCID50 of 20 ifu/ml on parallel cultures of the WGE cells. Human homologs contained the following changes in positions 242 and 244, corresponding to Y235 and E237 of the receptor, respectively: P242Y and V237E changes (H-40) (Yge), P242F and V237E changes (H-40F) (Fge), and P242W and V237E changes (H-40W) (Wge). Values are the average TCID50s from two experiments using virus generated from two independent transfections in endpoint dilution titration (n = 4), except that values for the FGE cells represent a single titration. No infections by W142M virus were observed on HGV, MGV, FGE, or WGE cells. Amino acids are indicated by the single-letter amino acid code.
DISCUSSION
In the candidate segment of ecotropic MLV SU that we analyzed, only tryptophan 142 appears to be involved in receptor binding. Replacement of tryptophan 142 with alanine or serine resulted in loss of receptor binding and infectivity, but we could not determine if the loss resulted from protein misfolding or from involvement in receptor binding. We reasoned that if we could find a change that affected entry but not folding, then we would be able to interpret infection and binding data. Once the coordinates for the crystal structure of the SU monomer were available, we saw that W142 was on the surface, raising the possibility that replacing it with hydrophobic residues, such as tyrosine and methionine, allows proper protein folding. The results of the analysis were that W142Y virions were infectious but that W142M virions were not. We performed virus binding assays to determine if the block to entry was at binding or at fusion. W142M virions showed a dramatic reduction in receptor binding. Although the W142M virions contained some misfolded molecules as evidenced by their lack of cleavage into mature SU and TM, the majority of molecules were mature SU, suggesting that they were correctly folded. Since the W142M particles consistently contained amounts of mature SU comparable to those in the W142Y virions (data not shown), they should have been as capable of receptor binding as the highly infectious W142Y virions. Instead, their binding was reduced to the level of W142S virions. Taken together, these data suggest that tryptophan 142 is involved in receptor binding as well as in SU folding. However, we cannot rule out the possibility that the mature SU molecules on W142M virions are subtly misfolded, allowing precursor cleavage but displacing critical binding site residues.
We focused our analysis on the W142Y virus, in which envelope protein folds properly and is processed normally, seeking genetic evidence to definitively determine if residue 142 plays a role in virus entry apart from its role in envelope protein folding. We asked if there was a significant difference between infection of W142Y virus and that of wild-type virus in cells expressing any receptor or human mutant receptor that varied only in the identity of residue 235 or of the corresponding human residue, 242. Although infection by W142Y virus paralleled that of wild-type virus in cells expressing mutant receptors, infection of cells expressing one of the human mutant receptors showed such a difference. We had previously shown that replacing the critical tryrosine with tryptophan in the human version of the ecotropic receptor (mutant H-40W; WGE) reduces the infection of wild-type virus by 100-fold (14). We now show that although a change in the candidate tryptophan in envelope protein does not reduce infection via the wild-type receptor or human mutant receptors containing tyrosine and glutamic acid or phenylalanine and glutamic acid, it does reduce infection via H-40W by 10,000-fold, a 100-fold greater reduction than that observed for wild-type virus via H-40W. Since W142Y virus used a human mutant receptor containing tyrosine or phenylalanine and glutamic acid as well as did wild-type virus, the 100-fold synergistic reduction of infection correlated with a change to tryptophan in the critical residue corresponding to position 235 in the receptor. Together, these virus binding and infection results provide strong genetic evidence supporting a direct interaction between tryptophan 142 and critical tyrosine 235 on the receptor.
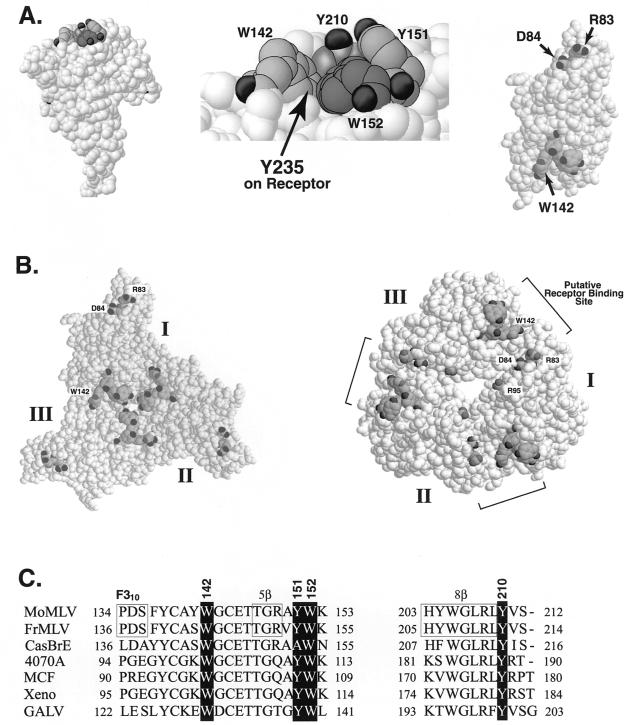
In the crystal structure of Friend MLV glycoprotein, the tryptophan that corresponds to Mo MLV position 142 lies on the surface of the folded SU (7) (Fig. 6A, left image). Strikingly, three other aromatic residues (tyrosines 151 and 210 and tryptophan 152) lie immediately adjacent to it. Together they form a shallow hydrophobic pocket or patch with slightly angled sides on the surface of SU (Fig. 6A, left and middle images). Several charged residues (aspartate 84 and arginines 83 and 95) in variable region A of SU have previously been shown to be involved in receptor recognition (2, 13). In Friend MLV SU, the corresponding residues (aspartate 86 and arginines 85 and 97) lie close together on a surface that has been designated the receptor binding surface (7). The hydrophobic patch is over 30 Å from these critical charged residues in monomeric SU (Fig. 6A, right image).
FIG. 6.
A model for the putative receptor binding site. All residues are represented as space-filled amino acids and are numbered according to the Mo MLV sequence starting with the N-terminal alanine on mature SU. Shaded residues represent those previously shown to be critical for receptor binding (D84, R83, and R95) and the residues in the hydrophobic patch (W142, W152, Y151, and Y210). (A) The left image shows the side view of SU. The middle image shows an enlargement of the hydrophobic patch from the same view as that shown in the left image. The arrow indicates the proposed binding site of tyrosine 235 in the receptor. The right image shows the top surface of the view seen in the left image. (B) The left image shows a diagram of the SU trimer model proposed by Fass and coworkers (6, 7); the right image shows a diagram of an alternative trimer model generated as described in Materials and Methods. (C) Alignment of the residues comprising the putative binding patch. Numbers to the right and left of the sequence indicate the amino acid positions beginning with the N-terminal residues of mature SU. Numbers above the sequence indicate the positions in Mo MLV. Boxes indicate the structural features in ecotropic MLV SU. Mo MLV, ecotropic Moloney MLV; Fr MLV, ecotropic Friend MLV; CasBrE, ecotropic MLV CasBrE; 4070A, amphotropic MLV; MCF, Mink cell focus-forming virus strain 1233; Xeno, xenotropic MLV strain NZB; GALV, gibbon ape leukemia virus strain SEATO.
Since the envelope protein is a trimer on virions, the question arises as to where the hydrophobic patch is located in the trimer with relation to the three critical charged residues. Although they did not show its structure, Fass and coworkers had reported generating a trimer packing model manually by placing carbohydrates toward the solvent, exposing variable sequences at the surface, and maximizing the contact area between SU monomers while avoiding steric clashes (7). The left image of Fig. 6B shows a view of this model from the side presumably facing the host cell (6). The hydrophobic patch is located at the center of the trimer, no closer to the charged residues than it is in monomer SU. Perhaps the initial interaction between SU and the receptor results in conformational changes that bring these two regions into proximity to form a site for a secondary interaction.
Because the model of Fass and coworkers made it difficult to reconcile the involvement of the charged residues with that of tryptophan 142 in virus entry and binding, we generated an alternative trimer model. We used the same criteria used by Fass and coworkers, except that we did not attempt to achieve maximal contact area between SU monomers but instead attempted to bring the three critical charged residues and the hydrophobic patch on adjacent monomers into proximity. We propose that the strong genetic evidence for involvement of these residues in virus binding makes this a reasonable modification of the criteria. In addition, maximization of the contact area between the SU fragments might not predict trimer conformation accurately since more than 200 residues of SU and all of the TM residues are absent from the crystal structure. The right panel of Fig. 6B shows a view of this alternative model from the same perspective as in the Fass model. The hydrophobic patch containing tryptophan 142 lies on the outside perimeter about 11 Å from aspartate 84 and 15 Å from arginine 83 on the adjacent monomer. This packing arrangement suggests a possible model: the shallow hydrophobic pocket binds the side chain of tyrosine 235 on the receptor, while arginine 83 and aspartate 84 in the adjacent monomer of SU interact with other critical receptor residues, possibly glutamate 237 or asparagine 232 and valine 233.
Both models have interesting similarities as well as some differences. Strikingly, both models place the hydrophobic patch at the interface between adjacent subunits. This brings up the intriguing possibility that the interaction between one patch on an envelope monomer and a single receptor molecule translates rapidly to the adjacent monomer, providing sufficient stimulus to trigger conformational changes in all subunits of the trimer. Notably, only the hydrophobic patch is adjacent to the monomer-monomer interfaces in the Fass model; the charged residues are distant from them. Both packing arrangements produce three variable lobes per trimer, each of which is made up of variable regions A and B from one monomer and variable region C from the adjacent monomer. However, the positions of the variable regions within a lobe were different because each monomer in the alternative model was turned counterclockwise and rotated around the x and y axes from its position in the Fass model to bring the hydrophobic patch as close to the charged residues as possible without producing steric clashes.
Davey, Zuo, and Cunningham recently proposed that serine 84, aspartate 86, and tryptophan 102 in Friend MLV (corresponding to serine 82, aspartate 84, and tryptophan 100 in Mo MLV) form the core of the binding site for both tyrosine 235 and glutamic acid 237 on the receptor (5). They based this proposal on three points: first, that binding and infection were reduced by certain changes in these residues (5, 13); second, that the three residues were adjacent on the surface of folded SU (7); and third, that the only critical receptor residues were tyrosine 235 and glutamic acid 237, from which erroneous assumption they concluded that any receptor binding site found on SU must be the site for these residues. However, Albritton and coworkers and Yoshimoto and coworkers had previously shown that at least two more receptor residues, asparagine 232 and valine 233, are involved in infection (1, 18). Moreover, Albritton and coworkers had shown that the presence of asparagine 232 and valine 233 in the receptor, in addition to tyrosine and glutamic acid, was critical for envelope protein binding (1). These analyses of the receptor indicated that the virus glycoprotein-receptor interaction consists of multiple contacts, suggesting that it involves a more extensive site on SU than the serine, aspartate, and tryptophan residues provide. More importantly, no evidence for a direct interaction between any of these three SU residues and tyrosine 235 or glutamic acid 237 was reported by Davey and coworkers (5). We propose instead that aspartate 84 on Mo MLV (aspartate 86 on Friend 57 MLV) interacts with a critical receptor residue(s) other than tyrosine 235, perhaps with asparagine 232 and valine 233, lysine 234, or possibly glutamic acid 237.
Davey, Zuo, and Cunningham also examined the effect of changes in Friend MLV at the residues corresponding to aspartate 135, phenylalanine 137, and tryptophan 142 examined in our study of Mo MLV. They found that replacement of aspartate 137 with alanine or of phenylalanine 139 with alanine in Friend MLV had no significant effect on virus entry (5), in agreement with our findings for the analogous D135A and F137A changes in Mo MLV. However, F139A did not result in detectable hyperglycosylation of SU in Friend MLV (5). Replacement of W144 with alanine markedly reduced infection (5), also in agreement with our observations for the analogous W142A change in Mo MLV. Davey and coworkers (5) did not examine other changes in that residue. Interestingly, replacing two other residues in the hydrophobic patch, tryptophan 154 and tyrosine 212, with alanine also reduced infection and produced virions lacking envelope protein (5). It is likely that alanine replacements of the corresponding residues, tryptophan 152 and tyrosine 210, in Mo MLV will give similar results. However, replacing them with other aromatic or hydrophobic residues might permit proper protein folding so that their role in virus entry might be examined. Replacement of the fourth member of the hydrophobic patch, tyrosine 153 (corresponding to tyrosine 151 in Mo MLV), with alanine did not affect virus infection or envelope protein incorporation (5). This last finding is not surprising since this residue is divergent among ecotropic MLV. For example, it is a tyrosine in Friend and Mo MLV, serine in strains AKV and RadLV, and alanine in strain CasBrE (10, 15, 16) (Fig. 6C).
A model for the specificity of receptor recognition.
Interestingly, three residues in the hydrophobic patch and the disulfide bridges that constrain it are conserved in the envelope proteins of other type C retroviruses (Fig. 6C). Why are these residues conserved in nonecotropic retroviruses? The most straightforward explanation is that their role in protein folding is essential. But their presence in the SU of nonecotropic MLV seems to imply that these other MLV should also bind the ecotropic receptor through an interaction with tyrosine 235. One possible explanation for this seeming paradox is that because tyrosine 235 is critical but not essential to virus binding and entry, an interaction with it alone is insufficient for virus attachment and entry. Several other retrovirus receptors contain critical but nonessential hydrophobic residues in their virus binding sites (14, 21). Perhaps the presence of the hydrophobic patch reflects a common solution for providing a binding site for the key hydrophobic receptor residues. If this is the case, then the specificity of the virus glycoprotein-receptor interaction would likely be dependent on additional contacts with divergent residues of SU adjacent to the hydrophobic patch. Without the contributions of these adjacent residues, the binding energy of any spurious interaction with a hydrophobic residue in a noncognate virus receptor would likely be too weak to proceed to entry.
ACKNOWLEDGMENTS
We thank Alan Rein for providing the anti-TM antiserum, Peter Kim for permission to show the Fass trimer model, Rob Davey for providing the coordinates for the Fass trimer model, and Nino Incardona for critical reading of the manuscript.
This work was supported by Public Health Service grant AI33410 from the National Institutes of Health (to L.M.A.).
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K L, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fass D. Ph.D. dissertation. Cambridge, Mass: Massachusetts Institute of Technology; 1997. [Google Scholar]
- 7.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 8.Guex N. Swiss-PdbViewer: a new fast and easy way to use PDB viewer for the Macintosh. Experientia. 1996;52:A26. [Google Scholar]
- 9.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 10.Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984;49:471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadan M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malhotra S, Scott A G, Zavorotinskaya T, Albritton L M. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J Virol. 1996;70:321–326. doi: 10.1128/jvi.70.1.321-326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merregaert J, Janowski M, Reddy E P. Nucleotide sequence of a radiation leukemia virus genome. Virology. 1987;158:88–102. doi: 10.1016/0042-6822(87)90241-8. [DOI] [PubMed] [Google Scholar]
- 16.Rassart E, Nelbach L, Jolicoeur P. Cas-Br-E murine leukemia virus: sequencing of the paralytogenic region of its genome and derivation of specific probes to study its origin and the structure of its recombinant genomes in leukemic tissues. J Virol. 1986;60:910–919. doi: 10.1128/jvi.60.3.910-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayle R A, Milner-White E J. RasMol: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 18.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zavorotinskaya T, Albritton L M. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. J Virol. 1999;73:5621–5629. doi: 10.1128/jvi.73.7.5621-5629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zavorotinskaya T, Albritton L M. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J Virol. 1999;73:5034–5042. doi: 10.1128/jvi.73.6.5034-5042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingler K, Belanger C, Peters R, Agard D, Young J A T. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J Virol. 1995;69:4261–4266. doi: 10.1128/jvi.69.7.4261-4266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]