Abstract
Hereditary spastic paraplegias (HSPs) a group of rare, clinically, and genetically heterogeneous disorders characterized by progressive degeneration of the corticospinal tract. Among these HSPs, SPG31 is due to autosomal dominant mutations in the receptor expression-enhancing protein 1 (REEP1) gene. Over 80 genes have been associated with HSPs, and the list is constantly growing as research progresses. This study is aimed to create a patient-derived human induced pluripotent stem cell (hiPSC) line with a specific nonsense mutation to better characterize the etiopathogenesis of the disease.
1. Introduction
Resource Table
| Unique stem cell line identifier | FSMi001-A |
|---|---|
| Alternative name(s) of stem cell line | FSM-SPG31-2 |
| Institution | Department of Neurobiology and Molecular Medicine, IRCCS Fondazione Stella Maris, Calambrone (PI), Italy |
| Contact information of distributor | Matteo Baggiani – matteo.baggiani@fsm.unipi.it |
| Devid Damiani – devid.damiani@fsm.unipi.it | |
| Type of cell line | hiPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC | Age: 47 years Sex: Female Ethnicity: Caucasian |
| Cell Source | Skin fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Episomal vectors encoding OCT4/sh-p53, SOX2/KLF4, EBNA-1, and L-MYC/LIN28 |
| Genetic Modification | NO |
| Type of Genetic Modification | None |
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | RT/qPCRs for plasmid-specific transcripts show no expression for reprogramming genes |
| Associated disease | SPG31 (Spastic Paraplegia 31) |
| Gene/locus | REEP1 (c.337C > T; p. Arg113*) |
| Date archived/stock date | 03 July 2013 |
| Cell line repository/bank | https://hspcreg.eu/cell-line/FSMi001-A |
| Ethical approval | Written informed consent was obtained from patient |
2. Resource utility
Generation of hiPSCs by means of somatic cell reprogramming from patients harboring different genetic variants offers an invaluable tool to understand disease mechanisms and explore novel treatment options for rescuing axonal defects and diverse cellular processes, especially in neurological diseases as SPG31 in which etiopathogenesis has been shown to be strictly dependent on the identity of mutations.
3. Resource details
Hereditary spastic paraplegias (HSPs) are inherited neurological disorders characterized by progressive degeneration of the corticospinal tract, leading to weakness and spasticity in the lower limb. Genetic alterations in more than 80 genes have been associated with HSPs (Elsayed et al., 2021). Among them, Spastic paraplegia type 31 (SPG31; MIM #610250) is an autosomal dominant form due to mutations in the REEP1 gene and characterized by lower limb spasticity, gait abnormalities, and muscle weakness. Most SPG31 patients are affected by a “pure” HSP form with onset between the second and the fourth decade of life. However, a not negligible percentage of the patients also display a “complex form”, presenting additional symptoms as sensory-motor peripheral neuropathy, bladder disturbances, white matter lesions, cerebellar ataxia, and dementia (Goizet et al., 2011, Toft et al., 2019). At the molecular level, several studies has involved the gene product in the regulation of lipid droplet metabolism and ER-mitochondrial membrane interface, with alteration of mitochondrial morphology, localization (Table 1), and functionality in case of heterozygous REEP1 mutations (Lavie et al., 2016, Naef et al., 2023).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
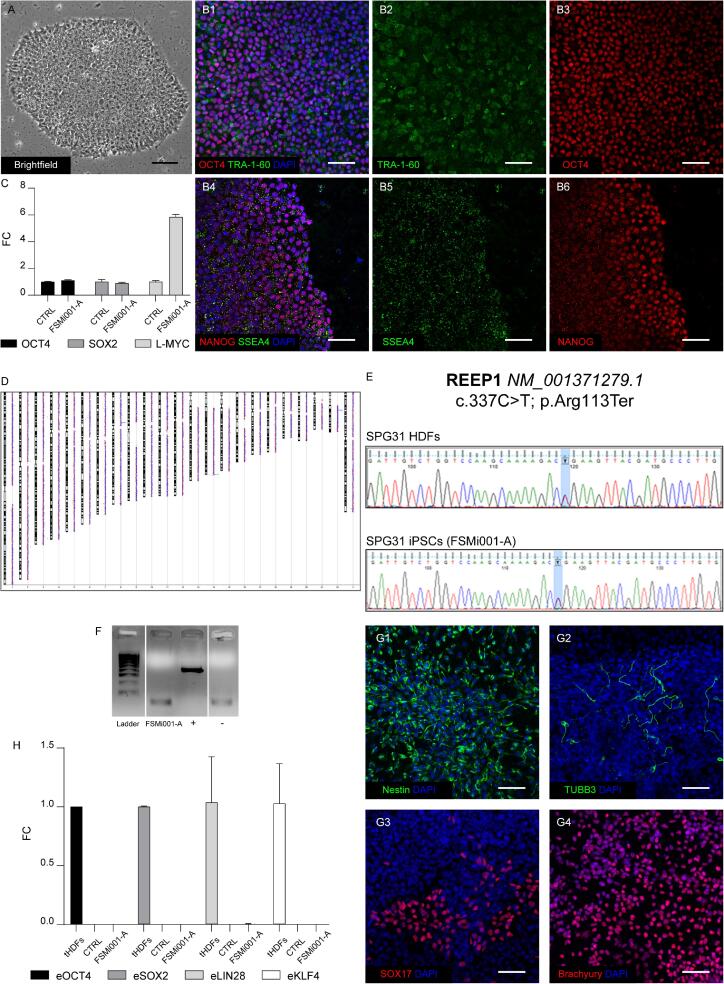
| Morphology | Photography Bright field | Normal morphology of human iPS cells | Fig. 1 A |
| Phenotype | Qualitative analysis by Immunocytochemistry |
hiPS cells express stemness markers: Oct4, Nanog, Tra 1–60, SSEA-4. | Fig. 1 B |
| Qualitative analysis by RT-qPCR |
Expression of stemness markers compared to ACS-1019 (CTRL) reference cell line/parental fibroblasts | Fig. 1 C | |
| Genotype | hPSC Genetic Analysis Kit; CGH array (60 kb) | No abnormalities detected. 46, XX. No abnormalities detected |
Fig. 1 D; Suppl. Fig. 1 B |
|
Identity |
Microsatellite PCR (mPCR) OR | Not performed | N/A |
| STR analysis | 16 markers tested- matched | submitted in archive with journal | |
|
Mutation analysis |
Sequencing | heterozygous, type of mutation. REEP1 (c.337C > T; p.Arg113Ter) | Fig. 1 E |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma detection by PCR | Negative | Fig. 1 F |
| Differentiation potential | Directed differentiation | Expression of layer-specific markers: Ectoderm (Nestin, β3- tubulin), Mesoderm (BRACHYURY) and Endoderm (SOX17). | Fig. 1 G |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
The purpose of this study was to generate a hiPSC line derived from a patient carrying a heterozygous nonsense mutation in exon 5 of REEP1, namely c.337C > T/(p. Arg113Ter). Our future goal is to use this line for in vitro differentiation in cortical tissue, in order to get a better characterization of etiopathogenesis of the disease, as previously done in other models of HSP (Damiani et al., 2024). A recent work has generated heterozygous and homozygous cell lines harboring a c.329-391del p.Lys112Serfs*54 variant in REEP1 by means of CRISPR-Cas9 technology (Korneck et al., 2024). Despite being very useful resources, this approach could not sufficient to fully describe the heterogeneity of the complex genetic background found in SPG31 patients. In particular, previous research identified numerous mutations hitting this gene. Insertions, deletions, and nucleotide changes were shown to result in frameshift, premature termination of translation, exon skipping and even changes in the 3′ UTR region, affecting the binding site of a known microRNAs, such as miR-140 and miR-691. In most cases, haploinsufficiency due to absence of protein produced by a single mutant allele seems to be the leading mechanism of the disease, as most terminating mutations are predicted to lead to nonsense-mediated decay. In other cases, variants of full length or truncated REEP1 protein could be produced, leading to alteration of subcellular targeting or even possible toxic gain-of-function. Most importantly, heterogeneity of genetic background in SPG31 patients directly reflects diversity in their symptomatology (Beetz et al., 2012, Beetz et al., 2008, Falk et al., 2014, Goizet et al., 2011, Hewamadduma et al., 2009, Maroofian et al., 2019, Toft et al., 2019, Züchner et al., 2006). In our case, the mutation was shown to result in haploinsufficiency due to nonsense-mediated decay of the transcript (Naef et al., 2023). Having received written informed consent, fibroblasts were derived and amplified from a skin-punch biopsy taken from a 47-year-old Italian woman. Cells were successively reprogrammed via episomal transfection of 3 plasmids encoding the classical Yamanaka factors. Pluripotent cell clones, emerging about 2 weeks after transfection, were picked, propagated as single cell lines, and passed at least 7 times before full immunocytochemical, molecular and genomic characterization. Among them, one clone was selected for further amplification and cryopreservation.
4. Materials and methods
4.1. Reprogramming of skin fibroblasts
A small sample of skin tissue was harvested from the patient via punch biopsy. Human dermal fibroblasts (HDFs) derived from the sample were cultured and amplified in HDF medium (DMEM High Glucose, 10 % Fetal Bovine Serum, L-Glutamine 2 mM, 100 U/ml PenStrep; Euroclone, Milan, Italy).
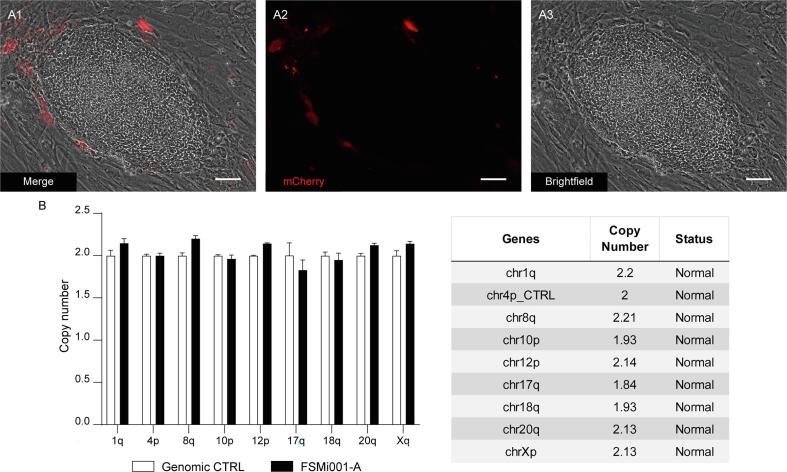
For reprogramming reaction, we followed a modified procedure based on protocols described in literature (Okita et al., 2013). HDFs were transfected with a mix of episomes for integration-free expression of human “Yamanaka factors” (L-MYC, LIN28, OCT3/4, SOX2, and KLF4). In particular, pCXLE-hOCT3/4-shp53-F + mCherry-2A-puro (gift from Ann Zovein, Addgene plasmid # 74947; Addgene, Watertown, MA), pCXLE-hUL (Addgene plasmid #27080), pCXWB-EBNA1 (Addgene, #37624), and pCXLE-hSK (Addgene, #27078, all gifts from Shinya Yamanaka) were nucleofected in a cell suspension via Amaxa Nucleofector 2b (Lonza, Tampa, FL), following manufacturer instructions. Transfected cells were plated on dishes coated with Geltrex (Thermo Fisher Scientific, Waltham, MA) and cultured with E6 containing 100 ng/ml FGF2 (named E7 medium), 0.5 mM sodium butyrate, 1 µM hydrocortisone. Successful transfection was checked after 2 days by positive expression of mCherry (Schmitt et al., 2017). To further increase the percentage of transfected cells, 0.5 µg/ml puromycin was added to the medium (PAC is encoded from Plasmid #74947). After 10 days, confluent cells were dissociated into single cell suspension with Trypsin (Euroclone), plated on Geltrex coated dishes at 5.000 cells/cm2, and cultured with E7 medium adding 0.5 mM sodium butyrate only for 2 days. hiPSC “islands” emerged after 14 days after transfection. At that time, medium was switched to Stemflex medium (Thermo Fisher Scientific) to grant iPSC survival and growth. hiPSC clones with uniform flat and round shaped morphology were picked in sterility conditions and propagated as single cell lines. To decrease the odds of integration of reprogramming sequences, clones were carefully checked to be completely free of mCherry-positive cells (Suppl Fig. 1A).
4.2. Culture of hiPSCs
hiPSCs were cultured at 37 °C and 5 % CO2 on Geltrex coated 6-well-plates in Stemflex medium, refreshing medium every other day. Cells were passed every 5–7 days with ReLeSR™ Passaging Reagent (Stem Cell Technologies, Vancouver, Canada), following manufacturer instruction (Fig. 1A).
Fig. 1.
4.3. Expression of stemness markers
Total RNA from hiPSCs and parental HDFs was extracted with All-prep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), and cDNA was synthesized with PrimeScript RT Reagent Kit (Takara Bio, Shiga, Japan). Relative expression of markers for the undifferentiated state OCT4, SOX2, and L-MYC was determined via RT-qPCR on a Mic qPCR Cycler (Bio Molecular Systems, Upper Coomera, Australia), normalizing values to expression of the established ACS-1019 hiPSC line (ATCC, VA), named CTRL, via 2−ΔΔCT method (Fig. 1 C). Human GAPDH was used as housekeeping gene. Moreover, to exclude the presence of episomes, the relative expression of eOCT4, eSOX2, eLIN28, and eKLF4 was measured via RT-qPCR on a Mic qPCR Cycler on tHDFs (transfected HDFs), CTRL, and FSMi001-A cell lines (Fig. 1 H). Primers used for qPCR reactions are listed in Table 2.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # | |
| Stemness Markers | Rabbit anti-OCT4 Rabbit anti-Nanog (D73G4) Mouse anti-TRA-1–60 Mouse anti-SSEA4 (clone MC813) |
1:500 1:200 1:500 1:500 |
Abcam Cat# ab19857 Cell Signaling Technology Cat# 4903 Cell Signaling Technology Cat # 4746 Cell Signaling Technology Cat #4755 |
| Differentiation Markers | Mouse anti-Nestin Mouse anti-Beta-3 Tubulin Rabbit anti-Brachyury (D2Z3J) Rabbit anti-SOX17 (D1T8M) |
1:500 1:1000 1:500 1:500 |
Abcam Cat# ab18102 Abcam Cat# ab7751 Cell Signaling Cat# 81694 Cell Signaling Cat# 81778 |
| Secondary antibodies | Goat Anti-Mouse Alexa Fluor 488 Goat Anti-Rabbit Alexa Fluor 555 |
1:500 1:500 |
Thermo Fisher Scientific Cat# a11029 Thermo Fisher Scientific Cat# a21429 |
| Primers | |||
| Target | Size of band | Forward/Reverse primer (5′-3′) | |
| Episomal Plasmids (qPCR) | eOCT4 eLIN28 eSOX2 eKLF4 |
70–150 bp 70–150 bp 70–150 bp 70– 150 bp |
Fwd: CAT TCA AAC TGA GGT AAG GG Rev: TAG CGT AAA AGG AGC AAC ATA G Fwd: AGC CAT ATG GTA GCC TCA TGT CCG C Rev: TAG CGT AAA AGG AGC AAC ATA G Fwd: TTC ACA TGT CCC AGC ACT ACC AGA Rev: TTT GTT TGA CAG GAG CGA CAA T Fwd: CCA CCT CGC CTT ACA CAT GAA GA Rev: TAG CGT AAA AGG AGC AAC ATA G |
| Stemness Markers (qPCR) | OCT4 L-MYC SOX2 |
70–150 bp 70–150 bp 70–150 bp |
Fwd: CCC CAG GGC CCC ATT TTG GTA CC Rev: ACC TCA GTT TGA ATG CAT GGG AGA GC Fwd: GCG AAC CCA AGA CCC AGG CCT GCT CC Rev: CAG GGG GTC TGC TCG CAC CGT GAT G Fwd: TTC ACA TGT CCC AGC ACT ACC AGA Rev: TCA CAT GTG TGA GAG GGG CAG TGT GC |
| House-Keeping Genes (qPCR) | GAPDH | 70–150 bp |
Fwd: GGA AGG ACT CAT GAC CAC AGT Rev: GGA TGA TGT TCT GGA GAG CCC |
| Genotyping | chr2:86,251,853–86,252,186 (spanning REEP1 Exon 5) | 334 bp |
Fwd: CCC AGC AAG AAC AAG GAT TG Rev: CCA CTG ATT GGT CCT TAG CC |
For immunofluorescence staining the hiPSCs were fixed with 4 % paraformaldehyde for 12 min at RT, permeabilized in PBS with 0.5 % Triton X-100 for 10 min at RT, blocked in PBS with 5 % Normal Goat Serum, 0.3 % Triton X-100 for 1 h at RT, and incubated with primary antibodies (Table 2) in PBS with 3 % Normal Goat Serum, 0.2 % Triton X-100 at 4 °C overnight. On the next day, the stained cells were washed 3 times with PBS and incubated in the same antibody solution with the corresponding secondary antibody (Table 2) for 1 h at RT. Nuclei were counterstained with DAPI (1 µg/ml; Merck, Darmstadt, Germany). All images were acquired with a Zeiss LSM 900 confocal microscope (Zeiss, Oberkochen, Germany) and processed with Fiji software.
4.4. Trilineage differentiation
With the purpose to have a formal demonstration of effective pluripotency of the generated cell line, we performed a series of differentiation protocols aimed to obtain the three different embryonic germ layers, namely ectoderm, endoderm, and mesoderm. To this end, hiPSCs were cultured in trilineage differentiation medium (DMEM High Glucose, Glutamax 2 mM, 100 U/ml PenStrep, MEM-NEAA 1x, β-mercaptoethanol 0.1 %; Thermo Fisher Scientific) to get spontaneous differentiation in neural progenitors (positive for neural stem cell marker Nestin, Fig. 1 G1), or neuronal marker β3-tubulin (Fig. 1 G2), as previously reported (Maria Turco et al., 2022). Modifying Lam et al., 2014 guidelines, to acquire an endodermal identity, 5 µM CHIR99021 (Stem Cell Technologies) for 1 day and then 100 ng/ml Activin A (Stem Cell Technologies,) for 3 days were added, generating cells positive to endodermal marker SOX17 (Fig. 1 G3). Instead, to induce mesodermal differentiation, 5 µM CHIR99021 (Stem Cell Technologies) was added for 2 days, obtaining cells positive to the mesodermal marker Brachyury (Fig. 1 G4). Medium was changed every day.
4.5. Mutation analysis
Genomic DNA from hiPSCs and their parental HDFs was extracted with All-prep DNA/RNA Mini Kit (Qiagen). The exon 5 of REEP1 was amplified by PCR (PCR primers are listed in Table 2). The identity of mutation was confirmed via Sanger sequencing (Fig. 1 E).
4.6. STR analysis
The identity of hiPSCs with parental HDFs was confirmed via gDNA typing with PowerPlex 16HS multiplex STR system (Promega, Madison, WI), including all 13 CODIS STR markers, Amelogenin for gender determination, Penta D and Penta E loci. The PCR products labeled with fluorescent dyes were detected with ABI-3500 Genetic Analyzer and data were analyzed with GeneMapper 5 (Thermo Fisher Scientific).
4.7. Genomic analysis via CGH array
High-resolution whole-genome array-based Comparative Genomic Hybridization (aCGH) analysis was conducted on genomic DNA extracted from patient derived hiPSCs, using the SurePrint G3 Human CGH Microarray 8 × 60 k (Agilent Technologies, Santa Clara, CA), a dual-color array containing 60-mer high-quality probes with 41 Kb genome-wide median probe spacing. Copy Number Variants (CNVs) were analyzed and mapped using the Human Genome Assembly GRCh37/hg19. Slides were scanned using an Agilent G2600D Microarray Scanner (Agilent Technologies) and processed using Feature Extraction software (v12.1.0.3). Agilent CytoGenomics software (v5.3.0.14) was used to analyze the results with default settings. Imbalances with at least three consecutive probes with abnormal log2 ratios for deletions and at least four consecutive probes for duplications were included into results. The Database of Genome Variants (https://dgv.tcag.ca/dgv/app/home), DECIPHER (https://www.deciphergenomics.org), UCSC genome browser (https://genome.ucsc.edu), Database of Human CNVs (https://gvarianti.homelinux.net/gvariantib37/index.php), SFARI (Simon’s Foundation Autism Research Initiative), and Gene Database (https://gene.sfari.org) databases were used in the interpretation of the results (Fig. 1D). Coincident results were obtained when the same experiment was performed on fibroblast patient’s genomic DNA.
Moreover, genomic instability was tested on SPG31 hiPSCs at P7 and was ruled out using hPSC Genetic Analysis Kit (Stem Cell Technologies) on 7500 Fast Real Time Pcr System (Thermo Fisher Scientific), capable of detecting the most common karyotypic abnormalities reported in human ES and iPS cells by comparing the copy number of loci in 9 different chromosomes via qPCR (Suppl Fig. 1B).
4.8. Mycoplasma testing
Absence of mycoplasma was assessed by PCR via Mycoplasma PCR Detection Kit (ABM, Richmond, Canada), according to the manufacturer instruction (Fig. 1 F).
Funding
This research was partly supported by RF-2019-12370112 (to AT), SG-2021-12375552 (to SM), and 2023 RC Linea 3 from Italian Ministry of Health, and Fondazione Telethon Grant GJC21131 (to FMS and DD). MB’s position is supported by the Fondazione Telethon grant GJC21131.
CRediT authorship contribution statement
Matteo Baggiani: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Filippo Maria Santorelli: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Funding acquisition, Data curation, Conceptualization. Serena Mero: Writing – review & editing, Visualization, Supervision, Investigation, Data curation. Flavia Privitera: Writing – review & editing, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Devid Damiani: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Alessandra Tessa: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2024.103472.
Contributor Information
Matteo Baggiani, Email: matteo.baggiani@fsm.unipi.it.
Filippo Maria Santorelli, Email: filippo.santorelli@fsm.unipi.it.
Devid Damiani, Email: devid.damiani@fsm.unipi.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
References
- Beetz C., Schüle R., Deconinck T., Tran-Viet K.-N., Zhu H., Kremer B.P.H., Frints S.G.M., van Zelst-Stams W.A.G., Byrne P., Otto S., Nygren A.O.H., Baets J., Smets K., Ceulemans B., Dan B., Nagan N., Kassubek J., Klimpe S., Klopstock T., Stolze H., Smeets H.J.M., Schrander-Stumpel C.T.R.M., Hutchinson M., van de Warrenburg B.P., Braastad C., Deufel T., Pericak-Vance M., Schöls L., de Jonghe P., Züchner S. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131:1078–1086. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetz C., Pieber T.R., Hertel N., Schabhüttl M., Fischer C., Trajanoski S., Graf E., Keiner S., Kurth I., Wieland T., Varga R.-E., Timmerman V., Reilly M.M., Strom T.M., Auer-Grumbach M. Exome sequencing identifies a REEP1 mutation involved in distal hereditary motor neuropathy Type V. Am. J. Hum. Genet. 2012;91:139–145. doi: 10.1016/j.ajhg.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D., Baggiani M., Della Vecchia S., Naef V., Santorelli F.M. Pluripotent Stem Cells as a Preclinical Cellular Model for Studying Hereditary Spastic Paraplegias. Int. J. Mol. Sci. 2024;25:2615. doi: 10.3390/ijms25052615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed L.E.O., Eltazi I.Z., Ahmed A.E., Stevanin G. Insights into Clinical, Genetic, and Pathological Aspects of Hereditary Spastic Paraplegias: A Comprehensive Overview. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.690899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J., Rohde M., Bekhite M.M., Neugebauer S., Hemmerich P., Kiehntopf M., Deufel T., Hübner C.A., Beetz C. Functional mutation analysis provides evidence for a role of REEP1 in lipid droplet biology. Hum. Mutat. 2014;35(4):497–504. doi: 10.1002/humu.22521. [DOI] [PubMed] [Google Scholar]
- Goizet C., Depienne C., Benard G., Boukhris A., Mundwiller E., Solé G., Coupry I., Pilliod J., Martin-Négrier M.-L., Fedirko E., Forlani S., Cazeneuve C., Hannequin D., Charles P., Feki I., Pinel J.-F., Ouvrard-Hernandez A.-M., Lyonnet S., Ollagnon-Roman E., Yaouanq J., Toutain A., Dussert C., Fontaine B., Leguern E., Lacombe D., Durr A., Rossignol R., Brice A., Stevanin G. REEP1 mutations in SPG31: Frequency, mutational spectrum, and potential association with mitochondrial morpho-functional dysfunction. Hum. Mutat. 2011;32:1118–1127. doi: 10.1002/humu.21542. [DOI] [PubMed] [Google Scholar]
- Hewamadduma C., McDermott C., Kirby J., Grierson A., Panayi M., Dalton A., Rajabally Y., Shaw P. New pedigrees and novel mutation expand the phenotype of REEP1-associated hereditary spastic paraplegia (HSP) Neurogenetics. 2009;10:105–110. doi: 10.1007/s10048-008-0163-z. [DOI] [PubMed] [Google Scholar]
- Korneck, M., Leonhardt, A., Schöls, L., Hauser, S., 2024. Generation of homozygous and heterozygous REEP1 knockout induced pluripotent stem cell lines by CRISPR/Cas9 gene editing. Stem Cell Res 2024 5, 77:103378. doi: 10.1016/j.scr.2024.103378. [DOI] [PubMed]
- Lam A.Q., Freedman B.S., Morizane R., Lerou P.H., Valerius M.T., Bonventre J.V. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J. Am. Soc. Nephrol. 2014;25:1211–1225. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie J., Serrat R., Bellance N., Courtand G., Dupuy J.-W., Tesson C., Coupry I., Brice A., Lacombe D., Durr A., Stevanin G., Darios F., Rossignol R., Goizet C., Bénard G. Mitochondrial morphology and cellular distribution are altered in SPG31 patients and are linked to DRP1 hyperphosphorylation. Hum. Mol. Genet. 2016;ddw425 doi: 10.1093/hmg/ddw425. [DOI] [PubMed] [Google Scholar]
- Maria Turco, E., Maria Giada Giovenale, A., Rotundo, G., Mazzoni, M., Zanfardino, P., Frezza, K., Torrente, I., Mary Carletti, R., Damiani, D., Santorelli, F.M., Luigi Vescovi, A., Petruzzella, V., Rosati, J., 2022. Generation and characterization of CSSi016-A (9938) human pluripotent stem cell line carrying two biallelic variants in MTMR5/SBF1 gene resulting in a case of severe CMT4B3. Stem Cell Res 65, 102946. Doi: 10.1016/j.scr.2022.102946. [DOI] [PubMed]
- Maroofian R., Behnam M., Kaiyrzhanov R., Salpietro V., Salehi M., Houlden H. Further supporting evidence for REEP1 phenotypic and allelic heterogeneity. Neurol Genet. 2019;5 doi: 10.1212/NXG.0000000000000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef V., Meschini M.C., Tessa A., Morani F., Corsinovi D., Ogi A., Marchese M., Ori M., Santorelli F.M., Doccini S. Converging Role for REEP1/SPG31 in Oxidative Stress. Int. J. Mol. Sci. 2023;24:3527. doi: 10.3390/ijms24043527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An Efficient Nonviral Method to Generate Integration-Free Human-Induced Pluripotent Stem Cells from Cord Blood and Peripheral Blood Cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Schmitt C.E., Morales B.M., Schmitz E.M.H., Hawkins J.S., Lizama C.O., Zape J.P., Hsiao E.C., Zovein A.C. Fluorescent tagged episomals for stoichiometric induced pluripotent stem cell reprogramming. Stem Cell Res Ther. 2017;8:132. doi: 10.1186/s13287-017-0581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft A., Birk S., Ballegaard M., Dunø M., Hjermind L.E., Nielsen J.E., Svenstrup K. Peripheral neuropathy in hereditary spastic paraplegia caused by REEP1 variants. J. Neurol. 2019;266:735–744. doi: 10.1007/s00415-019-09196-1. [DOI] [PubMed] [Google Scholar]
- Züchner S., Wang G., Tran-Viet K.-N., Nance M.A., Gaskell P.C., Vance J.M., Ashley-Koch A.E., Pericak-Vance M.A. Mutations in the Novel Mitochondrial Protein REEP1 Cause Hereditary Spastic Paraplegia Type 31. Am. J. Hum. Genet. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]