Abstract
Viral myocarditis affects an estimated 5 to 20% of the human population. The antiviral cytokine beta interferon (IFN-β) is critical for protection against viral myocarditis in mice. That is, nonmyocarditic reoviruses induce myocarditis in mice that lack IFN-α/β, and nonmyocarditic reoviruses both induce more IFN-β and are more sensitive to the antiviral effects of IFN-β than myocarditic reoviruses in primary cardiac myocyte cultures. Induction of IFN-β in certain cell types involves viral activation of the transcription factor interferon regulatory factor 3 (IRF-3). To address whether IRF-3 can induce IFN-β in cardiac myocytes, primary cardiac myocyte cultures and control L929 cells were transfected with a plasmid constitutively expressing IRF-3. Overexpression of IRF-3 resulted in induction of IFN-β in the absence of viral infection in both cell types. To address whether IRF-3 is required for viral induction of IFN-β, cell cultures were transfected with a plasmid constitutively expressing a dominant negative IRF-3 protein. The dominant negative IRF-3 reduced reovirus induction of IFN-β in control L929 cells and completely eliminated induction in primary cardiac myocyte cultures. This provides the first identification of a cardiac cellular factor required for viral induction of IFN-β and the first report of any cell type requiring IRF-3 for this response.
Viral myocarditis is an important human disease that affects an estimated 5 to 20% of the population (50). Viral myocarditis can be fatal in infants (4, 18) and, although usually resolved in older individuals, can progress to chronic myocarditis and/or dilated cardiomyopathy and cardiac failure (3, 17, 31). In recent years, antiviral agents have been tested in clinical trials in the hopes of improving disease outcome. Specifically, alpha interferon (IFN-α) and thymic agents that increase endogenous IFN levels and stimulate natural killer and T-cell activities have been shown to lower viral titer and improve cardiac function and survival rate in patients with viral myocarditis (13, 27, 28, 45). Neither of these therapies, however, completely restores cardiac function.
Enteroviruses (including coxsackieviruses) are the most frequently identified viruses associated with human myocarditis (17, 46); however, many other virus families have been implicated as well (19, 24, 39, 49). While enterovirus-induced myocarditis most likely reflects both immune system-mediated (6, 36) and direct cytopathic effects (5, 14), clinical studies of adenovirus (26) and human immunodeficiency virus (HIV)-associated (8) myocarditis suggest that the degree of cardiac inflammation does not correlate with the severity of cardiac dysfunction. Together, these data suggest that the innate response of cardiac cells to viral insult may be an important determinant of cardiac damage, yet the cardiac response to viruses remains largely unexplored.
Reovirus-induced myocarditis in mice is not mediated by the immune system and provides an excellent model for examining cardiac damage as a direct result of viral infection (42–44). Genes that encode viral core proteins involved in viral RNA synthesis are determinants of reovirus-induced myocarditis (41), and the rate of viral RNA synthesis in primary cardiac myocyte cultures correlates with viral myocarditic potential (40). In addition, myocarditic reoviruses spread through primary cardiac myocyte cultures more effectively and induce a greater cumulative cytopathic effect than nonmyocarditic reoviruses (40). One mechanism by which viral RNA synthesis can regulate viral spread is by induction of type I IFN (47). Indeed, nonmyocarditic reoviruses both induce more IFN-β and are more sensitive to the antiviral effects of IFN-α/β than myocarditic reoviruses in primary cardiac myocyte cultures (44). Moreover, a nonmyocarditic reovirus induces myocarditis in mice depleted of IFN-α/β (44).
IFN-β transcription is tightly regulated by the interactions of multiple positive and negative regulatory factors with the gene regulatory region (47). Positive regulatory domain III (PRDIII) contains an interferon regulatory factor (IRF)-binding element that can be bound by IRF-3 (38), a recently identified member of the IRF family (1). IRF-3 is constitutively expressed in all tissues examined thus far (1), thereby eliminating the need for de novo synthesis upon viral infection. Viral infection may (37) or may not (1) result in further induction of IRF-3, while IFN treatment does not induce expression of IRF-3 (1). In uninfected cells, IRF-3 is present in an autoinhibitory form (22). Viral infection can result in phosphorylation, activation, and homodimerization of IRF-3 (21, 22, 37, 48, 51). Such activation results in translocation of IRF-3 from the cytoplasm to the nucleus (37, 51), association with CREB binding protein (CBP) and/or p300 (37, 48, 51), and binding and induction of IFN-α and IFN-β genes (16, 37, 38, 51) and certain interferon-stimulated genes (ISGs) (1, 7, 32, 48).
What transcription factors are required for viral induction of IFN in the heart? Many viruses gain access to the heart, cardiac myocytes are not replenished, and yet the cardiac factors that mediate the cardiac IFN response to viral infection have not previously been investigated. Here, we examined the role of IRF-3 in reovirus induction of IFN-β in primary cardiac myocyte cultures. Our results provide the first identification of a transcription factor required for the cardiac IFN response to viruses and the first report, in any cell type, of a requirement for IRF-3 in viral induction of IFN-β.
MATERIALS AND METHODS
Cell cultures.
To generate primary cardiac myocyte cultures from Cr:NIH(S) mice (National Cancer Institute), term fetuses or 1-day-old neonates were sacrificed and the apical two-thirds of the hearts were removed, minced, and trypsinized (2). Cells were plated at a density of 1.25 × 106 cells per well in six-well clusters (Costar, Cambridge, Mass.) and incubated for 1.5 to 2 h to remove rapidly adherent cells (predominantly fibroblasts). The remaining cells (predominantly myocytes) were resuspended in Dulbecco’s modified Eagle medium (DMEM) (Gibco BRL, Gaithersburg, Md.) supplemented with 7% fetal calf serum (HyClone, Logan, Utah), 0.06% thymidine (Sigma Co., St. Louis, Mo.), and 10 μg of gentamicin (Sigma Co.) per ml. Primary cardiac myocyte cultures were plated at a density of 3.5 × 105 to 6 × 105 cells per well in 1 ml in 12-well tissue culture plates (Costar) and allowed to adhere for 1 day prior to transfection. Mouse L929 cells were maintained in suspension culture with minimal essential medium (MEM) (Gibco BRL) supplemented with 5% fetal calf serum (HyClone) and 2 mM l-glutamine (Gibco BRL). For transfection, cells were plated at 5 × 104 cells per well in 1 ml in 12-well tissue culture plates.
Plasmids.
pβLux was constructed by PCR to add HindIII restriction sites to bases 38 to 470 of the murine IFN-β regulatory region, containing all four PRDs (PRDI to IV) of pMUIFCAT-1200 (9) (generously provided by Hansjörg Hauser, Gesellschaft für Biotechnologische Forschung mbH, Braunschweig, Germany). The PCR fragment was gel purified and inserted into pGL3-Basic (containing the firefly luciferase gene but no promoter; Promega, Madison, Wis.) by using HindIII restriction sites. pGL3-Basic (lacking a promoter and therefore expressing baseline firefly luciferase), pGL3-Control (expressing firefly luciferase constitutively from a simian virus 40 promoter), and pRL-SV40 (expressing renilla luciferase constitutively from a simian virus 40 promoter) were all purchased (Promega). pEF-HAIRF-3 (expressing IRF-3) and pEF-HAIRF-358–427 (expressing a dominant negative IRF-3) contain the human EF-1α promoter for constitutive expression (30, 51) and were generously provided by Takashi Fujita (Kyoto University, Kyoto, Japan). pEF-BOS, a control plasmid, was constructed by removing IRF-3 from pEF-HAIRF-3 by EcoRI restriction digestion followed by ligation of the remaining fragment. pBOSLux was constructed by using PCR to add XbaI sites to the luciferase gene of pGL3-Control and then inserting the PCR product into pEF-BOS, using XbaI restriction sites. DNA was purified for transfection by using Qiagen’s maxiprep system (Qiagen Inc., Valencia, Calif.).
Transfection.
Transfection was performed 1 day postplating as previously described (33) by using FuGene6 according to the manufacturer’s protocol (Boehringer Mannheim/Roche Molecular Biomedicals, Indianapolis, Ind.). Unless noted otherwise in figures, the amount of DNA added to the indicated wells was as follows: pβLux, 1 μg; pGL3-Control, 0.5 μg; pRL-SV40, 0.02 μg; pEF-HAIRF-358–427, 6 μg; and pEF-HAIRF-3, 0.3 μg. FuGene6 was used in a volume equal to twice the total micrograms of plasmid DNA to be transfected per well (e.g., 2 μg of plasmid DNA per well required 4 μl of FuGene6 per well).
Infection.
Infection was performed 1 day posttransfection. L929 cells or primary cardiac myocyte cultures were washed twice with DMEM or MEM with supplements immediately prior to infection in order to remove residual FuGene6. The cells in two wells were trypsinized, and viable cells were counted by using trypan blue exclusion. Cells were infected with reovirus T3D (Dearing) at 25 PFU per cell in 300 μl of DMEM or MEM with supplements or were mock infected. After 1 h at 37°C in 5% CO2, 700 μl of MEM or DMEM with supplements was added. Cells were incubated at 37°C and 5% CO2 for 18 to 20 h.
Dual-luciferase assay.
The dual-luciferase assay was performed according to the manufacturer’s protocol (Promega) with the following exceptions: cells were washed twice with phosphate-buffered saline prior to the addition of lysis buffer, and cells were allowed to remain in lysis buffer at 4°C for at least 15 min, and then the surfaces of the wells were scraped with a Teflon cell lifter (Costar). Measurements were made by using a Lumat LB 9507 luminometer (EG&G Berthold, Oakridge, Tenn.) and autoinjection. Normalized luciferase activity was determined by dividing firefly luciferase activity by renilla luciferase activity.
Statistical analysis.
Statistical analysis was performed using a Student’s one-tailed t test and pooled variance. Results were considered significant at P of ≤0.05.
RESULTS
Reovirus induction of IFN-β can be monitored by using a luciferase reporter plasmid.
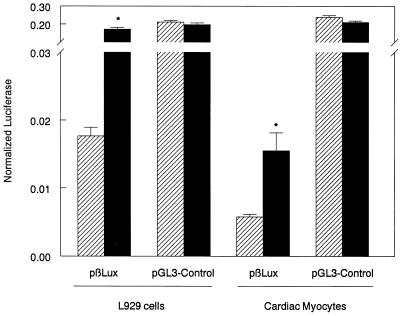
Using an IFN bioassay and reverse transcription-PCR, we previously demonstrated that reoviruses induce IFN-β in primary cardiac myocyte cultures between 10 and 20 h postinfection (44). To further investigate this induction and the cellular factors involved, a plasmid was constructed (pβLux) by inserting the murine IFN-β regulatory region upstream of a luciferase reporter gene. Transfection conditions were optimized previously (33), using a β-galactosidase reporter plasmid to demonstrate that myocytes in primary cardiac myocyte cultures were transfected (Fig. 1). L929 cells and primary cardiac myocyte cultures were transfected with pβLux. Cells were then virally or mock infected, and luciferase activity was analyzed 18 to 20 h postinfection. Viral infection induced pβLux but not pGL3-Control activity in both cell types (Fig. 2), confirming reovirus induction of IFN-β. Note that the lower level of induction in primary cardiac myocyte cultures most likely reflects the very low transfection efficiency achieved in these cultures (Fig. 1) (33). Addition of excess anti-IFN-α/β antibody did not affect viral induction of pβLux (data not shown), indicating that pβLux induction was a direct effect of the virus rather than an autocrine effect of virally induced IFN.
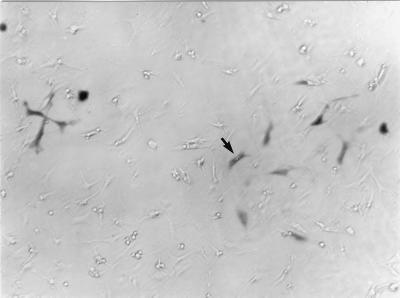
FIG. 1.
Myocytes in primary cardiac myocyte cultures are transfected. Primary cardiac myocyte cultures were transfected with a β-galactosidase reporter plasmid and stained for detection of β-galactosidase (33). The arrow indicates one example of a positively stained myocyte. Myocytes are distinguished from fibroblasts as described previously (2). Reprinted with permission from Roche Molecular Biochemicals.
FIG. 2.
Reovirus induces an IFN-β reporter construct, pβLux. L929 cells or primary cardiac myocyte cultures were transfected with the indicated plasmid and the normalization plasmid pRL-SV40 and infected 1 day posttransfection. The cells or cultures were mock infected (▨) or infected with reovirus T3D (■). Luciferase activity was measured 18 to 20 h postinfection. For each well, normalized luciferase activity was determined by dividing firefly luciferase activity by renilla luciferase activity. Each bar shows the mean of three wells (or four wells for L929 cells) (each error bar shows the standard error of the mean). Similar results were obtained in replicate experiments. Asterisks denote a significant increase between mock- and virus-infected cultures (for L929 cells, P < 0.001; for cardiac myocytes, P = 0.011).
Overexpression of IRF-3 can induce pβLux activity in the absence of reovirus infection.
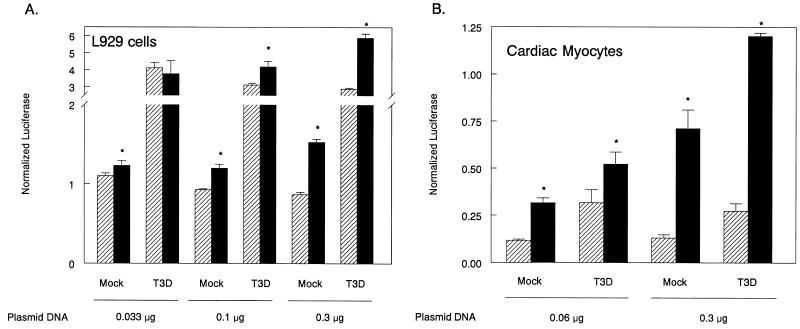
Overexpression of IRF-3 enhances viral induction of IFN-α and IFN-β in L929 and 293 cells (16, 51) and IFN-α in 3T3 cells (37) but fails to induce these genes in the absence of viral infection. In contrast, overexpression of IRF-3 induces IFN-β and IFN-α in REF cells independent of viral infection (16), suggesting that IRF-3 induction of type I IFN genes in the absence of viral infection may be cell type specific. To address whether IRF-3 can induce IFN-β in cardiac myocytes in the absence of viral infection, primary cardiac myocyte cultures and control L929 cells were transfected with a plasmid constitutively expressing IRF-3 or control DNA (pEF-BOS), infected, and harvested to assay pβLux activity as described above. IRF-3 overexpression increased pβLux activity in both L929 cells (Fig. 3A) and primary cardiac myocyte cultures (Fig. 3B) even in the absence of viral infection. In primary cardiac myocyte cultures, pβLux activity increased with increasing IRF-3 concentration. Synergy between IRF-3 overexpression and viral infection was detected with 0.3 μg of plasmid expressing IRF-3 in one experiment (Fig. 3B).
FIG. 3.
pβLux activity is induced in cells overexpressing IRF-3. L929 cells or primary cardiac myocyte cultures were transfected with pβLux, the normalization plasmid pRL-SV40, and the indicated amounts of either control DNA (pEF-BOS) (▨) or a plasmid expressing IRF-3 (■). Cells were infected 1 day posttransfection, and luciferase activity was measured 18 to 20 h postinfection. For each well, normalized luciferase activity was determined by dividing firefly luciferase activity by renilla luciferase activity. Each bar shows the mean of three wells (each error bar shows the standard error of the mean). Similar results were obtained in replicate experiments. Asterisks denote significant increases between cultures transfected with control DNA or IRF-3 (for L929 cells, from left to right, P = 0.046, 0.009, 0.002, <0.001, and < 0.001; for cardiac myocytes, from left to right, P = 0.001, 0.049, 0.002, and <0.001).
IRF-3 regulates reovirus induction of IFN-β in mouse L929 cells.
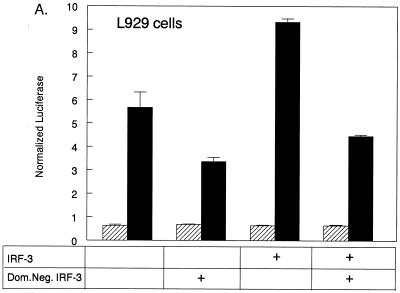
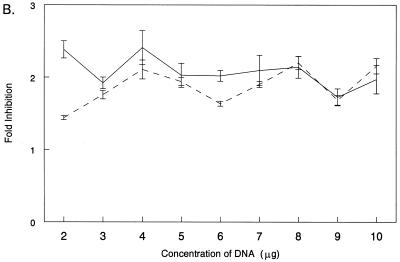
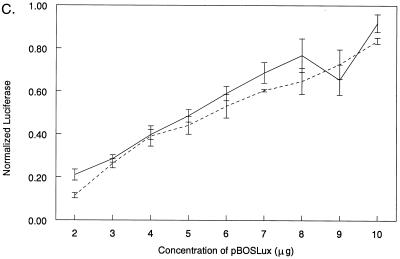
To investigate the role of IRF-3 in reovirus induction of IFN-β, L929 cells were transfected with pβLux and either control plasmid DNA (pEF-BOS) or a construct expressing a dominant negative IRF-3 protein (51). The dominant negative protein retains functional binding of the transcriptional cofactors CBP and p300 but lacks the DNA-binding domain and thus competes with endogenous IRF-3 for cellular CBP and p300. Overexpression of dominant negative IRF-3 using 6 μg of plasmid DNA partially inhibited viral induction of pβLux in L929 cells. The effect of the dominant negative protein on IRF-3 was confirmed by demonstrating that the dominant negative protein could reverse enhancement of viral induction of pβLux mediated by transfected IRF-3 (Fig. 4A). To test whether increasing quantities of dominant negative IRF-3 could result in full inhibition of viral induction of pβLux, the amount of dominant negative plasmid was increased in 1-μg increments from 2 to 10 μg. There was equivalent inhibition across the concentration range (Fig. 4B). To demonstrate that these higher plasmid concentrations resulted in increased protein expression, L929 cells were transfected with pBOSLux, a reporter constructed with luciferase, instead of dominant negative IRF-3, downstream of the EF-1α promoter. Increasing amounts of pBOSLux resulted in increasing luciferase expression across the concentration range (Fig. 4C). These results indicate that despite an apparent excess of dominant negative IRF-3, there was only partial inhibition of reovirus induction of IFN-β in L929 cells. Thus, while IRF-3 regulates reovirus induction of IFN-β in L929 cells, L929 cells may have other pathways for IFN-β induction in the absence of IRF-3.
FIG. 4.
IRF-3 is not required for but regulates pβLux induction in L929 cells. (A) L929 cells were transfected with pβLux, the normalization plasmid pRL-SV40, the indicated plasmid (IRF-3 and/or Dom.Neg. IRF-3), and/or control DNA (pEF-BOS) for a constant plasmid DNA concentration of 7.32 μg per well. Cells were infected 1 day posttransfection, and luciferase activity was measured 18 to 20 h postinfection. The cells were mock infected (▨) or infected with reovirus T3D (■). For each well, normalized luciferase activity was determined by dividing firefly luciferase activity by renilla luciferase activity. Each bar shows the mean of three wells (each error bar shows the standard error of the mean). (B) L929 cells were transfected with pβLux, the normalization plasmid pRL-SV40, and the indicated amounts of control DNA (pEF-BOS) or a plasmid expressing a dominant negative IRF-3 protein. Cells were infected, and luciferase activity was measured as described above. Fold inhibition was calculated by dividing normalized luciferase activity from virally infected cells transfected with control DNA by those transfected with dominant negative IRF-3. Each datum point is expressed as the average fold inhibition of three wells ± standard error of the mean. The results of two separate experiments are shown by the two lines, with average values of viral induction of pβLux of 9- and 11-fold. (C) L929 cells were transfected with the normalization plasmid pRL-SV40 and the indicated amounts of pBOSLux, mock infected, and harvested for luciferase as described above. For two separate experiments (depicted by the two lines), each datum point is expressed as 103 times the mean of three wells ± standard error of the mean.
IRF-3 is required for reovirus induction of IFN-β in primary cardiac myocyte cultures.
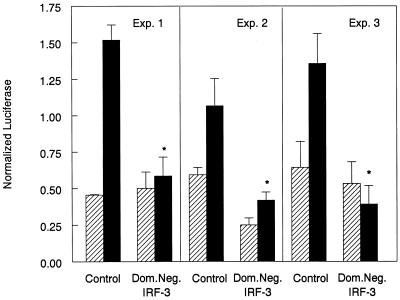
Primary cardiac myocyte cultures were transfected with pβLux and either control plasmid DNA (pEF-BOS) or a construct expressing a dominant negative IRF-3 protein (Fig. 5). Transfection with the dominant negative IRF-3 inhibited reovirus induction of pβLux with no significant effect on mock-infected baseline activity. Moreover, the dominant negative IRF-3 completely inhibited viral induction of pβLux activity to levels comparable to those of mock-infected cultures. Replicate experiments provided similar statistical results (Fig. 5), demonstrating that IRF-3 is required for reovirus induction of IFN-β in primary cardiac myocyte cultures.
FIG. 5.
IRF-3 is required for reovirus induction of pβLux in primary cardiac myocyte cultures. Primary cardiac myocyte cultures were transfected with pβLux, the normalization plasmid pRL-SV40, and control DNA (pEF-BOS) or a plasmid expressing a dominant negative (Dom.Neg.) IRF-3 protein. Cells were infected 1 day posttransfection, and luciferase activity was measured 18 to 20 h postinfection. The cells were mock infected (▨) or infected with reovirus T3D (■). Results from three independent experiments are shown. For each well, normalized luciferase activity was determined by dividing firefly luciferase activity by renilla luciferase activity. Each bar shows the mean of three wells (each error bar shows the standard error of the mean). Asterisks denote no significant difference between virus- and mock-infected cultures (P > 0.05 for all three experiments).
DISCUSSION
We have demonstrated that IRF-3 is required for reovirus induction of IFN-β in primary cardiac myocyte cultures, providing the first identification of a cardiac cellular factor required for viral induction of IFN-β and the first report in any cell type that IRF-3 is required for viral induction of IFN-β.
IRF-3 overexpression can induce IFN-β in the absence of viral infection.
IRF-3 overexpression induced IFN-β in the absence of viral infection in both L929 cells and primary cardiac myocyte cultures (Fig. 3). Our observation for L929 cells differs from previous reports (51); however, this may reflect transfection conditions. Indeed, when the total DNA concentration (including control plasmids) was increased, resulting in decreased transfection efficiency (data not shown), IRF-3 did not induce IFN-β in the absence of viral infection (Fig. 4A). The mechanism by which overexpression of IRF-3 results in activation of IFN-β in the absence of viral infection is unclear, but similar virus-independent effects have been seen for IRF-7 (25). It is possible that IRF-3 is minimally functional in the absence of viral activation and that overexpression allows detection of this low level of activity. Alternatively, IRF-3 may be inefficiently activated through a virus-independent mechanism(s), but such activation would be detectable only with IRF-3 overexpression.
IRF-3 regulates reovirus induction of IFN-β in L929 cells.
Overexpression of a dominant negative IRF-3 resulted in only partial inhibition of viral induction of IFN-β in L929 cells. Moreover, this partial inhibition was not increased when the dominant negative plasmid concentration was increased fivefold (Fig. 4B). These data suggest that reovirus induction of IFN-β in L929 cells uses both IRF-3-dependent and -independent pathways, although alternative interpretations of the data are possible. It is possible that endogenous IRF-3 or cofactors that interact with IRF-3 are expressed at higher levels in L929 cells than in cardiac myocytes, generating a requirement for greater concentrations of dominant negative IRF-3 in the former than in the latter cells. The constant fold inhibition in L929 cells despite increasing plasmid concentrations however, would argue that the dominant negative IRF-3 concentration is sufficient regardless of IRF-3 and IRF-3 cofactor levels. Moreover, transfection with higher concentrations of pBOSLux, expressed from the same promoter as the dominant negative IRF-3, resulted in higher normalized luciferase values (Fig. 4C), indicating that larger amounts of plasmid did increase protein expression. Together, the data demonstrate that increasing expression of dominant negative IRF-3 by fivefold in L929 cells does not increase inhibition of viral induction of pβLux, indicating that the amount of dominant negative IRF-3 is not limiting in the assay. In sum, the data suggest that reovirus induction of IFN-β in L929 cells can use IRF-3-independent pathways.
IRF-3 is required for reovirus induction of IFN-β in primary cardiac myocyte cultures.
Overexpression of a dominant negative IRF-3 resulted in complete inhibition of viral induction of IFN-β in primary cardiac myocyte cultures (Fig. 5). Previous characterization of the dominant negative IRF-3 demonstrated that, as for wild-type IRF-3, it is phosphorylated at the C terminus, translocates to the nucleus, and binds CBP and/or p300 (51). The N-terminal deletion, however, prevents DNA binding and therefore provides dominant negative function. Our data confirm that the dominant negative IRF-3 functions to inhibit IRF-3 (Fig. 4A). Could the dominant negative IRF-3 also inhibit IRF-1 or IRF-2, which can mediate viral regulation of IFN-β (10–12, 34), and could the full inhibition in primary cardiac myocyte cultures reflect this pleiotropic effect? IRF-1, which can induce IFN-β in other cell types (10, 23, 29), is not required for reovirus induction of IFN-β in primary cardiac myocyte cultures (unpublished data), indicating that the inhibitory effect of the dominant negative IRF-3 could not be due to interactions with IRF-1. IRF-2 represses IFN-β induction in other cell types (12, 23) and in primary cardiac myocyte cultures (unpublished data), indicating again that the dominant negative IRF-3 effects could not be mediated through this factor. Together, the data indicate that IRF-3 is required for reovirus induction of IFN-β in primary cardiac myocyte cultures. This provides the first identification of a cardiac cellular factor required for viral induction of IFN-β and the first report of any cell type requiring IRF-3 for this cell response. Moreover, given our evidence here that L929 cells may use IRF-3-independent pathways, the data suggest that cardiac myocytes may be uniquely dependent on IRF-3.
We previously demonstrated that nonmyocarditic reoviruses induce more IFN-β than do myocarditic reoviruses in primary cardiac myocyte cultures (44). This raises the intriguing possibility that myocarditic reoviruses differ from nonmyocarditic reoviruses in their activation or suppression of IRF-3. Recent investigations suggest that many viruses have mechanisms for interfering with IRF-3-mediated induction of IFN. Adenovirus has been implicated in cases of viral myocarditis (26) but mediates cardiac damage through an undetermined mechanism. Given that the heart requires IRF-3 for viral induction of IFN-β (this report) and that adenovirus protein E1A binds CBP and/or p300, thereby competing with IRF-3 for transcriptional cofactors required for induction of both IFN and ISG transcription (1, 16), it is possible that the mechanism of cardiac damage by adenovirus could be linked to its ability to interfere with the function of IRF-3. HIV Tat protein binds CBP and/or p300 to activate HIV transcription (15) and potentially competes with cellular IRF-3 for transcription cofactors. Although the impact on IFN induction has not been determined, expression of a dominant negative IRF-3 can inhibit the expression of the β chemokine RANTES (20) thereby lessening the cell’s innate antiviral response to HIV. Finally, the E6 oncoprotein of human papillomavirus 16 directly binds IRF-3 and interferes with the ability of Sendai virus to induce IFN-β (35). Our future investigations will compare IRF-3 activation by nonmyocarditic and myocarditic reoviruses.
We previously demonstrated that nonmyocarditic reoviruses are more sensitive to the antiviral effects of IFN-β (44). IFN induction of ISGs, which provide the antiviral effector functions, is mediated by activation of the JAK-STAT pathway (47). IRF-3, however, can directly induce ISGs (1, 7, 48) even in the absence of IFN (32). Therefore, it is possible that the differences in sensitivity to IFN-β between nonmyocarditic and myocarditic reoviruses are due, in part, to a difference in IRF-3 activation. Again, our future investigations will address this possibility.
Our results indicate that cardiac myocytes are dependent on IRF-3 for viral induction of IFN-β. Previous evidence indicates that the heart expresses a higher constitutive level of another IRF, IRF-1, than other tissues examined (29), but the effects of such expression remain unexamined. Together, these data suggest that the heart may provide a unique environment for IRF function. Further investigation of the cardiac IRF roles in viral induction of IFN-β will provide essential insight for more-effective IFN-based therapies for patients with viral myocarditis.
ACKNOWLEDGMENTS
We thank Kathleen Azzam for invaluable assistance and discussions.
This research was supported in part by NIH grant 1 R01 HL57161, grant 204743 from the North Carolina State University College of Veterinary Medicine, and a GAANN fellowship and North Carolina State University College of Veterinary Medicine stipend to D. L. Noah.
REFERENCES
- 1.Au W C, Moore P A, Lowther W, Juang Y T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baty C J, Sherry B. Cytopathogenic effect in cardiac myocytes but not in cardiac fibroblasts is correlated with reovirus-induced acute myocarditis. J Virol. 1993;67:6295–6298. doi: 10.1128/jvi.67.10.6295-6298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowles N E, Richardson P J, Olsen E G J, Archard L C. Detection of coxsackie B virus-specific RNA sequences in myocardial biopsies from cases of myocarditis and dilated cardiomyopathy. Lancet. 1986;i:1120–1122. doi: 10.1016/s0140-6736(86)91837-4. [DOI] [PubMed] [Google Scholar]
- 4.Cherry J D. Enteroviruses. In: Remington J S, Klein J O, editors. Infectious diseases of the fetus and newborn infant. 4th ed. Philadelphia, Pa: W. B. Saunders; 1995. pp. 404–446. [Google Scholar]
- 5.Chow L H, Beisel K W, McManus B M. Enteroviral infection of mice with severe combined immunodeficiency. Evidence for direct viral pathogenesis of myocardial injury. Lab Investig. 1992;66:24–31. [PubMed] [Google Scholar]
- 6.Cook D N, Beck M A, Coffman T M, Kirby S L, Sheridan J F, Pragnell I B, Smithies O. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science. 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 7.Daly C, Reich N C. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem. 1995;270:23739–23746. doi: 10.1074/jbc.270.40.23739. [DOI] [PubMed] [Google Scholar]
- 8.De Castro S, D’Amati G, Gallo P, Cartoni D, Santopadre P, Vullo V, Cirelli A, Migliau G. Frequency of development of acute global left ventricular dysfunction in human immunodeficiency virus infection. J Am Coll Cardiol. 1994;24:1018–1024. doi: 10.1016/0735-1097(94)90864-8. [DOI] [PubMed] [Google Scholar]
- 9.Dirks W, Mittnacht S, Rentrop M, Hauser H. Isolation and functional characterization of the murine interferon-beta 1 promoter. J Interferon Res. 1989;9:125–133. doi: 10.1089/jir.1989.9.125. [DOI] [PubMed] [Google Scholar]
- 10.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of the endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 11.Fujita T, Sakakibura J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1 mediating induction and silencing properties to human IFN-beta gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 13.Heim A, Stille-Siegner M, Kandolf R, Kreuzer H, Figulla H R. Enterovirus-induced myocarditis: hemodynamic deterioration with immunosuppressive therapy and successful application of interferon-alpha. Clin Cardiol. 1994;17:563–565. doi: 10.1002/clc.4960171010. [DOI] [PubMed] [Google Scholar]
- 14.Herzum M, Ruppert V, Kuytz B, Jomaa H, Nakamura I, Maisch B. Coxackievirus B3 infection leads to cell death of cardiac myocytes. J Mol Cell Cardiol. 1994;26:907–913. doi: 10.1006/jmcc.1994.1108. [DOI] [PubMed] [Google Scholar]
- 15.Hottinger M O, Nabel G J. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juang Y-T, Lowther W, Kellum M, Au W-C, Lin R, Hiscott J, Ptha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandolph R, Klingel K, Mertsching H, Canu A, Hohenadl C, Zell R, Reimann Y, Heim A, McManus B M, Foulis A K, Schultheiss H-P, Erdmann E, Riecker G. Molecular studies on enteroviral heart disease: patterns of acute and persistent infections. Eur Heart J. 1991;12:49–55. doi: 10.1093/eurheartj/12.suppl_d.49. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan M H, Klein S W, McPhee J, Harper R G. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev Infect Dis. 1983;5:1019–1032. doi: 10.1093/clinids/5.6.1019. [DOI] [PubMed] [Google Scholar]
- 19.Leslie K, Blay R, Haisch C, Lodge A, Weller A, Huber S A. Clinical and experimental aspects of viral myocarditis. Clin Microbiol Rev. 1989;2:191–203. doi: 10.1128/cmr.2.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteosome-mediated degredation. Mol Cell Biol. 1998;19:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivating and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. Mutational analysis of the interferon regulatory factors 1 and 2. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 24.Marboe C C, Fenoglio J J. Pathology and natural history of human myocarditis. Pathol Immunopathol Res. 1988;7:226–239. doi: 10.1159/000157119. [DOI] [PubMed] [Google Scholar]
- 25.Marie I, Durbin J, Levy D E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin A, Webber S, Fricker F, Jaffe R, Demmler G, Kearney D, Zhang Y-H, Bodurtha J, Gelb B, Ni J, Bricker T, Towbin J A. Acute myocarditis: rapid diagnosis by PCR in children. Circulation. 1994;90:330–339. doi: 10.1161/01.cir.90.1.330. [DOI] [PubMed] [Google Scholar]
- 27.Miric M, Miskovic A, Vasiljevic J D, Keserovic N, Pesic M. Interferon and thymic hormones in the therapy of human myocarditis and idiopathic dilated cardiomyopathy. European Heart J. 1995;16:150–152. doi: 10.1093/eurheartj/16.suppl_o.150. [DOI] [PubMed] [Google Scholar]
- 28.Miric M, Vasiljevic J, Bojic M, Popovic Z, Keserovic N, Pesic M. Long-term follow up of patients with dilated heart muscle disease treated with human leucocytic interferon alpha or thymic hormones. Initial results. Heart. 1996;75:596–601. doi: 10.1136/hrt.75.6.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that binds specifically to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morimoto S-I, Yamada K, Kubo N, Mizuno Y, Hasumi M, Hiramitsu S, Uemura A, Kimura K, Nishikawa T, Sekiguchi M. Clinical and pathological features of chronic myocarditis: four autopsy cases presenting as dilated cardiomyopathy in life. Am J Cardiovasc Pathol. 1992;4:181–191. [PubMed] [Google Scholar]
- 32.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomeglalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noah D L, Blum M A, Sherry B. Transfection of primary cardiac myocyte cultures with DNA and anti-sense oligonucleotides using FuGene6 transfection reagent. Biochemica. 1998;2:38–40. [Google Scholar]
- 34.Reis L F, Harada H, Wolchok J D, Taniguchi T, Vilcek J. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose N R, Hill S L. The pathogenesis of postinfectious myocarditis. Clin Immunol Immunopathol. 1996;80:S92–S99. doi: 10.1006/clin.1996.0146. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-β gene. FEBS Lett. 1998;452:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 38.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 39.See D M, Tilles J G. Viral myocarditis. Rev Infect Dis. 1991;13:951–956. doi: 10.1093/clinids/13.5.951. [DOI] [PubMed] [Google Scholar]
- 40.Sherry B, Baty C J, Blum M A. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J Virol. 1996;70:6709–6715. doi: 10.1128/jvi.70.10.6709-6715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherry B, Blum M A. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J Virol. 1994;68:8461–8465. doi: 10.1128/jvi.68.12.8461-8465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherry B, Li X, Tyler K L, Cullen J M, Virgin H W. Lymphocytes protect against and are not required for reovirus-induced myocarditis. J Virol. 1993;67:6119–6124. doi: 10.1128/jvi.67.10.6119-6124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherry B, Schoen F J, Wenske E, Fields B N. Derivation and characterization of an efficiently myocarditic reovirus variant. J Virol. 1989;63:4840–4849. doi: 10.1128/jvi.63.11.4840-4849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherry B, Torres J, Blum M A. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditic and are determined by viral core proteins. J Virol. 1998;72:1314–1323. doi: 10.1128/jvi.72.2.1314-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stille-Sieggener M, Heim A, Figulla H R. Subclassification of dilated cardiomyopathy and interferon treatment. Eur Heart J. 1995;16:147–149. doi: 10.1093/eurheartj/16.suppl_o.147. [DOI] [PubMed] [Google Scholar]
- 46.Tracy S, Wiegand V, McManus B, Gauntt C, Pallansch M, Beck M, Chapman N. Molecular approaches to enteroviral diagnosis in idiopathic cardiomyopathy and myocarditis. J Am Coll Cardiol. 1990;15:1688–1694. doi: 10.1016/0735-1097(90)92846-t. [DOI] [PubMed] [Google Scholar]
- 47.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–400. [Google Scholar]
- 48.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wenger N K, Abelmann W H, Roberts W C. Myocarditis. In: Hurst J W, Schlant R C, editors. The heart, arteries, and veins. New York, N.Y: McGraw-Hill Book Co.; 1990. pp. 1256–1277. [Google Scholar]
- 50.Woodruff J F. Viral myocarditis: a review. Am J Pathol. 1980;101:427–479. [PMC free article] [PubMed] [Google Scholar]
- 51.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor containing complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]