Abstract
Background and Aims
Hepatocellular carcinoma (HCC) incidence is increasing and correlated with metabolic dysfunction-associated steatotic liver disease (MASLD; formerly nonalcoholic fatty liver disease), even in patients without advanced liver fibrosis who are more likely to be diagnosed with advanced disease stages and shorter survival time, and less likely to receive a liver transplant. Machine learning (ML) tools can characterize large datasets and help develop predictive models that can calculate individual HCC risk and guide selective screening and risk mitigation strategies.
Methods
Tableau and KNIME Analytics were used for descriptive analytics and ML tasks. ML models were developed using standard laboratory and clinical parameters. Sci-kit learn algorithms were used for model development. Data from University of California (UC), Davis, were used to develop and train a pilot predictive model, which was subsequently validated in an independent dataset from UC San Francisco. MASLD and HCC patients were identified by International Classification of Diseases-9/10 codes.
Results
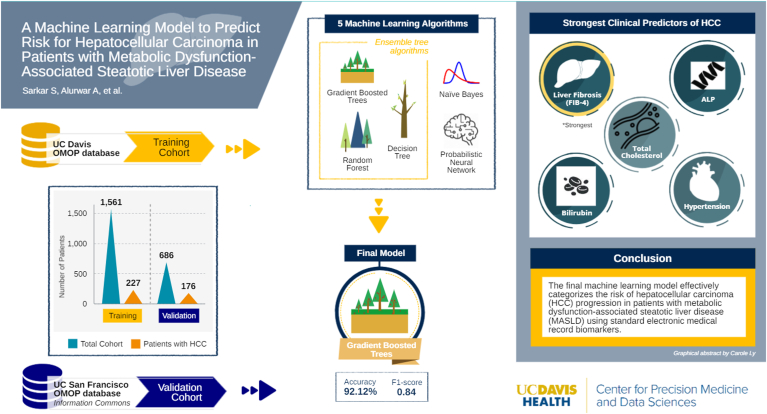
Of the patients diagnosed with MASLD (n = 1561 training; n = 686 validation), HCC developed in 14% (n = 227) of the UC Davis training cohort and 25% (n = 176) of the UC San Francisco validation cohort. Liver fibrosis determined by the noninvasive Fibrosis-4 score was the strongest single predictor for HCC in the model. Using the validation cohort, the model predicted HCC development at 92.06% accuracy with an area under the curve of 0.97, F1-score of 0.84, 98.34% specificity, and 74.41% sensitivity.
Conclusion
ML models can aid physicians in providing early HCC risk assessment in patients with MASLD. Further validation will translate to cost-effective, personalized care of at-risk patients.
Keywords: Machine Learning, Hepatocellular Carcinoma, Fatty Liver Disease, Metabolic Dysfunction-Associated Steatotic Liver Disease, Artificial Intelligence
Introduction
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-related deaths worldwide and is associated with chronic liver diseases of which nonalcoholic fatty liver disease (NAFLD) is a leading etiology.1, 2, 3, 4, 5 Due to the potential stigma associated with the term and its limitation in capturing the full range of disease etiologies, a new term—metabolic dysfunction-associated steatotic liver disease (MASLD)—has recently been proposed to replace NAFLD.6 The spectrum of MASLD encompasses the often benign and reversible condition of steatotic liver disease; and the more serious manifestation of metabolic dysfunction-associated steatohepatitis characterized by liver inflammation and damage that may advance to fibrosis, cirrhosis, and HCC. MASLD reportedly occurs in approximately 25% of the United States and global population1,7, 8, 9, 10 and is becoming the fastest-growing cause of HCC,11,12 driven in part by the rising worldwide prevalence of obesity and diabetes.1,2,12, 13, 14, 15
Typically asymptomatic, MASLD tends to be underdiagnosed, and its stealthy progression to HCC can go undetected until it is too late for intervention.12 MASLD patients who develop HCC are often diagnosed at more advanced disease stages and with shorter survival time and are less likely to receive a liver transplant than patients with other types of chronic liver disease leading to HCC.4,5,11 Furthermore, although HCC commonly develops in the presence of advanced fibrosis and cirrhosis, an estimated 20%–50% of all MASLD-related HCC cases arise in non-cirrhotic livers compared to 14% of HCC cases in other liver diseases,3,11,12,16 suggesting that the pathogenesis of HCC in non-cirrhotic livers occur by factors related to the pathogenesis of MASLD that are independent of the progression to advanced fibrosis and cirrhosis.11,12 In fact, many risk factors for MASLD are also independently associated with HCC.11 Currently, guidelines for HCC surveillance are informed primarily by histological evidence of cirrhosis and occasionally inclusive of level 3 advanced fibrosis.5,11 There is no consensus on screening or surveilling MASLD patients without advanced fibrosis/cirrhosis4,11 even in the presence of other determinants of HCC risk. The absence of HCC screening protocols for patients with MASLD without advanced fibrosis or cirrhosis further contributes to their late diagnosis and disease management.12
While universal biopsy screening for all MASLD patients is neither feasible nor advised, noninvasive prognostic models that can selectively identify MASLD patients who are at significant risk of developing HCC can help enable personalized screening strategies.17 Such tools could potentially facilitate cost-effective care and optimize clinical outcomes through early detection. Machine learning (ML)-based predictive/prognostic models have to date been used successfully to identify the likelihood of patients diagnosed with MASLD to develop non-alcoholic steatohepatitis and progress to cirrhosis.17, 18, 19, 20, 21 Many known risk factors for MASLD and HCC include demographic details, clinical and laboratory measures, and comorbidities found in the electronic medical record (EMR). ML models can be taught to identify relationships between such variables and discover the most relevant combination of which to predict a specific outcome. Recent studies have demonstrated that ML models developed using common laboratory indicators and demographic information are effective in screening for liver disease progression20,21 and are even superior to noninvasive tests such as Fibrosis-4 (FIB-4) in identifying clinically significant stages of MASLD and MASLD-related cirrhosis.21 However, more work is needed to develop a model based on risk factors common to both MASLD and HCC that is capable of predicting early and even in the absence of fibrosis.
In this study, we aimed to develop a ML model using EMR data to estimate the risk of HCC for MASLD patients at any stage of disease, but especially for those without evidence of advanced fibrosis or cirrhosis. Such a model would help to determine with sufficient accuracy, among millions of individuals with MASLD, those with high enough risk to justify further screening and surveillance and lead to more effective disease management and better outcomes.
Methods
Selection Criteria
The data used in this retrospective cohort study were drawn from 2 databases compiled from the EMR at 2 University of California health systems— University of California, Davis (UC Davis) and UC San Francisco (UCSF). The data were previously de-identified and standardized according to the Observational Medical Outcomes Partnership (OMOP) Common Data model.
The databases were queried using structured query language by applying International Classification of Diseases (ICD) Clinical Modification (CM) codes (Table A1) corresponding to a clinical diagnosis of MASLD to isolate records for patients with MASLD and deliberately excluding any codes associated with alcohol- or hepatitis-related liver disease. We also looked for alternate etiologies of liver disease to exclude them from our study, including but not limited to Wilson's disease, hemochromatosis, autoimmune hepatitis, primary sclerosing cholangitis, primary biliary cholangitis, and α1-antitrypsin deficiency but did not find any in our cohort. We separately pulled records for patients with HCC diagnosis using the associated ICD CM codes and subsequently joined them to the MASLD records to derive a composite dataset of all patients with MASLD who may or may not have developed HCC. An additional filter was then added to select only the first complete set of comorbidities recorded and laboratory values performed between 15 days before to one year after the recorded date of the initial MASLD diagnosis. We established this time frame to account for labs drawn within one year of initial diagnosis as HCC is a slow progressing disease. Patients lacking a full set of relevant data values within our specified time range were excluded from the cohort to avoid introducing bias through imputing missing values. Our aim is to predict the likelihood that a patient formally diagnosed with MASLD could progress to liver cancer based solely on the first set of laboratory values taken within the first year of the initial diagnosis. As such, patients with MASLD and comorbid HCC at the time of MASLD diagnosis were intentionally excluded. Additionally, our cohorts do not constitute a longitudinal examination of patients with MASLD but rather represent an isolated, critical time point from each patient’s record. Clinical notes and pathology and radiographic reports were not utilized in this analysis.
Study Cohorts
Based on our selection parameters, we identified a cohort of 1561 patients with a confirmed diagnosis of MASLD from the UC Davis OMOP database, comprising records on patients who received care at UC Davis Medical Center between 2010 and 2021. This cohort served as the training and initial testing dataset for teaching the ML algorithms. Using the same selection criteria, we drew a separate cohort of 686 patients from UCSF’s Information Commons OMOP database, comprising records on patients who received care at UCSF Medical Center between 2010 and 2021, to be used exclusively as a validation dataset to further validate the preliminary model developed from the UC Davis cohort. It is important to highlight that both cohorts are small since patients whose records were incomplete were excluded. It is likely that many of those excluded for incomplete records are considered low risk. Patients considered low risk for HCC and therefore without advanced fibrosis or cirrhosis would not undergo surveillance according to the current standard procedure; thus, a full laboratory mock-up would not be considered urgent or necessary. As such, our cohorts are not representative of the general population. Between the 2 health systems, we derived a total of 2247 patients with MASLD who had a complete set of laboratory values completed within one year of initial diagnosis. The 2 datasets were kept fully separate from each other to ensure that the model is trained and tested on one dataset and validated primarily on data from a separate population. The cohorts were loaded separately in the KNIME Analytics Platform (KNIME), and all categorical data points were then encoded for use in training the ML algorithms. This study was deemed exempt by the institutional review boards of UC Davis and UCSF.
Clinical Predictors
The data points used for the creation of the model are listed in Table 1 and include a combination of raw demographic details, comorbidities, and clinical and laboratory phenotypes pulled directly from the databases; and a calculated value, the FIB-4 Index for Liver Fibrosis, using the standard formula: Age ([year] x aspartate aminotransferase [U/L])/((platelet count [10(9)/L]) x (alanine transaminase [U/L]) (1/2)), where each numeric score is associated with the risk of advanced fibrosis specific to MASLD patients. These specific data points were selected under the expertise of 2 seasoned clinicians of hepatology and oncology in collaboration with data science specialists. This joint effort was important to identify and remove the obvious outliers (eg, lab error) for each data point, thus ensuring they are within the standards observed in clinical practice.
Table 1.
Patient Characteristics
| Characteristics | All training |
Non HCC training |
HCC training |
All validation |
Non HCC validation |
HCC validation |
|---|---|---|---|---|---|---|
| n = 1561 | n = 1334 | n = 227 | n = 686 | n = 510 | N = 176 | |
| Demographics, n (%) | ||||||
| American Indian/Alaska Native | 27 (2%) | 23 (2%) | 4 (2%) | 7 (1%) | 5 (1%) | 2 (1%) |
| Asian | 211 (14%) | 168 (12%) | 43 (19%) | 169 (25%) | 127 (25%) | 42 (24%) |
| Black/African American | 99 (6%) | 82 (6%) | 17 (7%) | 57 (8%) | 46 (9%) | 11 (6%) |
| Multi-race | 26 (2%) | 26 (2%) | - | 0 (0%) | 0 (0%) | 0 (0%) |
| Native Hawaiian/Pacific Islander | 11 (1%) | 10 (1%) | 1 (0.4%) | 6 (1%) | 5 (1%) | 1 (1%) |
| Other | 228 (15%) | 192 (14%) | 36 (16%) | 104 (15%) | 77(15%) | 27 (15%) |
| Unknown | 39 (3%) | 37 (3%) | 2 (1%) | 13 (2%) | 11 (2%) | 2 (1%) |
| White | 920 (59%) | 796 (60%) | 124 (55%) | 330 (48%) | 239 (47%) | 91 (52%) |
| Female, n (%) | 742 (48%) | 666 (50%) | 76 (33%) | 272 (40%) | 222 (44%) | 50 (28%) |
| Age, mean ± SD | 68 ± 12 | 67 ± 9 | 72 ± 9 | 70 ± 10 | 70 ± 8 | 72 ± 9 |
| Hispanic | 261 (17%) | 220 (17%) | 41 (18%) | 110 (16%) | 81 (16%) | 29 (16%) |
| Past medical history | ||||||
| Diabetes, n (%) | 942 (60%) | 814 (61%) | 128 (56%) | 412 (60%) | 312 (61%) | 100 (57%) |
| Hypertension, n (%) | 1362 (87%) | 1152 (86%) | 210 (93%) | 646 (94%) | 479 (94%) | 167 (95%) |
| Obesity, n (%) | 664 (43%) | 587 (44%) | 77 (34%) | 287 (42%) | 225 (44%) | 62 (35%) |
| Laboratory (mean ± SD) | ||||||
| Alkaline phosphatase, U/L | 86.46 ± 25.98 | 85.64 ± 25.73 | 91.31 ± 26.64 | 87.52 ± 27.46 | 85.35 ± 26.41 | 93.85 ± 29.23 |
| Creatinine, mg/dL | 0.94 ± 0.27 | 0.94 ± 0.21 | 0.94 ± 0.29 | 0.94 ± 0.27 | 0.94 ± 0.21 | 0.95 ± 0.30 |
| Blood urea nitrogen, mg/dL | 14.77 ± 4.89 | 14.78 ± 4.94 | 14.71 ± 4.62 | 14.79 ± 4.20 | 14.90 ± 4.14 | 14.48 ± 4.35 |
| Sodium, mEq/L | 137.58 ± 2.44 | 137.55 ± 2.47 | 137.79 ± 2.27 | 138.10 ± 2.22 | 138.11 ± 2.32 | 138.07 ± 1.95 |
| Potassium, mEq/L | 4.08 ± 0.34 | 4.08 ± 0.35 | 4.11 ± 0.33 | 4.09 ± 0.34 | 4.10 ± 0.34 | 4.09 ± 0.34 |
| Prothrombin time/international normalized ratio | 1.35 ± 0.17 | 1.13 ± 0.17 | 1.17 ± 0.15 | 1.13 ± 0.15 | 1.13 ± 0.16 | 1.15 ± 0.14 |
| Albumin, g/L | 3.67 ± 0.46 | 3.70 ± 0.45 | 3.52 ± 0.49 | 3.73 ± 0.45 | 3.77 ± 0.44 | 3.60 ± 0.45 |
| Chloride, mmol/L | 103.22 ± 3.01 | 103.09 ± 2.99 | 104.01 ± 3.02 | 103.70 ± 2.81 | 103.59 ± 2.8 | 104.03 ± 2.86 |
| Bilirubin, mg/dL | 1.07 ± 0.51 | 1.05 ± 0.51 | 1.20 ± 0.53 | 1.10 ± 0.53 | 1.07 ± 0.52 | 1.22 ± 0.55 |
| Cholesterol, mg/dL | 165.43 ± 35.52 | 166.89 ± 35.75 | 156.88 ± 32.90 | 167.07 ± 32.87 | 169.03 ± 33.40 | 161.37 ± 30.50 |
| FIB-4 score, n (%) | ||||||
| 0–1.3 | 251 (16%) | 243 (18%) | 8 (4%) | 73 (11%) | 65 (13%) | 8 (5%) |
| 1.3–2.67 | 536 (34%) | 484 (36%) | 52 (23%) | 239 (35%) | 200 (39%) | 39 (22%) |
| >2.67 | 774 (50%) | 607 (46%) | 167 (74%) | 374 (55%) | 245 (48%) | 129 (73%) |
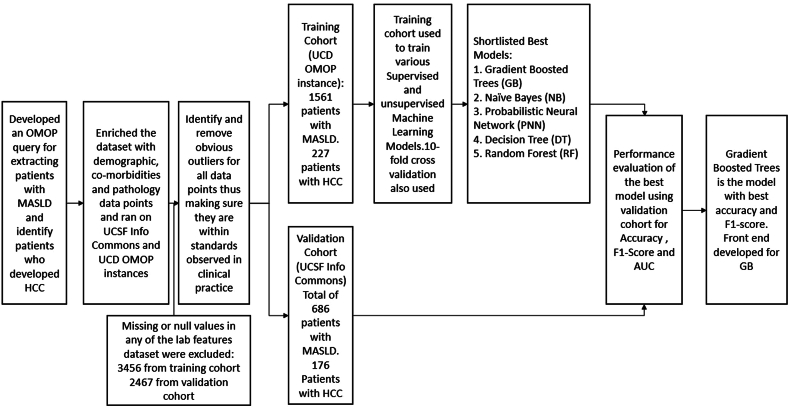
Model Development
Figure 1 shows the stepwise process of developing the HCC risk prediction model. We randomly split the UC Davis cohort into training (90%, n = 1404) and initial validation (10%, n = 157) sets. To develop the preliminary models, we strategically chose a combination of tree-based learning algorithms, Naïve Bayes (NB), stochastic gradient descent classifier, K- nearest neighbor (KNN), probabilistic neural network (PNN) and others22 (Figure A1A) due to their unique strengths and complementary attributes, and our familiarity with them. Tree-based algorithms, such as random forests and gradient boosting, are robust and versatile, capable of capturing complex relationships in the data while mitigating overfitting. Their ensemble nature allows for improved generalization performance and feature importance analysis, enhancing the interpretability of the model. NB, on the other hand, is particularly effective in handling high-dimensional data and is computationally efficient, making it suitable for medium-sized datasets. Its simplicity and assumption of feature independence make it a valuable choice for binary classification problems. Additionally, the stochastic gradient descent classifier excels at handling large-scale datasets and is well-suited for binary learning scenarios. KNN is a simple yet effective ML technique for classification where prediction is determined by the class of its KNNs in the feature space. It is a nonparametric and lazy learning algorithm that does not make explicit assumptions about the underlying data distribution during the training phase and defers learning until predictions are required. PNNs offer several strengths that make them attractive for general ML tasks. One of the primary advantages is their ability to provide probabilistic outputs, which means they can quantify uncertainty in predictions. This is crucial in applications, such as medical diagnosis, where knowing the model's confidence or uncertainty is as important as the prediction itself. PNNs inherently capture the uncertainty associated with the data, making them well-suited for real-world scenarios where uncertainty plays a significant role. Another strength of PNNs lies in their ability to model complex, nonlinear relationships in the data.
Figure 1.
HCC model development process.
By using these diverse algorithms, our motivation was to create a comprehensive and resilient ML model that can effectively address the intricacies of identifying HCC in patients with MASLD, providing both accuracy and interpretability. Each algorithm was taught through the training data to identify and learn the connections between the variables, independently and/or in combination, and link them to the known outcome of HCC among the patients in the cohort.
Overfitting is a common problem in ML that typically occurs when we try to create a complex model, and the model builds itself on irrelevant information rather than on the signal from the data. Whereas noise is the irrelevant information that holds no significant value to the outcome, the signal is the actual pattern in the dataset that we wish for the ML model to learn. An overfitted model performs very well on training data but poorly on unseen validation data. To avoid this problem, we employed k-fold cross-validation to iteratively train and validate in 10 folds based only on the training set.
Statistical Analysis
The performance of each of the 9 preliminary models (ie, how effectively it learned to and could predict HCC risk based on the relationships among the variables) was evaluated with the initial validation set from the UC Davis cohort (n = 157). The key metrics used to evaluate performance were accuracy, area under the curve (AUC), sensitivity, specificity, Cohen’s kappa, and F1-score, with specific emphasis on accuracy and F1-score. The F1-score was also used to covalidate the model as it encompasses both precision and recall outcomes of a model in the unbalanced outcome dataset where the number of patients without HCC outweighs those with HCC. All statistical analysis was performed on KNIME.
Of the 9 algorithms trained, 5 were shortlisted for performance comparison based on their high prediction accuracy and F1-score against the validation data (Table A2). These 5 models were subsequently validated with the full UCSF cohort (n = 686), representing data held back from the models during the training process. The deliberate separation of the UC Davis and UCSF cohorts ensures data independence between the training and validation phases of the project and prevents bias of each model toward the validation dataset.
Results
Cohort Demographics
Table 1 details the characteristics of the UC Davis training (n = 1561) and UCSF validation (n = 686) cohorts, both in the total cohorts and in the subsets that developed HCC (n = 227; 176). In the total cohorts, the mean age was 68 (standard deviation [SD] 10) in the training set and 70 (SD 12) in the validation set. The training cohort comprised 48% female, 59% White, 6% African American and 14% Asian. The validation cohort was more diverse with 40% female, 48% White, 8% African American, and 25% Asian. Hispanic ethnicity was present at 17% in the training cohort and 16% in the validation cohort. Compared to the total cohorts, among those who developed HCC, there was a higher percentage of females—33% training and 26% validation—and a higher mean age at 72 (SD 12) in both cohorts, which is in line with the higher prevalence of HCC among older male patients.
Scores on the FIB-4 demonstrated that the training cohort was overall at lower risk for advanced fibrosis based on the range for MASLD patients. Whereas the training cohort comprised 16% at low risk and 50% at high risk, the validation cohort had 11% at low risk and 55% at high risk. In the subsets of patients who developed HCC, a dramatically higher percentage scored within the range for high risk for advanced fibrosis; the distribution was also more similar between training and validation with 4% and 5% low risk and 74% and 73% high risk, respectively, with 23% and 22% in the indeterminate range. Since our cohorts come from primary and secondary care centers, the characteristics are more representative of the general population, and therefore the scores occur in the lower range.
Diabetes, hypertension, and obesity are well-observed in both cohorts, although there are clear differences in distribution between the total populations and the subsets that developed HCC. Among those who developed HCC in both cohorts, 93% and 95% of patients were also diagnosed with hypertension, 56% and 57% with diabetes, and 34% and 35% with obesity, suggesting a high prevalence of metabolic syndrome. Expectedly, hypertension was higher in the HCC subsets than in the total cohorts. However, obesity and diabetes were lower among those who developed HCC in both cohorts than in the total cohorts. Mean total cholesterol was within the normal range of <200 mg/dL between the training and testing cohorts in both datasets. Compared with the total cohorts (165.43 mg/dL training; 167.07 mg/dL testing), the HCC cohorts had lower means (156.88 mg/dL training; 161.37 mg/dL testing).
HCC Risk Prediction Model
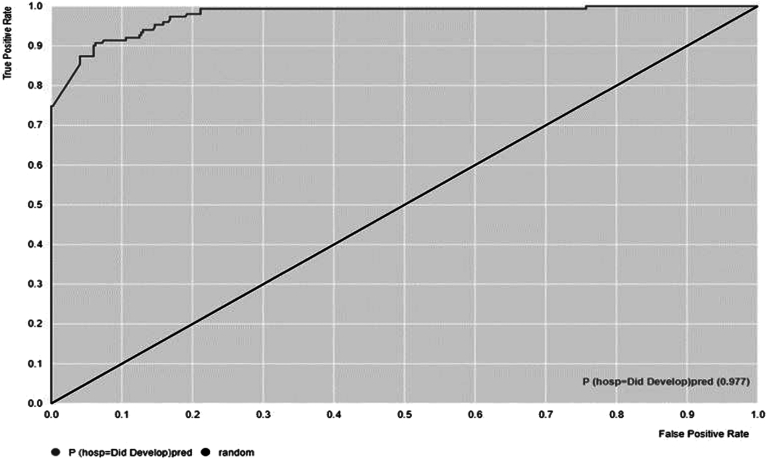
Table 2 lists the accuracy, AUC, specificity, sensitivity, and Cohen’s kappa for each of the 5 shortlisted models. Overall, the ensemble tree algorithms (gradient boosted [GB], decision tree, and random forest)—whose goal is to combine the predictions of several base estimators or tree methods to improve generalizability and robustness over a single estimator—performed better than PNN and NB. Figure A1B details a sample training process for the GB-based algorithm. The GB-based model performed with the highest overall accuracy (92.06%), AUC (0.97), F1-score (0.84), sensitivity (74.41), specificity (98.34), and Cohen’s kappa (0.78), which functions as a reliability indicator for a trained model (Figure 2). Table A3 displays the confusion matrix for this model, showing a larger proportion of type II errors but minimal type I errors.
Table 2.
Performance of All ML Models for HCC Prediction
| Metric | Value by algorithm |
||||
|---|---|---|---|---|---|
| GB | NB | PNN | DT | RF | |
| Accuracy | 92.12 | 68.08 | 81.92 | 87.17 | 90.29 |
| AUC | 0.97 | 0.64 | 0.91 | 0.87 | 0.97 |
| Specificity | 98.24 | 36.41 | 100.00 | 76.47 | 96.60 |
| Sensitivity | 74.42 | 79.01 | 29.54 | 84.31 | 74.23 |
| NPV | 91.76 | 83.41 | 80.28 | 93.38 | 91.57 |
| PPV | 93.92 | 30.01 | 91.06 | 55.27 | 88.27 |
| Cohen’s kappa | 0.78 | 0.15 | 0.38 | 0.64 | 0.51 |
| F1-score | 0.84 | 0.37 | 0.46 | 0.73 | 0.84 |
DT, decision tree; GB, gradient boosted; NB, naïve bayes; NPV, negative predictive value; PNN, probabilistic neural network; PPV, positive predictive value; RF, random forest.
Figure 2.
Area under receiver operating characteristics (AUROC) curve for HCC prediction model for patients with MASLD.
We evaluated the parameters that had the highest predictive accuracy for the determination of HCC risk. The variable importance score is an algorithm-based score for the relative influence of each parameter to the entire model.23,24 The noninvasive score for liver fibrosis, FIB-4, was the strongest predictive parameter for the development of HCC in the MASLD cohort, with a variable importance score of 53. Total cholesterol, ALP, bilirubin, and hypertension were other parameters with high influence on the model (Figure 3). This model was subsequently converted to predictive model markup language format, an open-source format that makes the model portable across various EMR systems and platform diagnostics.25
Figure 3.
Graphical model snapshot.
Discussion
Despite widespread lack of familiarity, ML as a subset of artificial intelligence is a rapidly evolving technology that is transforming every walk of life, including health care. In this study, we utilized a combination of open-source algorithms and analytical tools to develop our prediction model; and standardized ML methods and architecture to test all possible models, data for which we have presented above.
Our model directly predicts the risk of MASLD patients in developing HCC with strong specificity and sensitivity. As the model is trained on larger, more diverse, and multicentered cohorts, we expect its sensitivity to improve further. The liver fibrosis stage is the strongest clinical predictor for the worst clinical outcome in MASLD patients, including the development of HCC26,27 and that is reflected in our cohort. The other parameters—cholesterol, ALP, hypertension, and bilirubin—are also linked with a higher risk of MASLD.28, 29, 30 A combination of all these predictors in one model can thus help in predicting HCC risk.
We envision the model will be applicable in a clinical setting as a point-of-care tool as well as for population-level triaging. The tool can be configured to automatically generate a risk prediction score with requisite data from the EMR. The availability of such a score can help providers and patients effectively discuss screening strategies and institute modifiable measures31 to mitigate risks for the development of HCC. Health systems can also develop population-level tools using this or a similar model to establish protocols for effective screening strategies. Furthermore, reducing the risks for undiagnosed HCC can not only reduce suffering from progressed HCC but also minimize the higher costs of treatment for advanced HCC stages.
As we define ML models and their real-world clinical applications, it is essential to consider the ethical aspects of such predictive tools. In MASLD patients without advanced fibrosis, a high prediction HCC risk score would be beneficial for clinical management but can generate significant anxiety in otherwise asymptomatic patients. Additionally, a high risk score may affect a patient’s future ability to obtain insurance products. Thus, it is crucial to understand the clinical benefits and untoward effects when planning prospective studies with ML models that can predict cancer risk in patients.
Limitations
Our study is one of the earliest to use ML to predict HCC risk in MASLD patients with promising results, though a few limitations are worth considering. Our project, while using data from 2 separate institutions, featured relatively small cohorts that were limited to a single geographic region of Northern California. Nonetheless, our study provides a key foundation from which to build future ML studies. Although ICD CM codes are highly accessible and reproducible, reliance on their use for cohort and outcome identification can limit generalizability for patients with MASLD and HCC. Still, this remains one of the most effective means for large cohort studies. Furthermore, there is potential to include other types of liver cancer, although approximately 90% of primary liver cancer is HCC.2,32 Finally, while our model was created using several known clinical risk factors that are independently associated with MASLD and HCC, it did not incorporate environmental contributors such as evidence of exposure to fine particulate matter air pollution or genetic polymorphisms such as patatin-like phospholipase domain-containing protein 3 gene,11 both of which have been associated with increased risk of HCC. As we gain access to genetic data and larger and more varied datasets, the model can become even more personalized.
The lack of multimodal data is another drawback in our study. Our current model does not include clinical notes, liver biopsy results, or non-invasive test records such as ultrasound, magnetic resonance imaging, and elastography. We were also unable to include the NAFLD fibrosis score in our study as we did not have access to height, weight, and body mass index information through the de-identified databases to calculate the metric. The next phase of our project will be characterized by the inclusion of the NAFLD fibrosis score, clinical notes, and incrementally adding annotated images. In combining different modalities into one prediction model, a physician can be well equipped to be able to identify the patients who are most in need of additional care.
Conclusions
Use of ML models for predicting risk of future HCC development can help tailor resources to target screening and surveillance strategies to those most at risk. The ML model presented here uses readily available variables for accurate risk prediction, holding the promise for effective risk stratification of MASLD patients in the development of HCC.
Acknowledgments:
We thank Dr Sharat Israni for his efforts in facilitating our access to the UCSF Information Commons.
Authors' Contributions:
Aniket Alurwar developed the initial machine learning model and analysis, formulated the analytical portions of the paper, and provided technical expertise. Carole Ly assisted Aniket Alurwar with data analytics and analytical portions of the paper. Cindy Piao wrote the initial draft and edited all subsequent versions of the manuscript. Rajiv Donde offered expertise in model development and assisted with manuscript editing. Frederick J Meyers shared expertise in model creation and data analysis and edited the manuscript. Souvik Sarkar was instrumental in study initiation, data curation, and model development; manuscript writing and editing. Christopher Wang helped in study initiation, managing the project, and assisted in manuscript editing.
Footnotes
Conflicts of Interest: The authors disclose no conflicts.
Funding: The authors report no funding.
Ethical Statement: This study was deemed IRB Exempt by the University of California, Davis because of the use of de-identified data.
Data Transparency Statement: We can make open source tools available on request.
Reporting Guidelines: None.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2024.01.007.
Supplementary Materials
References
- 1.Kim E., Viatour P. Hepatocellular carcinoma: old friends and new tricks. Exp Mol Med. 2020;52(12):1898–1907. doi: 10.1038/s12276-020-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 3.Desai A., Sandhu S., Lai J.P., et al. Hepatocellular carcinoma in non-cirrhotic liver: a comprehensive review. World J Hepatol. 2019;11(1):1–18. doi: 10.4254/wjh.v11.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrysavgis L., Giannakodimos I., Diamantopoulou P., et al. Non-alcoholic fatty liver disease and hepatocellular carcinoma: clinical challenges of an intriguing link. World J Gastroenterol. 2022;28(3):310–331. doi: 10.3748/wjg.v28.i3.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loomba R., Lim J.K., Patton H., et al. AGA clinical practice update on screening and surveillance for hepatocellular carcinoma in patients with nonalcoholic fatty liver disease: expert review. Gastroenterology. 2020;158(6):1822–1830. doi: 10.1053/j.gastro.2019.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinella M.E., Lazarus J.V., Ratziu V., et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology. 2023;78(6):1966–1986. doi: 10.1097/HEP.0000000000000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabbany M.N., Conjeevaram Selvakumar P.K., Watt K., et al. Prevalence of nonalcoholic steatohepatitis-associated cirrhosis in the United States: an analysis of National Health and Nutrition Examination Survey Data. Am J Gastroenterol. 2017;112(4):581–587. doi: 10.1038/ajg.2017.5. [DOI] [PubMed] [Google Scholar]
- 8.Kanwal F., Shubrook J.H., Younossi Z., et al. Preparing for the NASH epidemic: a call to action. Gastroenterology. 2021;161(3):1030–1042.e8. doi: 10.1053/j.gastro.2021.04.074. [DOI] [PubMed] [Google Scholar]
- 9.Rinella M.E. Nonalcoholic fatty liver disease: a systematic review [published correction appears in JAMA 2015;314(14):1521] JAMA. 2015;313(22):2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 10.Younossi Z.M., Stepanova M., Ong J., et al. Nonalcoholic steatohepatitis is the most rapidly increasing indication for liver transplantation in the United States. Clin Gastroenterol Hepatol. 2021;19(3):580–589.e5. doi: 10.1016/j.cgh.2020.05.064. [DOI] [PubMed] [Google Scholar]
- 11.Ioannou G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J Hepatol. 2021;75(6):1476–1484. doi: 10.1016/j.jhep.2021.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18(4):223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazel Y., Koenig A.B., Sayiner M., et al. Epidemiology and natural history of non-alcoholic fatty liver disease. Metabolism. 2016;65(8):1017–1025. doi: 10.1016/j.metabol.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Younossi Z.M., Henry L. Epidemiology of non-alcoholic fatty liver disease and hepatocellular carcinoma. JHEP Rep. 2021;3(4) doi: 10.1016/j.jhepr.2021.100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Younossi Z.M., Otgonsuren M., Henry L., et al. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology. 2015;62(6):1723–1730. doi: 10.1002/hep.28123. [DOI] [PubMed] [Google Scholar]
- 16.Kanwal F., Kramer J.R., Mapakshi S., et al. Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology. 2018;155(6):1828–1837.e2. doi: 10.1053/j.gastro.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinani A.M., Kowdley K.V., Noureddin M. Application of artificial intelligence for diagnosis and risk stratification in NAFLD and NASH: the state of the art. Hepatology. 2021;74(4):2233–2240. doi: 10.1002/hep.31869. [DOI] [PubMed] [Google Scholar]
- 18.Kourou K., Exarchos T.P., Exarchos K.P., et al. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2014;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Wang X., Zhang J., et al. Applications of artificial intelligence (AI) in researches on non-alcoholic fatty liver disease (NAFLD): a systematic review. Rev Endocr Metab Disord. 2022;23(3):387–400. doi: 10.1007/s11154-021-09681-x. [DOI] [PubMed] [Google Scholar]
- 20.Ma X., Yang C., Liang K., et al. A predictive model for the diagnosis of non-alcoholic fatty liver disease based on an integrated machine learning method. Am J Transl Res. 2021;13(11):12704–12713. [PMC free article] [PubMed] [Google Scholar]
- 21.Chang D., Truong E., Mena E.A., et al. Machine learning models are superior to noninvasive tests in identifying clinically significant stages of NAFLD and NAFLD-related cirrhosis. Hepatology. 2023;77(2):546–557. doi: 10.1002/hep.32655. [DOI] [PubMed] [Google Scholar]
- 22.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit- learn: machine learning in Python. J Mach Learn Res. 2011;12(85):2825–2830. [Google Scholar]
- 23.H2O.ai. Variable Importance. https://docs.h2o.ai/h2o/latest-stable/h2o-docs/variable-importance.html Available at:
- 24.Guyon I., Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 25.Data Mining Group PMML 4.4.1 - general structure. https://dmg.org/pmml/v4-4-1/GeneralStructure.html Available at:
- 26.Angulo P., Kleiner D.E., Dam-Larsen S., et al. Liver fibrosis, but No other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149(2):389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unalp-Arida A., Ruhl C.E. Liver fibrosis scores predict liver disease mortality in the United States population. Hepatology. 2017;66(1):84–95. doi: 10.1002/hep.29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastberg S.E., Lund H.S., De Lucia-Rolfe E., et al. Hepatic steatosis is associated with anthropometry, cardio-metabolic disease risk, sex, age and urbanisation, but not with ethnicity in adult Kenyans. Trop Med Int Health. 2022;27(1):49–57. doi: 10.1111/tmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y., Lan J., Zhong Y., et al. Association of remnant cholesterol with nonalcoholic fatty liver disease: a general population-based study. Lipids Health Dis. 2021;20(1):139. doi: 10.1186/s12944-021-01573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinyopornpanish K., Khoudari G., Saleh M.A., et al. Hepatocellular carcinoma in nonalcoholic fatty liver disease with or without cirrhosis: a population-based study. BMC Gastroenterol. 2021;21(1):394. doi: 10.1186/s12876-021-01978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange N.F., Radu P., Dufour J.F. Prevention of NAFLD-associated HCC: role of lifestyle and chemoprevention. J Hepatol. 2021;75(5):1217–1227. doi: 10.1016/j.jhep.2021.07.025. [DOI] [PubMed] [Google Scholar]
- 32.Balogh J., Victor D., 3rd, Asham E.H., et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.