Abstract
Simian virus 40 (SV40) is an excellent model system for investigating the cis- and trans-acting factors involved in eukaryotic DNA replication because it uses host enzymes, with the exception of the virus-encoded T-antigen (T-ag), to replicate its genome. Although its origin of replication (ori) is essential for DNA replication, there are transcriptional promoters and enhancers that affect DNA replication efficiency. T-ag binds to sites I to III within and around ori with different affinities and induces structural changes. We were interested in determining if the position of the promoters relative to ori influences the binding of T-ag to these regions. Furthermore, we characterized the DNA structural changes that occur as a result of protein binding when the promoters are absent and also when the promoters are moved from their wild-type position upstream of ori to a position downstream of ori. Using sequence- and conformation-specific chemical probes, our data indicate that (i) the conformation of site I is influenced by T-ag binding and by flanking sequences, (ii) the conformation of the promoters after T-ag binding is dependent on their location, and (iii) unwinding of ori is influenced by the location of the promoters and their presence or absence. These differences in DNA conformation may help explain decreases in relative DNA replication efficiency that occur when the promoters are absent or located downstream of ori.
Simian virus 40 (SV40) is used as a paradigm for delineating the cis- and trans-acting factors involved in eukaryotic DNA replication (for reviews, see references 11 and 19). SV40 exploits its eukaryotic host by recruiting cellular proteins to replicate its own DNA (77). T antigen (T-ag) is the only virus-encoded protein required for SV40 DNA replication (12, 52, 60, 72). T-ag binds to a 64-bp bidirectional origin of replication (ori) (13, 18, 25, 74–76). ori is divided into three functional domains: an early palindrome (EP), a 27-bp pentanucleotide (PEN) region containing T-ag site II, and a 17-bp AT-rich region (2, 20, 21, 28, 31, 38, 44, 52, 68). Located adjacent to ori are three 21-bp promoters for transcription that bind T-ag (site III) weakly (33, 67, 74) and also bind the eukaryotic transcription factor Sp1 (24). In addition to the promoters, there are two 72-bp enhancers for transcription (1, 27, 30, 50) that bind several eukaryotic transcription factors (34, 36, 87). A diagram of the regulatory region of SV40 is shown in Fig. 1. Initially T-ag binds as a monomer to each of the four PEN repeats within ori (47), and subsequently a double hexamer is formed (6, 46, 54, 86). T-ag then induces structural alterations, including untwisting of the AT-rich region (3–5, 14, 53), and opens the core region by melting an 8-bp region within the EP (7, 15–17, 33, 53, 56, 84). T-ag’s helicase activity (29, 64, 66, 82), with the aid of replication protein A, bidirectionally unwinds the DNA (8, 32, 64, 81). In addition to binding to sites II and III, T-ag binds to site I, which is located immediately to the early-transcription side of ori (51, 58, 73, 75). The fact that DNA synthesis proceeds at a uniform rate after initiation suggests that initiation is the rate-limiting step. Therefore, those DNA molecules that are the most efficient at binding T-ag should replicate with the highest efficiency.
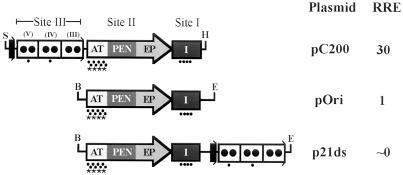
FIG. 1.
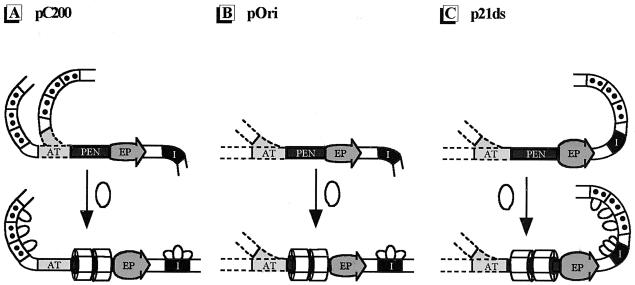
Organization of SV40 regulatory regions; relative DNA replication efficiencies of pC200, pOri, and p21ds; and target sites for adozelesin, bizelesin, and hedamycin. Shown are schematic representations of the regulatory regions of pC200, pOri, and p21ds and the relative DNA replication efficiencies (RRE) of these plasmids as determined in cotransfection assays (42). ori (represented by the arrows) consists of three functional domains as indicated: the EP, the PEN, and the AT-rich region (AT). Each 21-bp promoter is represented by a box containing two circles, and the binding site for the transcription factor AP-1 is indicated by a solid black rectangle. T-ag binds to the wild-type regulatory region at three sites: site I (represented by the black rectangle labeled I) is located on the early-transcription side of ori; site II is located within the PEN; and several weaker sites (labeled III, IV, and V), collectively called site III, are located within the 21-bp promoters. Restriction endonuclease sites are indicated as follows: BanI (B), EcoRI (E), HindIII (H), and SphI (S). The target sequences for adozelesin are indicated with solid circles. The target sequences for bizelesin are indicated with asterisks.
Our laboratory (41, 42) and others (2, 26, 35) have demonstrated that the organization of the SV40 regulatory region influences DNA replication efficiency. To determine relative DNA replication efficiencies, we transfected COS-1 cells with equimolar amounts of a test plasmid (pC200 or p21ds) and a control plasmid (pOri). After the plasmids were allowed to replicate for 48 h, the plasmid DNA was purified and quantified (42). pC200, a plasmid containing three 21-bp promoters and a binding site for the transcription factor AP-1 on the late-transcription side of ori and T-ag site I, had a 30-fold-higher DNA replication efficiency than pOri, a plasmid containing ori and T-ag site I. In contrast, replication was barely detectable in p21ds, a plasmid containing three 21-bp promoters and an AP-1-binding site on the early-transcription side of ori and T-ag site I (42). The rearranged regulatory regions of the plasmids and their relative DNA replication efficiencies are shown in Fig. 1. Rearranged regulatory regions are not simply products of artificial recombinant techniques. Serial passage of SV40 at high multiplicities of infection leads to the evolution of new viral DNA species, containing deletions and duplications of viral DNA and substitutions with cellular DNA (see reference 9 for a review). These new viral species often have an advantage in terms of DNA replication efficiency. By studying the organization of the regulatory regions of these naturally arising variants (10, 31, 37, 40, 41, 48, 57, 59, 65, 78, 85) and their impact on protein binding and DNA conformation, insights into which cis-acting sequences and trans-acting factors are important in DNA replication can be gained. Although cellular proteins, including Sp1 (70), also contribute to conformational changes, we have not investigated the roles of specific cellular proteins but have focused instead on T-ag.
When T-ag is limiting, differences in DNA replication efficiencies of rearranged regulatory regions are exaggerated (41). One hypothesis to explain this is that some regulatory regions bind T-ag more efficiently than others. Using gel mobility shift assays, we found that T-ag and/or the proteins from BSC-1 cell extracts were able to bind to the rearranged regulatory regions examined. However, using DNase I footprinting, we found that T-ag protects the sequences flanking ori with different affinities, and we observed no specific protection from DNase I in the rearranged regulatory regions when using only the proteins from BSC-1 cell extracts.
We also characterized protein-induced conformations by using chemical probes (adozelesin, bizelesin, and hedamycin) to determine whether any DNA structural changes could be correlated with DNA replication efficiency. Adozelesin targets specific bent DNA sequences, bizelesin targets specific sequences that are in a straight conformation, and hedamycin recognizes unwound DNA sequences (Fig. 1). Han and Hurley (33) presented a model of T-ag-induced conformational changes in the regulatory region of wild-type SV40 that used similar sequence- and conformation-specific probes. Their data suggest that T-ag-induced bending within the 21-bp promoters causes the naturally bent AT-rich region of ori to straighten and the EP to unwind. Our work extends their findings by characterizing conformations induced in rearranged SV40 regulatory regions. We also characterized conformations induced by T-ag and the proteins from BSC-1 cell extracts, a condition that more closely resembles the internal milieu of an SV40-infected cell. We found that protein-induced conformations of the regulatory regions of pC200, pOri, and p21ds are dependent on the organization of these regulatory regions. Also, the location of the 21-bp promoters affects the binding of T-ag to site III.
MATERIALS AND METHODS
DNA fragments.
Restriction endonucleases and polynucleotide kinase were used according to the manufacturer’s suggestions (New England Biolabs). The regulatory region of pC200 was isolated from pC342 (42) by digestion with HindIII and SphI and then 5′-end labeled at the HindIII site. The regulatory regions of pOri and p21ds were isolated from pOri and p21ds (42), respectively, by digestion with EcoRI and BanI and were 5′-end labeled at the EcoRI site. The DNA fragments were dephosphorylated with calf intestinal alkaline phosphatase (Boehringer Mannheim) prior to 5′-end labeling with [γ-32P]ATP (ICN) and eluted from acrylamide gels. Figure 1 provides an illustration of the organization of the regulatory regions.
Cell extracts.
BSC-1 cells, a continuous and permissive African green monkey kidney cell line, were seeded in Nunclon culture dishes (10 cm diameter; Nunc) and grown to 90% confluency. The cells were washed two times with 3 ml (per dish) of cold Tris-salts buffer (20 mM Tris-HCl [pH 7.4], 137 mM NaCl, 5 mM KCl, 1 mM CaCl2), and 2 ml of cold hypotonic buffer (5 mM potassium acetate, 0.5 mM MgCl2, 0.5 mM dithiothreitol, 20 mM HEPES [pH 7.8]) was added to each dish prior to incubation at 25°C for 5 min. The cells were collected and lysed with a Dounce homogenizer. The lysate was incubated on ice for 1 h. Cell debris and chromosomal DNA were removed by centrifugation at 10,000 × g and 4°C in a JA-20 rotor in a J-21C centrifuge (Beckman Instruments) for 10 min. The supernatant, containing the cellular proteins, was stored at −70°C.
T-ag.
T-ag was purified by the procedure of Simanis and Lane (61) after expression in Sf9 insect cells infected with a recombinant baculovirus containing the T-ag gene. The Sf9 cells, baculovirus, and pAB419 cells that supply T-ag antibody were gifts from Bruce Stillman, Cold Spring Harbor Laboratory.
Gel mobility shift assay.
Approximately 0.2 ng of the γ-32P-labeled regulatory-region DNA of pC200, pOri, or p21ds was incubated with 1 μg of T-ag and/or 6 μg of proteins from BSC-1 cell extract at 25°C for 40 min in binding buffer (8 mM ATP, 60 mM HEPES [pH 7.5], 14 mM MgCl2, 80 mM creatine phosphate, 2 mM dithiothreitol, 40 μg of creatine kinase/ml, 0.2% [wt/vol] bovine serum albumin, 40% [vol/vol] glycerol). The DNA-protein complexes were separated on a 4% acrylamide gel and visualized by autoradiography.
DNase I footprinting.
Approximately 0.1-ng quantities of the γ-32P-labeled regulatory-region DNA of pC200, pOri, or p21ds were incubated in binding buffer with various amounts of T-ag or proteins from BSC-1 cell extract in a reaction volume of 26 μl at 25°C for 40 min. After addition of 4 μl of 37.5 mM CaCl2–75 mM MgCl2, the DNA was digested with 0.005 μg of DNase I (Worthington) at 25°C for 1 min. The EDTA concentration was adjusted to 40 mM, 10 μg of Escherichia coli tRNA was added, and the DNA was precipitated with ethanol. The DNA was dissolved in 0.3 M sodium acetate (pH 6), precipitated and washed with ethanol, dried, and dissolved in sequencing dye. After being denatured, the DNA was fractionated on a 6% acrylamide gel containing 8 M urea and TBE (0.1 M Tris-borate, 2 mM EDTA [pH 8.3]) and visualized by autoradiography.
Chemical probes.
Adozelesin and bizelesin were gifts from the Pharmacia and Upjohn Company (Kalamazoo, Mich.), and hedamycin was a gift from the National Cancer Institute (Bethesda, Md.). The probes were dissolved in dimethylformamide (adozelesin, 2 μM; bizelesin, 0.1 μM; and hedamycin, 0.05 μM).
Strand breakage assay.
Approximately 0.1 ng of the γ-32P-labeled regulatory-region DNA of pC200, pOri, or p21ds was incubated with 1 μg of T-ag and/or 6 μg of proteins from BSC-1 cell extract in binding buffer in a reaction volume of 20 μl at 37°C for 40 min. The reaction mixtures were treated with 2 μl of each chemical probe at 37°C for various periods of time. The reactions were quenched by adding 15 μg of sonicated calf thymus DNA, and the reaction mixtures were brought to a final volume of 100 μl with Tris buffer (1 mM EDTA, 10 mM Tris-HCl [pH 8]). The reaction mixtures were boiled for 5 min before and after addition of 10 μl of 1 M piperidine. Approximately 0.01 ng of a γ-32P-labeled 522-bp DNA fragment was added as a control for DNA recovery. The samples were extracted with phenol, phenol-chloroform (1:1), and chloroform, back extracted with H2O, and precipitated with ethanol. The DNA was dissolved in 0.3 M sodium acetate (pH 6), precipitated and washed with ethanol, dried, and dissolved in sequencing dye. The DNA was denatured and fractionated on a 6% acrylamide gel containing 8 M urea and TBE and visualized by autoradiography. Quantification of the cleavage products was performed by image analysis (NIH Image 1.52, developed at the National Institutes of Health and available on the Internet [52a]).
RESULTS
DNA-protein binding.
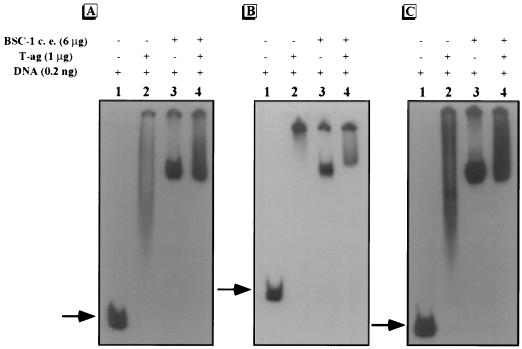
Gel mobility shift assays were done to determine whether the organization of the SV40 regulatory region influences protein binding. γ-32P-labeled regulatory-region DNA of pC200, pOri, or p21ds was incubated with T-ag and/or the proteins from BSC-1 cell extract, and DNA-protein complexes were separated by gel electrophoresis. Mobility shifts were seen with all of the rearranged regulatory regions examined after incubation with purified T-ag and/or the proteins from BSC-1 cell extract, indicating that these proteins were able to interact with each of the regulatory regions, regardless of their organization (Fig. 2).
FIG. 2.
Gel mobility shift assays of the regulatory regions of pOri (A), pC200 (B), and p21ds (C). Approximately 0.2 ng of γ-32P-labeled regulatory-region DNA was incubated without (−) protein (lane 1), with (+) T-ag (lane 2), with the proteins from BSC-1 cell extract (c. e.) (lane 3), or with T-ag plus the proteins from BSC-1 cell extract (lane 4). The regulatory elements are as indicated in Fig. 1. The arrows indicate unbound DNA.
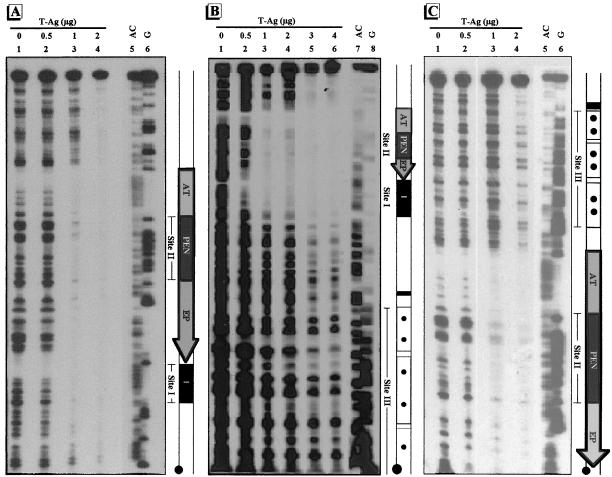
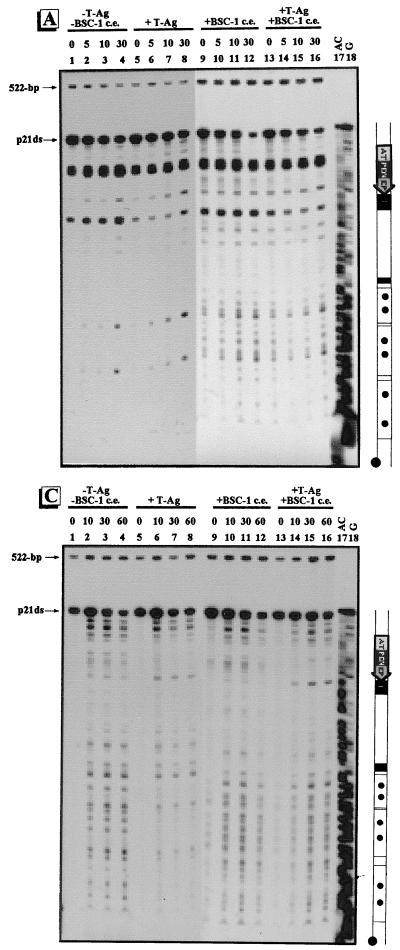
DNase I footprinting was used to identify the site at which T-ag or the proteins from BSC-1 cell extract bind to the regulatory regions of pC200, pOri, and p21ds. When each of the γ-32P-labeled regulatory-region DNAs was incubated with 0.5 μg of T-ag, site I was protected from DNase I digestion (Fig. 3A and B, lanes 1 and 2, and data not shown). As more T-ag was added, protection of ori was detected (Fig. 3, lanes 3 and 4). Therefore, regardless of the organization of the regulatory region, T-ag binds sites I and II. In pC200, the 21-bp promoters were partially protected after incubation with 2 μg of T-ag (Fig. 3C, lane 4). These results with pC200 are consistent with previous findings (54, 74). In contrast, slight protection of the 21-bp promoters in p21ds was seen only after addition of 3 μg of T-ag (Fig. 3B, lane 5), and even after addition of 4 μg of T-ag (lane 6), the 21-bp promoters were only slightly protected. The organization of the regulatory region also affected the binding of T-ag to the region immediately 3′ to ori. For example, in pOri, upon addition of 2 μg of T-ag, protection was seen across the entire fragment (Fig. 3A, lane 4). However, in pC200, when the region immediately 3′ to ori was occupied by the 21-bp promoters, full protection was not seen (Fig. 3C). In p21ds, the region immediately 3′ to ori was slightly protected only after addition of 3 μg of T-ag. Essentially the entire regulatory regions of pOri and pC200, but not that of p21ds (especially within the 21-bp promoters), were at least partially protected from DNase I digestion when incubated with 2 μg of T-ag (Fig. 3, lanes 4). We saw no protection when the regulatory-region DNAs were individually incubated with 6 μg of proteins from BSC-1 cell extract (data not shown).
FIG. 3.
DNase I footprints of the regulatory regions of pOri (A), p21ds (B), and pC200 (C). The regulatory regions of pOri and p21ds are labeled at the 5′ end of the EcoRI site, and the regulatory region of pC200 is labeled at the 5′ end of the HindIII site. Approximately 0.1 ng of γ-32P-labeled regulatory-region DNA was incubated with various amounts of T-ag for 40 min at 25°C and digested with 0.005 μg of DNase I for 1 min at 25°C. Maxam-Gilbert sequencing reactions for the fragments are shown in the rightmost lane of each panel. To the right of each autoradiogram is a schematic illustration of the regulatory region. The regulatory elements are as indicated in Fig. 1.
Characterization of conformational changes by using three chemical probes.
Adozelesin recognizes the sequences 5′-(A/T)(A/T)A*-3′ and 5′-(A/T)(G/C)(A/T)A*-3′ (39, 79) and alkylates the adenine followed by the asterisk if the target sequence is in a bent conformation (55). These target sequences are in the AT-rich region of ori, site I, and the 21-bp promoters. Bizelesin cross-links the adenines (indicated by the asterisks) of the target sequences 5′-TAATTA*-3′/3′-A*TTAAT-5′ and 5′-TAAAAA*-3′/3′-A*TTTTT-5′ (22, 69), which are located within the AT-rich region. Such cross-linking occurs if the target sequence is in a straight conformation (49). Finally, hedamycin alkylates the guanines in unwound DNA at the N7 position (69). Target sequences are shown in Fig. 1. All experiments using chemical probes were done a minimum of three times for each probe and for each construct. Bands were quantified by densitometry, and representative gels are shown in Fig. 4 to 6.
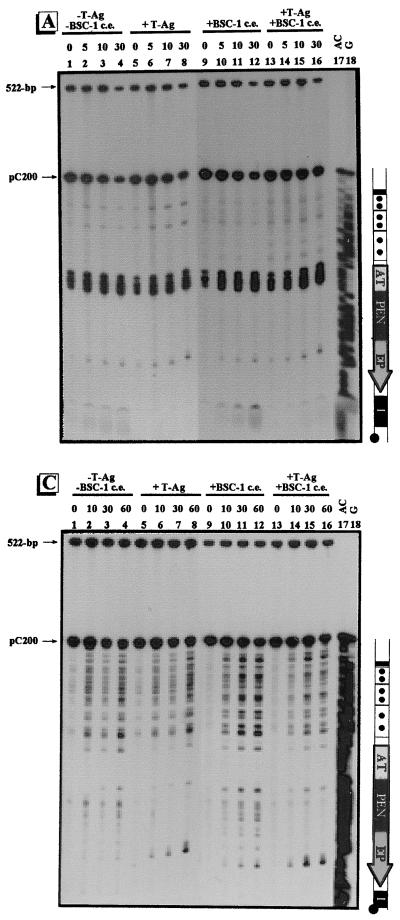
FIG. 4.
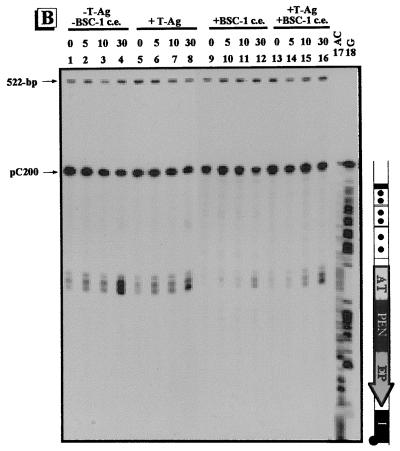
Conformational changes within the γ-32P-labeled regulatory region of pC200. The regulatory region of pC200 was treated with adozelesin (A), bizelesin (B), or hedamycin (C). Approximately 0.1 ng of the γ-32P-labeled regulatory-region DNA of pC200 was incubated without (−) proteins (lanes 1 to 4), with (+) 1 μg of T-ag (lanes 5 to 8), with 6 μg of the proteins from BSC-1 cell extract (c.e.) (lanes 9 to 12), and with 1 μg of T-ag plus 6 μg of the proteins from BSC-1 cell extract (lanes 13 to 16). Reaction times for adozelesin and bizelesin were 0, 5, 10, and 30 min; reaction times for hedamycin were 0, 10, 30, and 60 min. The Maxam-Gilbert sequencing reactions are shown in lanes 17 to 18. To the right of each autoradiogram is a schematic illustration of the regulatory region of pC200. The regulatory elements are as indicated in Fig. 1. The positions of the unmodified pC200 regulatory fragment and the 522-bp control fragment are indicated by arrows.
FIG. 6.
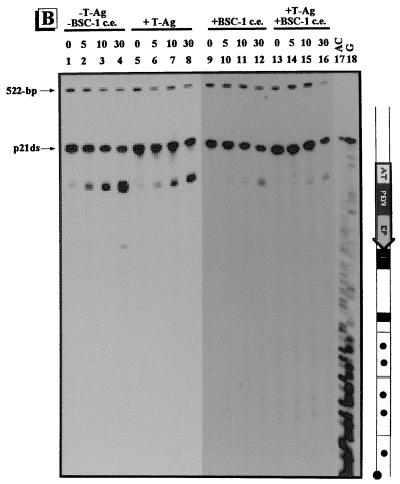
Conformational changes within the γ-32P-labeled regulatory region of p21ds. The regulatory region of p21ds was treated with adozelesin (A), bizelesin (B), and hedamycin (C). Approximately 0.1 ng of the γ-32P-labeled regulatory-region DNA of p21ds was incubated without (−) proteins (lanes 1 to 4), with (+) 1 μg of T-ag (lanes 5 to 8), with 6 μg of the proteins from BSC-1 cell extract (c.e.) (lanes 9 to 12), or with 1 μg of T-ag plus 6 μg of the proteins from BSC-1 cell extract (lanes 13 to 16). Reaction times for adozelesin and bizelesin were 0, 5, 10, and 30 min; reaction times for hedamycin were 0, 10, 30, and 60 min. The Maxam-Gilbert sequencing reactions are shown in lanes 17 and 18. To the right of each autoradiogram is a schematic illustration of the regulatory region of p21ds. The regulatory elements are as indicated in Fig. 1. The positions of the unmodified p21ds regulatory fragment and the 522-bp control fragment are also indicated.
(i) pC200.
We characterized the conformation of the regulatory region of pC200 after incubation with T-ag and/or proteins from BSC-1 cell extract, using sequence- and conformation-specific chemical probes. No alkylations were detected in the γ-32P-labeled regulatory-region DNA of pC200 when adozelesin was absent (data not shown). Prior to incubation with proteins, alkylation of the AT-rich region was detected, indicating that this region was bent (Fig. 4A, lanes 1 to 4). Incubation with T-ag and/or proteins from BSC-1 cell extract caused no significant difference in the amount of alkylation of the AT-rich region (Fig. 4A, lanes 5 to 16). Site I was also alkylated prior to incubation with proteins; however, the addition of T-ag prevented alkylation of site I (lanes 5 to 8 and 13 to 16). Incubation with the proteins from BSC-1 cell extract was not sufficient to cause a significant decrease in the alkylation of site I, as determined by quantification of the band intensities (lanes 9 to 12). T-ag also played a role in the conformation of the 21-bp promoters. The 21-bp promoters were alkylated in the presence (lanes 5 to 16) or absence (lanes 1 to 4) of proteins. Upon incubation with T-ag, either alone or in combination with the proteins from BSC-1 cell extract, there was an increase in alkylation of the 21-bp promoters (lanes 5 to 8 and 13 to 16). This is consistent with previous data (33). Although there was an increase in the alkylation within the 21-bp promoters when both T-ag and the proteins from BSC-1 cell extract were added, the effect on alkylation was not as dramatic as that occurring when T-ag alone was added. This suggests that when T-ag binds the 21-bp promoters it holds or stabilizes the naturally bent 21-bp promoters in a bent conformation, while the proteins from BSC-1 cell extract have a slightly inhibitory effect.
We also probed the regulatory region of pC200 with bizelesin, which alkylates the AT-rich region if it is in a straight conformation. No alkylations were detected in the γ-32P-labeled regulatory region when bizelesin was omitted from the reaction mixture (data not shown). In the absence of proteins, the AT-rich region was alkylated by bizelesin (Fig. 4B, lanes 1 to 4) and adozelesin (Fig. 4A), suggesting that the AT-rich region is flexible. Incubation with T-ag resulted in a slight decrease in alkylation (Fig. 4B, lanes 5 to 8), and the proteins from BSC-1 cell extract caused an even further decrease in alkylation of the AT-rich region (lanes 9 to 12); however, we have no direct evidence for the involvement of specific proteins from BSC-1 cell extract. Finally, when T-ag and the proteins from BSC-1 cell extract were added (lanes 13 to 16), the results were comparable to those obtained with T-ag alone after 30 min (lanes 5 to 8).
Hedamycin, which alkylates guanines at the N7 position, where DNA is unwound, was used to probe the γ-32P-labeled regulatory-region DNA of pC200. No alkylations were detected when hedamycin was omitted from the reaction mixture (data not shown). Prior to incubation with proteins, guanines throughout the regulatory region were alkylated by hedamycin, indicating that the DNA was unwound (Fig. 4C, lanes 1 to 4). T-ag had no significant effect on the amount of alkylation within the 21-bp promoters. Although unwinding of the AT-rich region could not be monitored due to a lack of guanines, the regions immediately adjacent to the AT-rich region were alkylated (lanes 1 to 4). In the absence of proteins, alkylation of the PEN was detected; however, addition of T-ag prevented alkylation within the PEN (lanes 5 to 8). In comparison to when no T-ag was present, there was an increase in alkylation of the EP (lane 6 to 8). In the presence of the proteins from BSC-1 cell extract (lanes 9 to 12), guanines in the 21-bp promoters, PEN, and areas immediately adjacent to the AT-rich region were alkylated by hedamycin, and alkylation of the EP was detected. When the γ-32P-labeled regulatory-region DNA was incubated with T-ag and the proteins from BSC-1 cell extract, the 21-bp promoters were unwound similarly to when only T-ag or proteins from BSC-1 cell extract were present (lanes 13 to 16). Compared to when no proteins were added, there was decreased alkylation of guanines in the PEN and regions flanking the AT-rich region, while there was an increase in alkylation in the EP. These data suggest that T-ag plays a positive role in unwinding of the EP region while it plays a negative role in unwinding of the PEN.
(ii) pOri.
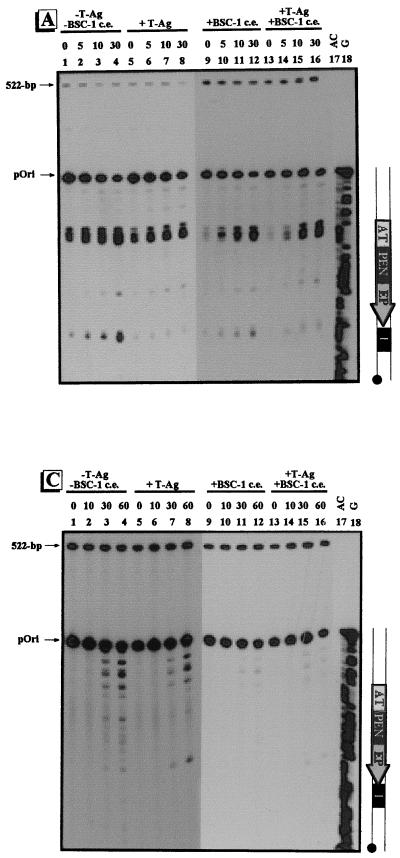
Representative gels of strand breakage assays of the γ-32P-labeled regulatory-region DNA of pOri are shown in Fig. 5. The AT-rich region was alkylated by adozelesin prior to incubation with proteins, and no significant difference was seen after incubation with T-ag and/or the proteins from BSC-1 cell extract (Fig. 5A, lanes 1 to 16). In the absence of proteins (lanes 1 to 4) or in the presence of proteins from BSC-1 cell extract (lanes 9 to 12), site I was alkylated, indicating that it was in a bent confirmation. Incubation with T-ag, either alone or with the proteins from BSC-1 cell extract, resulted in a decrease in alkylation of site I (lanes 5 to 8 and 13 to 16). After incubation with adozelesin for 30 min (lane 8) or 60 min (data not shown), alkylation of site I was barely detectable. Although there was a slight decrease in alkylation of site I with the addition of the proteins from BSC-1 cell extract (lanes 9 to 12), T-ag appears to be the major contributor to the conformation of site I. As with pC200 (Fig. 4A), there was a significant decrease in alkylation of site I in the presence of T-ag (lanes 5 to 8 and 13 to 16).
FIG. 5.
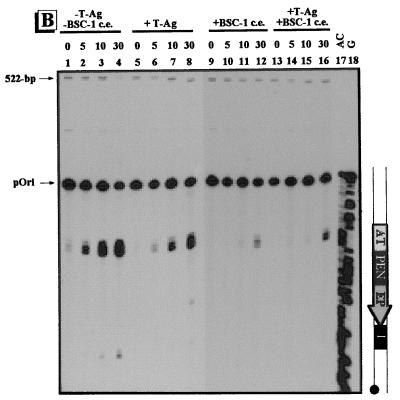
Conformational changes within the γ-32P-labeled regulatory region of pOri. The regulatory region of pOri was treated with adozelesin (A), bizelesin (B), and hedamycin (C). Approximately 0.1 ng of γ-32P-labeled regulatory-region DNA of pOri was incubated without (−) proteins (lanes 1 to 4), with (+) 1 μg of T-ag (lanes 5 to 8), with 6 μg of the proteins from BSC-1 cell extract (c.e.) (lanes 9 to 12), or with 1 μg of T-ag plus 6 μg of the proteins from BSC-1 cell extract (lanes 13 to 16). Reaction times for adozelesin and bizelesin were 0, 5, 10, and 30 min; reaction times for hedamycin were 0, 10, 30, and 60 min. The Maxam-Gilbert sequencing reactions are shown in lanes 17 and 18. To the right of each autoradiogram is a schematic illustration of the regulatory region of pOri. The regulatory elements are as indicated in Fig. 1. The positions of the unmodified pOri regulatory fragment and the 522-bp control fragment are indicated by arrows.
The γ-32P-labeled regulatory region of pOri was also probed with bizelesin. In the absence of proteins the AT-rich region was alkylated, indicating that it was in a straight conformation (Fig. 5B, lanes 1 to 4). After incubation with T-ag, there was a decrease in alkylation of the AT-rich region (lanes 5 to 8); however, significantly less reactivity with the AT-rich region was detected when the proteins from BSC-1 cell extract were present (lane 9 to 12). When incubated with the proteins from BSC-1 cell extract, either alone or in combination with T-ag, essentially no alkylation of the AT-rich region was detected until the 30-min time point (lanes 9 to 16). Unlike that of pC200, the AT-rich region of pOri was less able to straighten in the presence of both T-ag and proteins from BSC-1 cell extract than it was when only T-ag was present.
Hedamycin was used to probe for unwound regions in the γ-32P-labeled regulatory region of pOri. Prior to incubation with proteins, guanines throughout the fragment were accessible to hedamycin (Fig. 5C, lanes 1 to 4). When T-ag was added, the PEN, site I, and regions immediately adjacent to the AT-rich region were accessible to hedamycin (lanes 5 to 8). The EP was the only region to exhibit an increased reactivity with hedamycin when T-ag was present. Incubation with the proteins from BSC-1 cell extract resulted in a decrease in alkylation of all regions (lanes 9 to 12). Furthermore, the increased unwinding of the EP seen in pC200 in the presence of T-ag was not seen in pOri when the proteins of BSC-1 cell extract were also present (lanes 13 to 16).
(iii) p21ds.
Upon probing of the γ-32P-labeled regulatory region of p21ds with adozelesin, no significant differences in alkylation of the AT-rich region before and after incubation with proteins were detected (Fig. 6A, lanes 1 to 16). Also, prior to incubation with proteins (lanes 1 to 4) and after incubation with the proteins from BSC-1 cell extract (lanes 9 to 12), site I was alkylated by adozelesin, indicating that it was in a bent conformation. In contrast to the regulatory regions of pC200 and pOri, site I of p21ds was alkylated after incubation with T-ag (lanes 5 to 8). The 21-bp promoters of p21ds were alkylated (lanes 1 to 4); however, unlike for pC200, there was no significant increase in modification after incubation with T-ag (lanes 5 to 8). Proteins from BSC-1 cell extract had no significant effect on the reactivity with the 21-bp promoters, either alone (Fig. 6A, lanes 9 to 12) or in combination with T-ag (lanes 13 to 16).
The AT-rich region was detected in a straight conformation prior to incubation with proteins (Fig. 6B, lanes 1 to 4). Fewer molecules of the γ-32P-labeled regulatory-region DNA of p21ds were detected in a straight conformation when T-ag was present (lanes 5 to 8) than when no proteins were present. Incubation with the proteins from BSC-1 cell extract, either alone (lanes 9 to 12) or in combination with T-ag (lanes 13 to 16), resulted in a decrease in alkylation of the AT-rich region compared to that occurring when neither the BSC-1 cell extract proteins nor T-ag was present.
When the γ-32P-labeled regulatory region of p21ds was probed with hedamycin, unwound regions were detected throughout the PEN, the EP, the region flanking the AT-rich region, and the 21-bp promoters prior to incubation with proteins (Fig. 6C, lanes 1 to 4). As with pC200, there was an increase in alkylation of the EP of p21ds after incubation with T-ag as well as a decrease in alkylation of the PEN and the 21-bp promoters (lanes 5 to 8). After incubation with the proteins from BSC-1 cell extract (lanes 9 to 12), the PEN, the EP, and the 21-bp promoters were unwound similarly to when no proteins were present. After incubation with T-ag and the proteins from BSC-1 cell extract (lane 13 to 16), the reactivity with the EP increased compared to that occurring when only the proteins from BSC-1 cell extract were present. The decrease in reactivity that was detected with the PEN and the 21-bp promoters suggests that T-ag has an inhibitory effect on unwinding of the regulatory region of p21ds (lanes 5 to 8).
DISCUSSION
The goal of this work was to characterize protein-induced conformations in rearranged SV40 regulatory regions. By using gel mobility shift assays, DNase I footprinting, and sequence- and conformation-specific chemical probes, we have provided insights into the importance of the organization of the regulatory region for DNA conformation and have discussed possible implications of this region’s organization with regard to DNA replication efficiency.
The location of the 21-bp promoters affects binding of T-ag to site III.
Using gel mobility shift assays, we found no gross differences in the ability of T-ag and/or proteins from BSC-1 cell extract to bind the regulatory regions of pC200, pOri, and p21ds. We used DNase I footprinting to identify specific interactions between the rearranged regulatory regions and T-ag or the proteins from BSC-1 cell extract. We observed no protection when the regulatory regions were incubated with the proteins from BSC-1 cell extract. In addition, T-ag binding to the 21-bp promoters (site III) was dependent on their location. In pC200, 2 μg of T-ag partially protected the 21-bp promoters from DNase I (Fig. 3C). In p21ds, the 21-bp promoters were not significantly protected even by 4 μg of T-ag (Fig. 3B). The data suggest that the ability of site III to bind T-ag is position dependent, since more T-ag is required to protect the 21-bp promoters in p21ds than is needed to protect those in pC200. Also, in p21ds, T-ag binding to site I could induce a conformation that hinders T-ag binding to the 21-bp promoters. One disadvantage to DNase I footprinting is that AT tracts, such as the AT-rich region of ori, are generally not sensitive to DNase I digestion, even in the absence of protein binding (Fig. 3). Therefore, we were not able to determine whether binding of T-ag to the PEN also protected the AT-rich region. It was interesting that T-ag protected the region 3′ to ori and 5′ to site I in pOri. Perhaps the bent nature of the 21-bp promoters is slightly inhibitory to T-ag binding.
Flanking sequences influence the conformation of site I.
Alkylation of site I by adozelesin is dependent on the organization of the regulatory region. Prior to T-ag binding, site I in each of the regulatory regions was bent (Fig. 4A, 5A, and 6A). Alkylation of site I in pC200 and pOri was barely detectable when T-ag was present. This could be explained in either of two ways: (i) T-ag binding to site I could sterically hinder alkylation by adozelesin, or (ii) T-ag binding to site I could cause a conformational change eliminating it as a target for adozelesin. Our results support the latter theory. If the reduced alkylation in site I of pC200 and pOri were caused by steric hindrance, there should be an equivalent reduction in alkylation in site I of p21ds. However, alkylation of site I in p21ds was detected after T-ag binding. In addition, we showed that T-ag binds site I in all of the regulatory regions examined (Fig. 3). Therefore, we conclude that the reduced alkylation in site I of pC200 and pOri is most probably due to a change in DNA conformation. The data also suggest that in order for site I to straighten, only one of the flanking regions can be bent, as in pC200 (Fig. 7A) and pOri (Fig. 7B). In p21ds, site I is flanked by both a bent AT-rich region and the bent 21-bp promoters (Fig. 7C).
FIG. 7.
Proposed conformations of pC200, pOri, and p21ds. (A) Model for pC200. Site I, the AT-rich region (AT) of ori, and the 21-bp promoters are in a naturally bent conformation, and the EP is naturally unwound. Because the AT-rich region was alkylated by both adozelesin and bizelesin in the absence of T-ag, both conformations are shown. Upon T-ag binding, site I and the AT-rich region straighten but the 21-bp promoters remain bent. More regulatory regions are detected with an unwound EP (indicated by the larger bubble at the EP) after T-ag binding. (B) Model for pOri. Site I and the AT-rich region of ori are in a naturally bent conformation, and the EP is naturally unwound. Because the AT-rich region was alkylated by both adozelesin and bizelesin in the absence of T-ag, both conformations are shown. Upon T-ag binding, the AT-rich region reacts with both adozelesin and bizelesin, suggesting that it is a flexible region. More regulatory regions are detected with an unwound EP (indicated by the larger bubble at the EP) after T-ag binding. (C) Model for p21ds. Site I, the AT-rich region of ori, and the 21-bp promoters are in a naturally bent conformation, and the EP is naturally unwound. Because the AT-rich region was alkylated by both adozelesin and bizelesin in the absence of T-ag, both conformations are shown. Upon T-ag binding, site I and the 21-bp promoters remain in a bent conformation and the AT-rich region reacts with both adozelesin and bizelesin, suggesting that it is a flexible region. More regulatory regions are detected with an unwound EP (indicated by the larger bubble at the EP) after T-ag binding.
The T-ag-induced conformation of the 21-bp promoters is dependent on the organization of the regulatory region.
In pC200, T-ag binding results in increased alkylation of the 21-bp promoters by adozelesin, which is consistent with previous data (33). Therefore, T-ag may stabilize the 21-bp promoters in a bent conformation. T-ag does not increase the bending of this region in p21ds, and 2 μg of T-ag was not enough to protect the 21-bp promoters of p21ds from DNase I digestion (Fig. 3B). If p21ds and pC200 were in the same cell, p21ds might not recruit T-ag as efficiently as does pC200. There is competition for other cellular proteins in addition to T-ag, such as polymerase α-primase, which also interacts with T-ag (23, 80), and topoisomerase I (23, 62, 63). As a consequence of p21ds not effectively recruiting T-ag, it may not effectively recruit other replication proteins. In support of this idea, it has been shown that the differences in DNA replication efficiencies are exaggerated when there is competition for T-ag (41). An alternative, but not necessarily mutually exclusive, explanation for the barely detectable replication efficiency of p21ds involves steric effects. Regardless of whether T-ag bound to the 21-bp promoters, the EP would be trapped between a double hexamer of T-ag at the PEN, a trimer of T-ag at site I, and a bent 21-bp promoter region (Fig. 7C). This is consistent with the data which suggest that the conformation of the bent 21-bp promoters in p21ds does not significantly change when T-ag is present. This “trapping” of the replication bubble could have a negative effect on initial movement of the replication fork.
Unwinding of a regulatory region is influenced by its organization.
Previous studies have shown that the EP of ori opens first to initiate SV40 DNA replication (7, 15–17, 33, 53, 56, 84). The DNA replication efficiency of plasmids containing ori and site I but lacking 21-bp repeats is lower than that of plasmids containing the 21-bp promoters and an AP-1 binding site on the late-transcription side of ori and site I (42). We hypothesized that the unwinding efficiency of pOri could contribute to its decreased replication efficiency. The amounts of EP unwinding in the three regulatory regions differed. In pC200, T-ag enhanced the unwinding of EP. The ease of opening an already-unwound region (compare lanes 1 to 4 of Fig. 4C with the corresponding lanes of Fig. 5C) could contribute to a replication efficiency that is 30-fold higher than that of pOri. Little unwinding was detected in the regulatory region of pOri compared to that of pC200, suggesting that the 21-bp promoters aid in unwinding of the fragment, as shown by Gutierrez et al. (32). The unwinding of the EP in p21ds is more difficult to explain (Fig. 6C). The barely detectable replication efficiency of p21ds (42) cannot be explained entirely by an inability of the regulatory region to unwind, since the EP is unwound under all protein conditions tested. This suggests that DNA replication efficiency is not solely dependent on the ability of the DNA fragment to unwind. Conformations of the regions flanking the unwound regions as well as proteins bound to the flanking regions could contribute to DNA replication efficiency. Although the regulatory region of p21ds is able to unwind at the EP, the regions flanking the EP (i.e., the 21-bp promoters) bind T-ag and could inhibit initiation of DNA replication. This is supported by the studies by Smelkova and Borowiec (64). Using purified T-ag to investigate the interaction of T-ag with DNA during unwinding, they demonstrated that the binding of T-ag is strongly dependent on the length of the single-stranded DNA of the replication bubble, suggesting that a topological change occurs in the T-ag–DNA complex to allow T-ag helicase activity.
Models for conformational changes in pC200, pOri, and p21ds.
We propose models for the conformational changes occurring in each of the regulatory regions. For simplicity, we have included only the contribution of T-ag. Our model for pC200 (Fig. 7A) is similar to the model presented by Han and Hurley (33). In the first conformational change, site I goes from a bent to a straight conformation upon T-ag binding. Next, T-ag binds to the PEN. Although we could not detect a conformational change in the AT-rich region with adozelesin after T-ag binding, others have shown that the AT-rich region becomes hypersensitive to chemical modification (3) and that the sequence of the AT-rich region is important (2, 20, 21, 28, 31, 38, 44, 52, 68). We do have evidence, obtained by using bizelesin, that a straight AT-rich region is detected in the presence of T-ag; however, there are fewer molecules detected in a straight conformation in the presence of the proteins from BSC-1 cell extract than there are in the presence of T-ag. Possibly the bent conformation of the flexible AT region is stabilized by cell extracts. Finally, the 21-bp promoters are slightly more bent after T-ag binding, and the EP unwinds. Han and Hurley (33) showed that site I was modified by adozelesin when T-ag was absent and was not modified in the presence of T-ag; however, site I straightening was not incorporated into their model.
The model for pOri is similar to the model for pC200 (Fig. 7B). Again, site I is bent and straightens upon T-ag addition. There seems to be no dominant conformation in the AT-rich region when probing is performed with adozelesin or bizelesin, suggesting that the conformation of this region may be greatly influenced by the 21-bp promoters. Finally, the EP unwinds as a result of T-ag binding.
We also propose a model for the conformational changes occurring in p21ds (Fig. 7C). As with pC200 and pOri, T-ag binds to site I. However, in p21ds, site I does not straighten. Again, there is no dominant conformation in the AT-rich region, suggesting that it is flexible. Even though the 21-bp promoters are present in this regulatory region, they are not in a location that would allow them to impact the conformation of the AT-rich region. T-ag has no significant effect on the already-bent 21-bp promoters. The ability of the EP to unwind does not seem to promote DNA replication efficiency in p21ds; replication is barely detectable relative to that of pOri. Smelkova and Boroweic (64) have suggested that even though the EP unwinds in wild-type SV40, this may be insufficient to allow the hexamers of T-ag to facilitate DNA replication.
SV40 has served as a model to identify the molecular events leading to the initiation of DNA replication in eukaryotes (11, 19, 43, 67, 83). Because DNA replication in any organism is highly regulated and complex, understanding the mechanisms controlling initiation is essential. Protein-induced conformational changes in SV40 regulatory regions have been studied extensively (3–8, 14–17, 28, 29, 32, 33, 53, 54, 56, 63, 64, 70, 71, 81, 82, 84). T-ag is both an initiator protein and a helicase, and it induces conformational changes within ori and flanking regions (7, 15–17, 29, 53, 64, 82). Upon T-ag binding to ori, the EP and the AT-rich region of ori become hypersensitive to chemical modification (7), and an 8-bp sequence in the distal arm of the EP denatures (5, 7, 53, 54). AT-rich tracts are a common feature in a wide variety of replication origins in prokaryotes, eukaryotes, and viruses; however, in SV40, it is the EP that is initially melted, not the AT-rich region (56, 84). Single-base substitutions within the AT-rich region, which result in modified bending, have deleterious effects on SV40 DNA replication (16). However, the exact mechanism by which T-ag deforms the AT-rich region is unclear (6, 45). Protein-induced conformational changes are not limited to T-ag binding; replication protein A facilitates DNA unwinding (8), Sp1 induces DNA bending within the 21-bp promoters (70), and other proteins may have similar roles. DNA distortion is also important for recruiting the replication machinery, such as DNA polymerase α, which is provided by the host cell and binds T-ag (80).
This study provided evidence that the organization of the regulatory region influences T-ag binding and DNA conformation. These data are in agreement with the studies by Han and Hurley (33) and provide additional information about the consequences of the presence of the 21-bp promoters and their location within those regulatory regions that promote differences in DNA replication efficiency. Our data indicate that the layout of protein binding sites within the SV40 regulatory region has an impact on the conformational changes that occur when proteins bind this region. These probes will continue to be powerful reagents in characterizing DNA structural changes that occur as a result of protein binding. In addition, we are currently mapping, at the nucleotide level, the site(s) of initiation of DNA replication to determine if it is influenced by the organization of the regulatory region (34a).
ACKNOWLEDGMENTS
We thank Robert Kelly at Upjohn Pharmacia Co. for providing adozelesin and hedamycin and Jill Johnson at the National Cancer Institute for providing hedamycin.
This work was partially funded by Sigma Xi, The Scientific Research Triangle (P.J.W.), and Miami University (P.J.W.).
REFERENCES
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Bergsma D J, Olive D M, Hartzell S W, Subramanian K N. Territorial limits and functional anatomy of the simian virus replication origin. Proc Natl Acad Sci USA. 1982;79:381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya S, Lorimer H E, Prives C. Murine polyomavirus and simian virus 40 large T antigens produce different structural alterations in viral origin DNA. J Virol. 1995;69:7579–7585. doi: 10.1128/jvi.69.12.7579-7585.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boroweic J A. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J Virol. 1992;66:5248–5255. doi: 10.1128/jvi.66.9.5248-5255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boroweic J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 7.Borowiec J A, Dean F B, Hurwitz J. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J Virol. 1991;65:1228–1235. doi: 10.1128/jvi.65.3.1228-1235.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 9.Brockman W W. Evolutionary variants of simian virus 40. Prog Med Virol. 1977;23:69–95. [PubMed] [Google Scholar]
- 10.Brockman W W, Gutai M W, Nathans D. Evolutionary variants of simian virus 40: characterization of cloned complementary variants. Virology. 1975;66:36–52. doi: 10.1016/0042-6822(75)90177-4. [DOI] [PubMed] [Google Scholar]
- 11.Bullock P. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 12.Chou J Y, Avila J, Martin R G. Viral DNA synthesis in cells infected by temperature-sensitive mutants of simian virus 40. J Virol. 1974;14:116–124. doi: 10.1128/jvi.14.1.116-124.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danna K J, Nathans D. Bidirectional replication of simian virus DNA. Proc Natl Acad Sci USA. 1972;69:3097–3100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean F B, Hurwitz J. Simian virus 40 large T antigen untwists DNA at the origin of DNA replication. J Biol Chem. 1991;266:5062–5071. [PubMed] [Google Scholar]
- 15.Deb S, DeLucia A L, Baur C-P, Koff A, Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986;6:1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deb S, DeLucia A L, Koff A, Tsui S, Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986;6:4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deb S, Tsui S, Koff A, DeLucia A L, Parsons R, Tegtmeyer P. The T-antigen-binding domain of the simian virus 40 core origin of replication. J Virol. 1987;61:2143–2149. doi: 10.1128/jvi.61.7.2143-2149.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLucia A L, Lewton B A, Tjian R, Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983;46:143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePamphilis M L, editor. Concepts in eukaryotic DNA replication. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- 20.DiMaio D, Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980;140:129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- 21.DiMaio D, Nathans D. Regulatory mutants of simian virus 40: effect of mutations at a T antigen binding site on DNA replication and expression of viral genes. J Mol Biol. 1982;156:531–548. doi: 10.1016/0022-2836(82)90265-0. [DOI] [PubMed] [Google Scholar]
- 22.Ding Z-M, Harshey R M, Hurley L H. Dextro CC-1065 as a structural probe of mu transposase-induced bending of DNA: overcoming limitations of hydroxyl-radical footprinting. Nucleic Acids Res. 1993;21:4281–4287. doi: 10.1093/nar/21.18.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dornreiter I, Erdile L F, Gilbert I U, von Winker D, Kelly T J, Fanning E. Interactions of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dynan W S, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 25.Fareed G C, Garon C F, Salzman N P. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol. 1972;10:484–491. doi: 10.1128/jvi.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fromm M, Berg P. Deletion mapping of DNA region required for SV40 early promoter function in vivo. J Mol Appl Genet. 1982;1:457–481. [PubMed] [Google Scholar]
- 27.Fromm M, Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983;3:991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerard R, Gluzman Y. Functional analysis of the role of the A+T-rich region and upstream flanking sequences in simian virus 40 DNA replication. Mol Cell Biol. 1986;6:4570–4577. doi: 10.1128/mcb.6.12.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goetz G S, Dean F B, Hurwitz J, Matson S W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 30.Gruss P, Dhar R, Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci USA. 1981;78:943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutai M W, Nathans D. Evolutionary variants of simian virus 40: nucleotide sequence of a conserved SV40 DNA segment containing the origin of viral DNA replication as an inverted repetition. J Mol Biol. 1978;126:259–274. doi: 10.1016/0022-2836(78)90362-5. [DOI] [PubMed] [Google Scholar]
- 32.Gutierrez C, Guo Z-S, Roberts J, DePamphilis M L. Simian virus 40 origin auxiliary sequences weakly facilitate T-antigen binding but strongly facilitate DNA unwinding. Mol Cell Biol. 1990;10:1719–1728. doi: 10.1128/mcb.10.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han F X, Hurley L H. A model for the T-antigen-induced structural alteration of the SV40 replication origin based upon experiments with specific probes for bent, straight, and unwound DNA. Biochemistry. 1996;35:7995–8001. doi: 10.1021/bi960251d. [DOI] [PubMed] [Google Scholar]
- 34.Herr W, Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate from one another. Cell. 1986;45:461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- 34a.Hu, B., and M. E. Woodworth. Unpublished data.
- 35.Innis J W, Scott W A. DNA replication and chromatin structure of simian virus 40 insertion mutants. Mol Cell Biol. 1984;4:1499–1507. doi: 10.1128/mcb.4.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones N C, Rigby P W J, Ziff E B. trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988;2:267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- 37.Khoury G, Fareed G C, Berry K, Martin M A, Lee T N H, Nathans D. Characterization of a rearrangement in viral DNA: mapping of the circular simian virus 40-like DNA containing a triplication of a specific one-third of the genome. J Mol Biol. 1974;87:289–301. doi: 10.1016/0022-2836(74)90150-8. [DOI] [PubMed] [Google Scholar]
- 38.Learned R M, Meyers R M, Tjian R, editors. Replication in monkey cells of plasmid DNA containing the minimal SV40 origin. New York, N.Y: Academic Press, Inc.; 1981. [Google Scholar]
- 39.Lee C S, Pfeifer G P, Gibson N W. Mapping of DNA alkylation sites induced by adozelesin and bizelesin in human cells by ligation-mediated polymerase chain reaction. Biochemistry. 1994;33:6024–6030. doi: 10.1021/bi00185a043. [DOI] [PubMed] [Google Scholar]
- 40.Lee T N H, Brockman W W, Nathans D. Evolutionary variants of simian virus 40: cloned substituted variants containing multiple initiation sites for DNA replication. Virology. 1975;66:53–69. doi: 10.1016/0042-6822(75)90178-6. [DOI] [PubMed] [Google Scholar]
- 41.Lee-Chen G-J, Woodworth-Gutai M. Evolutionarily selected replication origins: functional aspects and structural organization. Mol Cell Biol. 1986;6:3077–3085. doi: 10.1128/mcb.6.9.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee-Chen G-J, Woodworth-Gutai M. Simian virus 40 DNA replication: functional organization of regulatory elements. Mol Cell Biol. 1986;6:3086–3093. doi: 10.1128/mcb.6.9.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Kelly T. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J J, Peden K W C, Dixon R A F, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986;6:1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S, Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastrangelo I A, Hough P V C, Wall L S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 47.Mastrangelo I A, Hough P V C, Wilson V, Wall J, Hainfeld J, Tegtmeyer P. Monomers through trimers of large tumor antigen bind in region I and monomers through tetramers bind in region II of simian virus 40 origin of replication DNA as stable structures in solution. Proc Natl Acad Sci USA. 1985;82:3626–3630. doi: 10.1073/pnas.82.11.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mertz J E, Carbon J, Herzberg M, Davies R W, Berg P. Isolation and characterization of individual clones of simian virus 40 mutants containing deletion, duplications and insertion in their DNA. Cold Spring Harbor Symp Quant Biol. 1974;39:69–84. doi: 10.1101/sqb.1974.039.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell M A, Kelly R C, Wicnienski N A, Hatzenbuhler N T, Williams M G, Tetzold G C, Slighton J L, Siemieniak P R. Synthesis and DNA cross-linking by a rigid CPI dimer. J Am Chem Soc. 1991;113:8994–8995. [Google Scholar]
- 50.Moreau P, Hen R, Wasylyk B, Everett R, Gaub M P, Chambon P. The SV40 72 bp repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6069. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myers R M, Rio D C, Robbins A K, Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981;25:373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- 52.Myers R M, Tjian R. Construction and analysis of SV40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci USA. 1980;77:6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.National Institutes of Health. Revision date, 14 April 1999. NIH Image 1.52. National Institutes of Health, Bethesda, Md. [Online.] http://rsb.info.nih.gov/nih-image. [12 October 1999, last date accessed.]
- 53.Parsons R, Anderson M E, Tegtmeyer P. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J Virol. 1990;64:509–518. doi: 10.1128/jvi.64.2.509-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds V L, Molineux I J, Kaplan D J, Swenson D H, Hurley L H. Reaction of the antitumor antibiotic CC-1065 with DNA. Location of the site of thermally induced strand breakage and analysis of DNA sequence specificity. Biochemistry. 1985;24:6228–6237. doi: 10.1021/bi00343a029. [DOI] [PubMed] [Google Scholar]
- 56.Roberts J M. Simian virus 40 (SV40) large tumor antigen causes stepwise changes in SV40 origin structure during initiation of DNA replication. Proc Natl Acad Sci USA. 1989;86:3939–3943. doi: 10.1073/pnas.86.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg M, Segal S, Kuff E L, Singer M F. The nucleotide sequence of repetitive monkey DNA found in defective simian virus 40. Cell. 1977;11:845–857. doi: 10.1016/0092-8674(77)90296-3. [DOI] [PubMed] [Google Scholar]
- 58.Shalloway D, Kleinberger T, Livingston D M. Mapping of SV40 DNA replication binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980;20:411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- 59.Sheflin L, Celeste A, Woodworth-Gutai M. Recombination of simian virus 40-infected cells: structure of naturally arising variants ev-2114, ev-2101, and ev-1110. J Biol Chem. 1983;258:14315–14321. [PubMed] [Google Scholar]
- 60.Shortle D R, Margolskee R F, Nathans D. Mutational analysis of the simian virus 40 replicon: pseudorevertants of mutants with a defective replication origin. Proc Natl Acad Sci USA. 1979;76:6128–6131. doi: 10.1073/pnas.76.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 62.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 63.Simmons D T, Roy R, Chen L, Gai D, Trowbridge P W. The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. J Biol Chem. 1998;273:20390–20396. doi: 10.1074/jbc.273.32.20390. [DOI] [PubMed] [Google Scholar]
- 64.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sol C J A, Hassing I, Maris W, Walig C, van der Noordaa J. Evolutionary variants of simian virus 40 which are impaired in early lytic functions but transform nonpermissive cells. J Virol. 1981;37:395–410. doi: 10.1128/jvi.37.1.395-410.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 68.Subramanian K N, Shenk T. Definition of the boundaries of the origin of DNA replication in SV40. Nucleic Acids Res. 1978;5:3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun D, Hurley L H. Analysis of the monoalkylation and cross-linking sequence specificity of bizelesin, a bifunctional alkylation agent related to (+CC)-1065. J Am Chem Soc. 1993;115:5925–5933. [Google Scholar]
- 70.Sun D, Hurley L H. Cooperative bending of the 21-base-pair repeats of the SV40 viral early promoter by human Sp1. Biochemistry. 1994;33:9578–9587. doi: 10.1021/bi00198a025. [DOI] [PubMed] [Google Scholar]
- 71.Sun D, Hurley L H. TBP binding to the TATA box induces a specific downstream unwinding site that is targeted by pluramycin. Chem Biol. 1995;2:457–469. doi: 10.1016/1074-5521(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 72.Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972;10:591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tegtmeyer P, Andersen B, Shaw S B, Wilson V G. Alternative interactions of the SV40 A protein with DNA. Virology. 1981;115:75–87. doi: 10.1016/0042-6822(81)90090-8. [DOI] [PubMed] [Google Scholar]
- 74.Tegtmeyer P, Lewton B A, DeLucia A L, Wilson V G, Ryder K. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J Virol. 1983;46:151–161. doi: 10.1128/jvi.46.1.151-161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tjian R. The binding site on SV40 DNA for a T-antigen related protein. Cell. 1978;13:165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- 76.Tjian R. Protein-DNA interactions at the origin of simian virus 40 DNA replication. Cold Spring Harbor Symp Quant Biol. 1978;43:655–662. doi: 10.1101/sqb.1979.043.01.073. [DOI] [PubMed] [Google Scholar]
- 77.Tooze J. DNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- 78.Wakamiya T, McCutchan T, Rosenberg M, Singer M F. Structure of simian virus 40 recombinants that contain both host and viral DNA sequence. I. The structure of variant CVP8/1/P2 (Eco RI res) J Biol Chem. 1979;254:3584–3591. [PubMed] [Google Scholar]
- 79.Weiland K L, Dooley T P. In vitro and in vivo DNA binding by the CC-1065 analog U-73975. Biochemistry. 1991;30:7559–7565. doi: 10.1021/bi00244a027. [DOI] [PubMed] [Google Scholar]
- 80.Weisshart K, Bradley M K, Weiner B M, Schneider C, Moarefi I, Fanning E, Arthur A K. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase α and replicate SV40 DNA in vitro. J Virol. 1996;70:3509–3516. doi: 10.1128/jvi.70.6.3509-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiekowski M, Schwarz M W, Stahl H. Simian virus 40 large T antigen DNA helicase. Characterization of the ATPase-dependent DNA unwinding activity and its substrate requirements. J Biol Chem. 1988;263:436–442. [PubMed] [Google Scholar]
- 83.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wold M, Li F, Kelly T. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci USA. 1987;84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woodworth-Gutai M, Celeste A, Sheflin L, Sclair M. Naturally arising recombinants that are missing portion of the simian virus 40 regulatory region. Mol Cell Biol. 1983;3:1930–1936. doi: 10.1128/mcb.3.11.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wun-Kim K, Upsom R, Young W, Melendy T, Stillman B, Simmons D T. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zenke M, Grundstrom T, Matthes H, Wintzerith M, Schatz C, Wildeman A, Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986;5:387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]