Abstract
There are natural mutations in the coding and noncoding regions of the human immunodeficiency virus type 1 (HIV-1) CC-chemokine coreceptor 5 (CCR5) and in the related CCR2 protein (the CCR2-64I mutation). Individuals homozygous for the CCR5-Δ32 allele, which prevents CCR5 expression, strongly resist HIV-1 infection. Several genetic polymorphisms have been identified within the CCR5 5′ regulatory region, some of which influence the rate of disease progression in adult AIDS study cohorts. We genotyped 1,442 infants (1,235 uninfected and 207 HIV-1 infected) for five CCR5 and CCR2 polymorphisms: CCR5-59353-T/C, CCR5-59356-C/T CCR5-59402-A/G, CCR5-Δ32, and CCR2-64I. The clinical significance of each genotype was assessed by measuring whether it influenced the rate of perinatal HIV-1 transmission among 667 AZT-untreated mother-infant pairs (554 uninfected and 113 HIV-1 infected). We found that the mutant CCR5-59356-T allele is relatively common in African-Americans (20.6% allele frequency among 552 infants) and rare in Caucasians and Hispanics (3.4 and 5.6% of 174 and 458 infants, respectively; P < 0.001). There were 38 infants homozygous for CCR5-59356-T, of whom 35 were African-Americans. Among the African-American infants in the AZT-untreated group, there was a highly significant increase in HIV-1 transmission to infants with two mutant CCR5-59356-T alleles (47.6% of 21), compared to those with no or one mutant allele (13.4 to 14.1% of 187 and 71, respectively; P < 0.001). The increased relative risk was 5.9 (95% confidence interval, 2.3 to 15.3; P < 0.001). The frequency of the CCR5-59356-T mutation varies between population groups in the United States, a low frequency occurring in Caucasians and a higher frequency occurring in African-Americans. Homozygosity for CCR5-59356-T is strongly associated with an increased rate of perinatal HIV-1 transmission.
The CC-chemokine receptor 5 (CCR5) is the major coreceptor for the most commonly transmitted (R5) strains of human immunodeficiency virus type 1 (HIV-1) (1, 5, 8, 10, 11). Over the past few years, several mutations in CCR5 have been identified as natural genetic polymorphisms able to influence the probability of acquiring HIV-1 infection or to affect the rate of disease progression, in infected adults (7, 12, 17, 22, 31, 42) or infants (3, 24, 32). A 32-nucleotide deletion in CCR5, the CCR5-Δ32 allele which encodes a defective CCR5 protein, has been identified as a natural genetic polymorphism reducing the risk of acquiring HIV-1 infection and the rate of disease progression in infected adults. The complete absence of CCR5 expression in CCR5-Δ32 homozygotes is strongly protective against infection by R5 HIV-1 variants (7, 12, 17, 31), although a few such individuals have been infected with CXCR4-using (X4) strains (2, 23, 28, 36). Adults and children heterozygous for the CCR5-Δ32 allele are not protected against HIV-1 infection but progress more slowly to AIDS and death, compared to CCR5 wild-type individuals (3, 7, 12, 13, 22, 24, 32, 42). However, the magnitude of CCR5 expression on blood CD4+ T cells varies substantially among individuals with one or two wild-type CCR5-coding alleles (25, 30, 37), leading to suggestions that polymorphisms in the CCR5 untranslated (regulatory) region (5′-UTR) might influence HIV-1 transmission and disease progression (25, 27, 30, 39). Several genetic variations have been identified within the CCR5 5′-UTR (14, 19, 21, 26, 27), some of which have been reported recently to affect the rate of disease progression in adults (19, 21, 26). One such polymorphism (CCR5-59653-T) is genetically associated with a coding region mutation in CCR2 (an isoleucine instead of valine at amino acid 64; CCR2-64I). Adults who possess the CCR2-64I allele are not protected against HIV-1 infection but progress less rapidly to disease once infected (13, 14, 34), although the mutation does not affect CCR2 protein function (16).
No studies on the CCR5 5′-UTR have yet been conducted in the setting of mother-infant HIV-1 transmission, which accounts for a significant fraction of new HIV-1 infections worldwide. Thus, we have now assessed whether genetic variation in the CCR5 5′-UTR, and whether possession of the CCR5-Δ32 and CCR2-64I alleles, influences perinatal HIV-1 transmission. To do this, we have used four well-characterized cohorts of HIV-1-infected and uninfected infants from the United States. Since CCR5-Δ32 and CCR5 5′-UTR polymorphisms exist at different frequencies among different ethnic groups (13, 18–20, 26, 27, 35), we also subdivided the cohorts into Caucasian, Hispanic, and African-American groupings. In addition, because of the significant reduction in perinatal HIV-1 transmission that is associated with zidovudine (AZT) prophylaxis (6, 37), we examined the relationship between the CCR5 and CCR2 polymorphisms and HIV-1 perinatal transmission separately for AZT-treated and AZT-untreated mother-infant pairs.
MATERIALS AND METHODS
Clinical samples.
Pediatric clinical samples were obtained from four cohorts: the Women and Infants Transmission Study (WITS) cohort, the New York City-Western New England (NY-WNE) cohort, the Newark Perinatal Cohort (NPC), and the ARIEL Project cohort. All pediatric clinical samples were derived from patients in the United States. For this study, only infants followed from birth were included. Infants first observed later in life were excluded to avoid introducing a strong selection bias resulting in an apparently higher fraction of infected infants; infected infants are far more likely than uninfected ones to be brought to the attention of pediatricians in early life. HIV-1 infection was defined as two positive viral cultures and/or HIV-1 DNA PCR assays on two separate occasions. The race of the infant was taken to be the self-described race of the mother.
Treated and untreated groups represent the infants who, along with their mothers, received an AZT prophylaxis regimen and those infants who did not receive prophylactic AZT or were born to mothers who received no AZT during pregnancy. Mothers and infants in the AZT-treated group received a variety of treatment regimens (i.e., different antiretroviral agents were given to mothers at different periods, sometimes intravenously during delivery and sometimes not; not all infants were treated the same).
The WITS and ARIEL cohort designs are described elsewhere (4, 33). In the WITS cohort, non-HIV-1 infection was defined as two negative cultures at 1 month or older, one of which was at 6 months or older, and no positive cultures. Any case not meeting the standard definition of infected or noninfected was examined by a committee and assigned to one or the other group, if possible, by considering the totality of the clinical evidence. For the WITS cohort, the distribution of infants among the ethnic groups was as follows: 355 African-Americans, 130 Caucasians, 311 Hispanics, and 30 of other ethnic groups. The ARIEL cohort used a similar strategy for confirming each infant’s HIV-1 status. In this cohort, there were 134 African-Americans, 16 Caucasians, and 41 Hispanics. The NY-WNE cohort consists of infants and children born to HIV-1-seropositive women who were followed prospectively at the following consortium sites: NYU Medical Center/Bellevue Hospital, New York, N.Y.; University of Massachusetts Medical School, Worcester; Baystate Medical Center, Springfield, Mass.; University of Connecticut Medical School, Farmington; and Connecticut Children’s Hospital, Hartford. The cohort includes children enrolled in perinatal transmission protocols and followed from birth. Informed consent for these Institutional Review Board-approved protocols was obtained in the individual participating institutions. HIV-1 infection was defined as in the WITS cohort. For the combined NY-WNE cohort, the distribution of infants among the ethnic groups was 60 African-Americans, 35 Caucasians, 124 Hispanics, and 34 of other ethnic origins. The NPC enrolled HIV-1-infected pregnant women and their newborns either during pregnancy or at delivery. The entrance criterion was that all infants enrolled were born to HIV-1-infected women.
Information concerning demographics, HIV-1-specific risk factors, medical history, and delivery variables were collected through interview and medical record review for participating mothers. The NPC used a strategy similar to that used by the WITS cohort for confirming each infant’s HIV-1 status. The ethnic distribution of the infants was 142 African-Americans, 13 Caucasians, 16 Hispanics, and 1 of other ethnic origin.
Identification of CCR5-59356-C/T polymorphic site.
Genomic DNA was extracted from peripheral blood mononuclear cells (QIAamp Blood kit; Qiagen) from 132 uninfected Caucasian, Hispanic, Asian, or African-American adult volunteers resident in the United States. A 688-nucleotide sequence spanning the CCR5 5′-UTR and the two noncoding exons (26, 27) was amplified from each sample by PCR (Fig. 1). The primers were LK84 (5′-AAGTCCAGGATCCCCCTCTA-3′; positions 59043 to 59064) and LK87 (5′-CATTCCAAACTGTGACCCTTTCC-3′; positions 59709 to 59732; GenBank accession no. U95626). Forty cycles of amplification were performed in a 96-well thermocycler (model 9600; Perkin-Elmer) under conditions previously described (14). Each PCR product was purified and sequenced, using LK84, LK85 (5′-GTGTAGTGGGATGAGCAGAGA-3′; positions 59290 to 59310), and LK86 (5′-CAGAAGAGCTGAGACATCCGT-3′; positions 59530 to 59550) as overlapping sequencing primers. The CCR5-59356-C/T, CCR5-59353-T/C, and CCR5-59402-A/G polymorphisms were identified by analysis of the aligned DNA sequences, using standard procedures (data not shown).
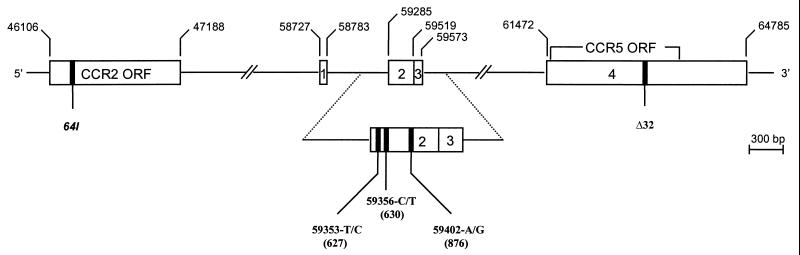
FIG. 1.
Schematic representation of the genomic organization of the CCR2 and CCR5 genes on chromosome 3 and locations of polymorphic sites in the regulatory region of CCR5 (59029-G/A, 59353-T/C, 59356-C/T, and 59402-A/G) and in the coding regions of CCR5 (Δ32) and CCR2 (64I) genes. Open boxes indicate noncoding exons and open reading frames (ORF); lines signify introns. Exons and mutations are numbered based on the nucleotide position of the unpublished sequence with GenBank accession no. U95626. For each CCR5 polymorphism, the first letter indicates the wild-type nucleotide and the second indicates the mutant nucleotide. Numbers in parentheses indicate positions of the polymorphic sites in an alternative numbering system (19).
Spectral genotyping.
To genotype the five polymorphisms (CCR5-Δ32, CCR2-64I, CCR5-59353-T/C, CCR5-59356-C/T, and CCR5-59402-A/G) in each infant from all four cohorts, we used both DNA sequencing and spectral genotyping (4, 15, 33). The spectral genotyping assays for CCR5-Δ32 and CCR2-64I have been previously described (14, 15). To genotype the CCR5-59402-A/G polymorphism, we designed a new assay based on the general operational principle described previously (14, 15). One molecular beacon, labeled with 6-carboxyfluorescein (FAM), recognizes the wild-type CCR5-59402-A allele (A at position 59653); the other, labeled with hexachlorofluorescein (HEX), is for the mutant CCR5-59402-G allele (G at position 59402). The nucleotide sequences were FAM-5′-CGGCGTTCTTCTTTTTAAGTTGAGGCCG-3′-DABCYL and HEX-5′-CGCGGTCAGTGAA CAGTTCTTCCTTTTAAGTCCGCG-3′- DABCYL, respectively. The primers were LK85 and LK112 (5′-GCTGTGCAAATCAATCATATAGAG-3′; positions 59497 to 59520). The real-time PCR conditions and data analyses were as previously described (14, 15) except that the annealing and data collection temperature was 55°C instead of 60°C.
For the CCR5-59353-T/C and CCR5-59356-C/T mutations, we designed two separate multiplex spectral genotyping assays (four allele-specific molecular beacons in each assay). This approach was necessary because the two polymorphic sites are separated by only two nucleotides and the hybridization of molecular beacons can be severely affected. To genotype the CCR5-59353-T/C polymorphism, we designed four molecular beacons; two (HEX-5′-CGGGCCTGGTCTGAAAGTTTATTTAGCCCG-3′-DABCYL and HEX-5′-CGGGCCTGGTCTAAAAGTTTATTTAGCCCG-3′-DABCYL) recognized the CCR5-59353T/CCR5-59356C and CCR5-59353T/CCR5-59356T allele haplotypes, and two (FAM-5′-CGGGCCTGGTCTGAAGGTTTATTTAGCCCG-3′-DABCYL and FAM-5′-CGGGCCTGGTCTAAAGGTTTATTTAGCCCG-3′-DABCYL) recognized the CCR5-59353C/CCR5-59356C and CCR5-59353C/CCR5-59356T allele haplotypes. To genotype the CCR5-59356-C/T polymorphism, we designed four molecular beacons; two (HEX-5′-CGGGCCTGGTCTGAAAGTTTATTTAGCCCG-3′-DABCYL and HEX-5′-CGGGCCTGGTCTGAAGGTTTATTTAGCCCG-3′-DABCYL) recognized the CCR5-59353T/CCR5-59356C and CCR5-59353C/CCR5-5935C6C allele haplotypes, and two (FAM-5′-CGGGCCTGGTCTAAAAGTTTATTTAGCCCG-3′-DABCYL and FAM-5′-CGGGCCTGGTCTAAAGGTTTATTTAGCCCG-3′-DABCYL) recognized the CCR5-59353T/CCR5-59356T and CCR5-59353C/CCR5-59356T allele haplotypes. The primers were LK85 and LK112. The real-time PCR conditions were as previously described (14, 15) except that fluorescence spectra were recorded at 52°C. The emission spectra were decomposed into the individual spectral contributions of FAM and HEX, which allowed the determination of all possible CCR5-59353/CCR5-59356 allele haplotypes.
Statistical analysis.
The Pearson chi-square test for 3 × 2 contingency tables was used to determine the statistical significance of differences in genotype distributions among different populations (e.g., infected and uninfected, different ethnic groups). Fisher’s exact test was applied to 2 × 2 contingency tables and used to determine the statistical significance (two sided) of differences in perinatal transmission rates between different subgroups (e.g., between different genotypes). Logistic regression analysis was used to evaluate the relative odds of perinatal transmission, the 95% confidence interval, and the statistical significance for each variable in the regression equation (e.g., between different genotypes). The Hardy-Weinberg equilibrium for each genetic polymorphism was assessed by the Pearson chi-square test with a 3 × 2 contingency table (1 df) of the observed genotype distribution in comparison with the expected distribution based on the observed allele frequency. Mantel-Haenzel analyses were used to verify that the results presented were not influenced by the year of enrollment or study cohorts. Multivariate logistic regression was performed to determine if unions and intersections of polymorphism genotypes were associated with the risk of transmission. Logistic regressions for these analyses were done by a step-down procedure that started with the entire collection of variables (intersections done separately from unions). The univariate genotypes were included in the analysis set of independent variables. The step-down procedure used an alpha level of 0.01 for a variable to stay in the regression. Logistic regression was also used to determine if a univariate association of a polymorphism genotype with HIV-1 perinatal transmission remained, after adjusting for other known predictors of transmission, for the subset of infants in the WITS and ARIEL cohorts (n = 656). The adjustment variables included in this analysis were as follows: African-American race, mother and infant AZT use, mother and infant CD4 counts, premature birth, birth after February 1994, and CCR5-59356T homozygosity.
RESULTS
A total of 1,442 infants born to HIV-1-infected mothers in four pediatric cohorts met the inclusion criteria for this study. Of these infants, 1,235 were uninfected and 207 were infected, producing an overall transmission rate of 14.4% (Table 1). The racial distributions for the combined cohorts were 47.9% (n = 691) African-Americans, 13.5% (n = 194) Caucasians, 34.1% (n = 492) Hispanics, and 4.5% (n = 65) other racial groups. In total, 667 infants were in the AZT-untreated group and 473 were in the AZT-treated group. For 302 mother-infant pairs, treatment data were missing or prophylaxis was only partial; these 302 cases were excluded from analyses. The racial frequencies among the 302 excluded infants, the AZT-treated group, and the AZT-untreated group were not significantly different. In the AZT-treated category, the HIV-1 transmission rate was significantly lower (8.5%) than in the untreated category (17.1%; P < 0.001), confirming that AZT prophylaxis is associated with a lower risk of transmission (0.45; 95% confidence interval, 0.31 to 0.66) (P < 0.001). A lower transmission rate for mother-infant pairs receiving AZT was observed in all four individual cohorts, for all ethnic groups and all coreceptor genotypes. There was no evidence in this data set for a time-associated decrease in transmission among mother-infant pairs that did not receive AZT.
TABLE 1.
Baseline characteristics of pediatric AIDS study cohorts and HIV-1 infection status
| Variable | HIV-1-infected infants/totala (% HIV-1-infected)
|
||
|---|---|---|---|
| Overall | AZT given to mother and infant | No AZT given to mother or infant | |
| Cohort | |||
| WITS | 127/826 (15.4) | 29/284 (10.2) | 64/362 (17.7) |
| NY-WNE | 35/253 (13.8) | 3/54 (5.6) | 24/152 (15.8) |
| NPC | 30/172 (17.4) | 0/6 (0.0) | 19/112 (17.0) |
| ARIEL | 15/191 (7.9) | 8/128 (6.3) | 6/42 (14.3) |
| Total | 207/1442 (14.4) | 40/473 (8.5) | 113/667 (16.9) |
| Race | |||
| African-Americans | 98/691 (14.2) | 18/214 (8.4) | 52/323 (16.1) |
| Caucasians | 30/194 (15.5) | 6/61 (9.8) | 18/92 (19.6) |
| Hispanics | 71/492 (14.4) | 11/170 (6.5) | 42/231 (18.2) |
| Other | 8/65 (12.3) | 5/28 (17.9) | 1/21 (4.8) |
| Genetic analyses performed | |||
| CCR5-Δ32 | 207/1442 (14.4) | 40/473 (8.5) | 113/667 (16.9) |
| CCR2-64I | 206/1425 (14.5) | 40/471 (8.5) | 113/659 (17.1) |
| CCR5-59353-T/C | 195/1382 (14.1) | 39/465 (8.4) | 106/635 (16.7) |
| CCR5-59356-C/T | 183/1248 (14.7) | 33/382 (8.6) | 101/601 (16.8) |
| CCR5-59402-A/G | 203/1420 (14.3) | 40/468 (8.5) | 110/655 (16.8) |
Total number of infants first seen at birth for which there were samples accessible for genetic analysis.
We analyzed the effect of each of five coreceptor genetic polymorphisms (CCR5-Δ32, CCR2-64I, CCR5-59353-T/C, CCR5-59356-C/T, and CCR5-59402-A/G), and of composite genotypes, on perinatal HIV-1 transmission. Separate analyses were performed on both the AZT-treated and untreated groups, because the substantial effect of AZT might in principle obscure subtle genetic influences on transmission. The treated group comprises a complex set of therapeutic regimens and treatment histories, and so we considered the untreated mother-infant pairs to be the optimal subset for discerning genetic associations with HIV-1 transmission. Figure 1 shows the positions on chromosome 3 of all of the studied mutations, including the CCR5-59356-C/T polymorphism that we identified in this study. Each mutation was evaluated by (i) comparing the genotype and mutant allele frequencies between HIV-1-infected and uninfected groups; (ii) comparing the fraction of HIV-1-infected children among the different genotypes; and (iii) evaluating the relative risks by regression analyses. The full set of results for the AZT-untreated group is shown in Table 2; the significant observations are further analyzed and presented in Fig. 2 and 3.
TABLE 2.
CCR5 polymorphisms on HIV-1 vertical transmission among the AZT-untreated groupa
| Polymorphism | Group | Wt/Wt | Wt/Mut | Mut/Mut | Allele (%) | P valuee |
|---|---|---|---|---|---|---|
| CCR5-Δ32b | Uninfected | 520 | 34 | 0 | 3.1 | 0.85 |
| HIV-1 infected | 108 | 5 | 0 | 2.2 | 0.96 | |
| Transmission rate | 17.2% | 12.8% | N/A | 0.48 | ||
| CCR2-64Ic | Uninfected | 406 | 125 | 15 | 14.2 | 0.37 |
| HIV-1 infected | 81 | 30 | 2 | 15.0 | 0.92 | |
| Transmission rate | 16.6% | 19.4% | 11.8% | 0.61 | ||
| CCR5-59353-T/Cd | Uninfected | 174 | 250 | 105 | 43.5 | 0.68 |
| HIV-1 infected | 37 | 43 | 26 | 44.8 | 0.18 | |
| Transmission rate | 17.5% | 14.7% | 19.8% | 0.39 | ||
| CCR5-59356-C/T | Uninfected | 403 | 85 | 12 | 10.9 | 0.02 |
| HIV-1 infected | 70 | 21 | 10 | 20.3 | 0.002 | |
| Transmission rate | 14.8% | 19.8% | 45.5% | <0.001 | ||
| CCR5-59402-A/G | Uninfected | 328 | 167 | 50 | 24.5 | 0.001 |
| HIV-1 infected | 74 | 32 | 4 | 18.2 | 0.97 | |
| Transmission rate | 18.4% | 16.1% | 7.4% | 0.12 |
Only infants first seen at birth are considered. Wt, wild type; Mut, mutant; NA, not applicable.
For the NY-WNE cohort, the results are based on unpublished data of Pollack et al.
In the AZT-treated group, there was a trend toward reduced transmission among CCR2-64I heterozygotes (4.1% of 121) relative to wild-type homozygotes (10.0% of 338; P = 0.06).
In the AZT-treated group, there is a significantly lower rate of transmission among CCR5-59353-C/T heterozygotes (5.4%; P = 0.004) and CCR5-59353-C mutant homozygotes (6.1%; P = 0.04) compared to CCR5-59353-T wild-type homozygotes (14.0%).
P values corresponding to uninfected and HIV-1-infected infants denote the Hardy-Weinberg equilibrium values, whereas those for transmission rates denote the significance of the difference in genotype distribution between the HIV-1-infected and uninfected groups (Pearson 2 × 3 chi-square values). The low Hardy-Weinberg value for CCR5-59402-A/G is due to different patterns of genotype distribution in the various racial groups; the genotype distribution of CCR5-59402-A/G is in Hardy-Weinberg equilibrium (P > 0.1) for each of the racial groups separately.
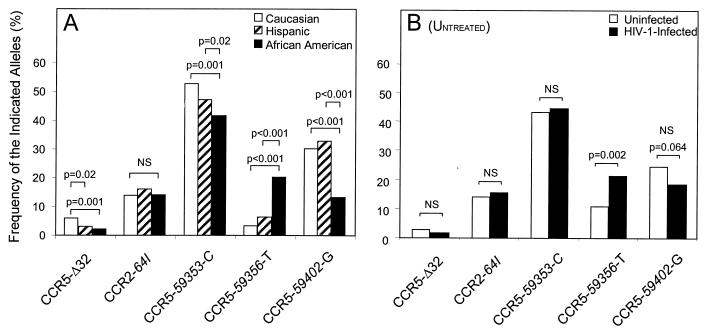
FIG. 2.
(A) Frequency of CCR2 and CCR5 mutations among Caucasians, Hispanics, and African-Americans from all cohorts. Categorization into racial groups was determined by self-reporting. (B) Frequencies of mutant CCR5-Δ32, CCR2-64I, CCR5-59353-C, CCR5-59356-T, and CCR5-59402-G alleles in HIV-1-infected and uninfected infants (untreated category). The frequency of the CCR5-59356-T allele is significantly higher in HIV-1-infected than in uninfected infants (P = 0.002), whereas there was a trend in the association of the CCR5-59402-G allele with a decreased rate of perinatal transmission (P = 0.064). There is no significant difference (NS) in the frequencies of the CCR5-Δ32, CCR2-64I, and CCR5-59353-C alleles between the two groups.
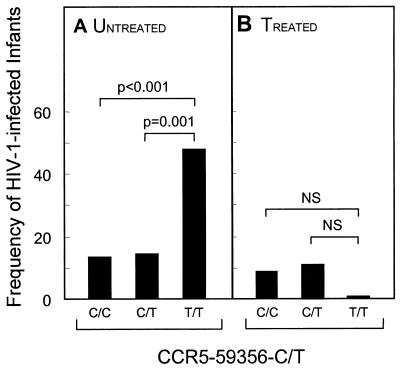
FIG. 3.
(A) Untreated African-American CCR5-59356-T mutant homozygotes have a highly significantly (P < 0.001) increased rate of HIV-1 transmission (47.6% of 21 individuals) compared to CCR5-59356-C wild-type homozygotes (13.4% of 187) or heterozygotes (14% of 71). (B) The enhancing effect of the CCR5-59356-T mutation on HIV-1 transmission was not observed in the AZT-treated group. NS, not significant.
The CCR5-Δ32 deletion is significantly more prevalent in Caucasians (allele frequency, 5.9% in 194 individuals) than in African-Americans (2.0% in 691; P = 0.001) and Hispanic individuals (2.4% in 492; P = 0.02) (Fig. 2A), consistent with previous report (5, 10, 20). Because of the smaller number of Caucasians in this study, no CCR5-Δ32/Δ32 homozygotes were present, and so the effect of this genotype on perinatal transmission could not be determined. Untreated CCR5-Δ32 heterozygotes have lower HIV-1 transmission rates than CCR5 wild-type homozygous infants (Table 2), but this was not statistically significant (P > 0.3).
The distribution of the CCR2-64I genotypes is similar (allele frequency, 13.4 to 16.5%; P = 0.05) among the three racial groups (Fig. 2A), consistent with previous reports (14, 34). In the untreated group, there is no effect of CCR2-64I on HIV-1 transmission (Fig. 2B and Table 2), but in the AZT-treated group there is a trend (P = 0.06) for reduced transmission to CCR2-64I heterozygotes compared to CCR2 wild-type homozygotes (Table 2, footnote c).
The CCR5-59356-T mutant allele is significantly (P < 0.001) more prevalent among African-Americans (20.6% of 552 individuals) than in the Hispanic (5.6% of 458) or Caucasian (3.4% of 174) group (Fig. 2A). In fact, CCR5-59356-T mutant homozygotes are most often found among African-Americans: 35 of the 38 mutant homozygotes in this study are in African-Americans. Therefore, we analyzed the CCR5-59356 alleles in the African-American group.
Among the 552 African-Americans, 6.3% were homozygotes for the mutant CCR5-59356-T allele. In the untreated group, the genotype and allele distributions of the CCR5-59356-T mutation are significantly (P < 0.001) different between HIV-1-infected and uninfected infants (Table 2 shows data for the AZT-untreated group, all races combined). Among the 45 HIV-1-infected African-American infants, there is a significantly (P < 0.002) higher CCR5-59356-T allele frequency (33.3%) (Fig. 2B), and a CCR5-59356-T mutant homozygote genotype frequency (22.2%), than among the 234 uninfected infants (17.7 and 4.7%, respectively). By comparing the fractions of untreated, HIV-1-infected infants in the three CCR5-59356 genotypes (Fig. 3A), we found a significantly higher rate of transmission among CCR5-59356-T mutant homozygotes (47.6% of 21 CCR5-59356-T mutant homozygote infants) compared with both CCR5-59356-A wild-type homozygotes (13.4% of 187; P < 0.001) and CCR5-59356 heterozygotes (14.1% of 71; P = 0.001). In other words, about half of the untreated infants in the four combined cohorts who are homozygotes for the CCR5-59356-T mutation are infected with HIV-1, an unexpectedly large fraction. Infants who are CCR5-59356-T mutant homozygotes are associated with an increased relative risk for HIV-1 infection of 5.9 (95% confidence interval, 2.3 to 15.3) (P < 0.001). The enhancing effect of the CCR5-59356-T mutation on HIV-1 transmission was not observed in the AZT-treated group (Fig. 3B).
A logistic regression was performed on data from the WITS and ARIEL cohorts (only these two cohorts were included, as the other studies did not have detailed information on all of the covariables to determine if the HIV-1 transmission resistance demonstrated by infants homozygous for CCR5-59356T remained after adjusting for other known predictors of HIV transmission). The variables used are included in Table 3, with the exception of the log of the mothers’ HIV-1 RNA count at the time of delivery. There was only a small number of WITS mothers for whom the RNA count was available (n = 146, HIV-1 infected = 33), and so the full results including this variable are not presented here; results of the analysis including this variable were similar to those presented for homozygous CCR5-59356T (odds ratio = 12, P < 0.04). The results in Table 3 indicate that the presence of the CCR5-59356T homozygous genotype was a significant independent predictor of transmission. The AZT variable and the time indicator of birth are highly correlated. The time indicator function of a baby’s delivery date indicates whether the birth was before or after February 1994, when it first became clear that AZT could dramatically reduce mother-infant HIV-1 transmission and AZT became widely used. AZT use by the mother is not significant in this regression analysis due to the inclusion of the time variable. The statistical finding in favor of the time variable probably reflects other changes in treatment combinations after 1994 that may be more successful than AZT alone.
TABLE 3.
Logistic regression analysis of the combined WITS and ARIEL cohorts
| Variablea | Parameter estimate | SE | Pr > chi squareb | Odds ratio |
|---|---|---|---|---|
| African-American ethnic group | −0.46 | 0.25 | 0.06 | 0.63 |
| Mother received AZT | −0.49 | 0.33 | 0.13 | 0.61 |
| Infant received AZT | 0.82 | 0.47 | 0.08 | 2.28 |
| Log of infant’s CD4 count | −0.51 | 0.24 | 0.04 | 0.60 |
| Premature birth | 0.37 | 0.28 | 0.18 | 1.45 |
| Birth after February 1994 | −1.25 | 0.46 | 0.01 | 0.29 |
| Homozygous CCR5-59356T | 1.35 | 0.59 | 0.02 | 3.84 |
| Log of mother’s CD4 count | −0.55 | 0.14 | 0.00 | 0.57 |
Logistic regression analysis of the combined WITS and ARIEL cohorts (total of 656; 94 HIV-1 infected) based 1 df for each variable. Variables that were thought to influence transmission were included. The African-American ethnic group was included as a variable because the allele frequency for CCR5-59356T is far greater in that group.
Probability that under the null hypothesis of no correlation with transmission, the observed data are attributable to chance alone.
We investigated whether the higher frequency of the CCR5-59356-T mutation among African-Americans would significantly increase the relative risk of HIV-1 perinatal transmission in this group by several methods, including a Mantel-Haenzel analysis and a comparison of transmission frequencies in African-Americans versus non-African-Americans. We could, however, find no evidence for an association between race and HIV-1 transmission (Table 1). Although homozygosity for CCR5-59356-T significantly increases the risk of transmission, and this genotype is most commonly found in African-Americans, its frequency is too low to profoundly affect the overall transmission rate among African-Americans. How CCR5 5′-UTR polymorphisms affect perinatal transmission in pediatric cohorts from Africa is currently being investigated.
The frequency of the CCR5-59353-C allele is high (44.8%) in the combined population (Fig. 2A) but is significantly lower among African-Americans (40.3% of 656 individuals) than in Hispanic individuals (48.2% of 477; P = 0.02) and Caucasians (53.8% of 185; P = 0.001). In the AZT-untreated group, the CCR5-59353-C allele had no observable effect on transmission (Fig. 2B), but in the treated group, the CCR5-59353 genotype distribution was different (P = 0.006) between HIV-1-infected and uninfected infants (Table 2, footnote d).
The frequency of the CCR5-59402-G mutant allele is significantly lower (P < 0.001) among African-Americans (13.7% of 681 individuals) than in the Caucasian (29.1% of 189) and Hispanic groups (33.2% of 486) (Fig. 2A). In the untreated group, there is a trend (P = 0.05) for a reduced HIV-1 transmission rate to infants who are CCR5-59402-G mutant homozygotes compared to CCR5-59402-A wild-type homozygous infants (Table 2). The relative risk of 0.35 (95% confidence interval, 0.12 to 1.0) is also reduced (P = 0.05). There was no significant effect of the CCR5-59402 genotype on HIV-1 transmission rates in the AZT-treated group.
In a systematic study of linkage disequilibrium between the different loci to search for any effects of combinations of alleles on HIV-1 transmission, we observed indications of an additive protective effect against HIV-1 transmission of the CCR5-Δ32, CCR5-59353-C, and CCR5-59402-G alleles in combination. The P values obtained in this analysis should be interpreted cautiously, as we performed multiple tests to look for combinations of alleles of particular interest. Given this caveat, there was a significant difference (P = 0.01) in the linked distribution of the CCR5-59353 and CCR5-59402 genotype distributions between HIV-1-infected and uninfected infants who are CCR5-Δ32 heterozygotes, an effect not seen in infants who are CCR5 wild-type homozygotes. In the untreated group, infants who are CCR5-Δ32 heterozygotes, and who also have two or more CCR5-59353-C, CCR5-59402-G, and CCR5-59356-T mutant alleles (n = 25), have a significantly lower transmission rate (0% of 25 infants) than the 642 other infants who do not have this genotype combination (17.6% transmission rate; 113 of 642 infants). Moreover, in the full cohort (AZT treated or untreated), none of the 46 infants in the combined groups who are CCR5-Δ32 heterozygotes, and who also have two or more CCR5 5′-UTR mutations, was HIV-1 infected.
DISCUSSION
Our studies on coreceptor gene polymorphisms in infants exposed to HIV-1 in vitro or during delivery have revealed that possession of the CCR5-59356-T/T genotype can significantly increase the rate of perinatal HIV-1 transmission. It is conceivable that the CCR5 genotype of the mother will also affect her capacity to transmit HIV-1 to her infant, but we have not yet completed studies of this. There are no previous reports of the influence of CCR5 regulatory region polymorphisms on perinatal HIV-1 transmission, although it has been shown that some such polymorphisms affect the rate of disease progression in HIV-1-infected adults (19, 21). Among these reports is one by Martin et al., who found no association between the possession of any of the CCR5 regulatory region alleles (CCR5P1 to CCR5P4 based on their nomenclature) and protection from sexual HIV-1 transmission (19). However, HIV-1-infected adults who were CCR5P1/P1 homozygotes (CCR5-59353-C/C and CCR5-59356-C/C homozygotes based on our nomenclature) progressed to AIDS more rapidly than those with other CCR5 promoter haplotypes. It is important to note that the infants who were CCR5-59356-T/T homozygotes (n = 38) in our study also had the CCR5-59353-T/T-, CCR5-59402-A/A-, and CCR5-coding wild-type genotypes. In a different study, McDermott et al. found that another mutation within the CCR5 regulatory region, a G-to-T variant at position 59029 (CCR5-59029-G/T polymorphism), is associated with relatively slow progression to AIDS in adults (21). We have not investigated whether this polymorphism affects perinatal transmission. We also do not yet know whether any of the CCR5 5′-UTR polymorphisms that we have studied influence disease progression in infected infants; these studies are complicated by the relative lack of long-term clinical follow-up in pediatric cohorts before use of antiretroviral agents became common, compared to the longer-established adult sexual transmission cohorts that have been studied by others (19, 21).
The central finding of the present study is that homozygosity for a genetic mutation in the CCR5 5′-UTR (CCR5-59356-T) is associated with an increased rate of HIV-1 perinatal transmission. This mutation is found at a higher frequency among African-Americans than in Hispanic or Caucasian populations. Conversely, mutations associated with a reduced rate of perinatal transmission (CCR5-Δ32 and CCR5-59402-G) are less common among African-Americans. However, the frequency of the CCR5-59356-T genotype among African-Americans was not sufficient to cause an overall increase in the rate of perinatal transmission in this group. It remains possible that these various genetic factors contribute to the relatively high rate of perinatal HIV-1 transmission reported to occur in Africa, although many other influences on transmission efficiency are also likely to be relevant (38).
As yet, we have only limited information on the origin of the CCR5 5′-UTR mutations we have studied and on their global distribution. What we have observed among present-day African-Americans probably reflects founder effects; certain CCR5 alleles may be enriched among African-Americans because they were prevalent in their African ancestors who arrived in United States during the 16th to 19th centuries. Over time, the distribution of these alleles will change because of population mixing; the CCR5-Δ32 allele has been introduced into present-day African-American and Hispanic populations by contact with the northern Europeans in whom it originated (18, 20). It will therefore be valuable to determine the frequencies of the CCR5 5′-UTR alleles among modern African populations. This may well not be uniform; an analogy is the distribution of the CCR5-Δ32 allele in Caucasian populations, which is high in northern Europe and less common in central and southern Europe, probably reflecting population migration patterns over the past few thousand years (18, 20).
We do not know how the effects of the CCR5 5′-UTR polymorphism are manifested biologically. It is a reasonable assumption, however, that they influence the expression of CCR5 in relevant immune system cells. This could be achieved by modifying the extent of CCR5 expression or the cells in which CCR5 is expressed. The levels of CCR5 on CD4+ T cells vary significantly among different individuals with wild-type CCR5-coding alleles (19, 25, 30, 39). We are presently exploring whether such variation is directly linked to polymorphisms in the CCR5 5′-UTR. However, it is also possible that CCR5 promoter usage varies, absolutely or quantitatively, between tissues and that the 5′-UTR polymorphisms affect this difference. The CCR5 5′-UTR has twin promoters, a feature often associated with tissue-dependent protein expression patterns (27). Of note is that CCR5 expression has been detected in multiple cell types in the female genital tract (40). Furthermore, the extents of CCR5 expression in the ectocervix and in CD4+ T cells of the peripheral blood are not correlated (29). This is potentially relevant to the efficiency of HIV-1 perinatal transmission, since CCR5-using viruses are the most commonly transmitted strains, both perinatally and sexually (7, 12, 22, 41, 42).
In principle, mutations affecting CCR5 expression or function might have different influences on perinatal transmission, depending on the time at which transmission occurs. For instance, HIV-1 transmission in utero may take place by a process that is less dependent on CCR5, compared to transmission mechanisms that operate at or soon after birth. There is some evidence that HIV-1 infections which occur despite AZT therapy result primarily from ante partum transmission, based on positive HIV-1 culture and DNA or RNA PCR results from blood samples obtained within 48 h of birth (10). In the WITS cohort, a significantly higher proportion of HIV-1 perinatal infections were antepartum (P < 0.05) after March 1994, when the ACTG 076 results became publicly known, versus before March 1994. We have observed that among the AZT-treated, HIV-1-infected group in the combined cohorts, the deleterious effect of the CCR5-59356-T/T mutant genotype is not evident. However, future meta-analyses may fruitfully address the issue of whether CCR5 polymorphisms have an effect on perinatal transmission rates that is conditional on the timing or route of transmission. Likewise, meta-analysis will be able to confirm whether certain CCR5-genotype combinations provide partial protection from HIV-1 perinatal transmission, as indicated by our study.
Finally, whatever genetic influences contribute to HIV-1 perinatal transmission, a consistent finding of this and other studies (6, 37) is that AZT prophylaxis reduces the probability of transmission. There is, therefore, every reason to support the provision of AZT or relevant drugs to HIV-1-infected, pregnant women worldwide.
ACKNOWLEDGMENTS
We are indebted to the staff members of the hospitals listed below who provided clinical data and samples for this study. The participants and funding sources were as follows: for the WITS, C. Diaz and E. Pacheco-Acosta (University of Puerto Rico, San Juan; U01 AI 34858), R. Tuomala, E. Cooper, and D. Mestehene (Boston/Worcester Site, Boston, Mass.; U01 AI 34856), J. Pitt and A. Higgins (Columbia Presbyterian Hospital, New York, N.Y.; U01 AI 34842), S. Landesman, H. Mendez, and G. Moroso (State University of New York, Brooklyn; HD-8-2913 and RO-1-HD-25714), K. Rich and D. Turpin (University of Illinois at Chicago; U01 AI 34841), W. Shearer, C. Hanson, and N. Cooper (Baylor College of Medicine, Houston, Tex.; U01 AI 34840), R. Nugent (National Institute of Child Health and Human Development, Bethesda, Md.), and V. Smeriglio (National Institute on Drug Abuse, Rockville, Md.); for the NY-WNE, M. McManus (University of Massachusetts, Worcester), B. Stechenberg (Baystate Medical Center, Springfield, Mass.); P. Krause (University of Connecticut Medical School, Farmington), and K. Krasinski, M. Rigaud, A. Kaul, R. Lawrence, W. Hoover, and S. Chandwani (New York University Medical School, New York, N.Y.); for the NPC, T. Denny (University of Medicine and Dentistry of New Jersey, Newark); for the ARIEL, C. Hanson (Texas Children’s Hospital/Baylor, Houston), W. T. Shearer (Baylor College of Medicine), R. B. Van Dyke (Tulane University School of Medicine, New Orleans, La.), S. M. Widmayer (Children’s Diagnostic and Treatment Center, Fort Lauderdale, Fla.), A. Wiznia (Bronx Lebanon Hospital Center, Bronx, N.Y.), A. Ammann (Center for AIDS Prevention, University of California San Francisco, San Francisco), I. S. Y. Chen (University of California Los Angeles, Los Angeles), R. Koup (University of Texas, Dallas), P. Krogstad (University of California Los Angeles, Los Angeles), J. Mullins (University of Washington, Seattle), and B. D. Walker (Massachusetts General Hospital, Boston). We are indebted to M. G. Fowler, A. J. Amman, C. Wilfert, and T. Divine for assistance with sample access and to M. Small for secretarial help.
Financial support was provided by the National Institutes of Health (grants RO1 AI43868 [to L.G.K.] and RO1 AI41420 [to J.P.M.]). Additional support was provided by the Irene Diamond Fund; the Columbia-Rockefeller Center for AIDS Research; the Committee for the Advancement of Research and the Gonda-Goldschmied Medical Diagnostic Center at Bar-Ilan University (A.U.N. and L.D.); the National Institutes of Health (grant NO1-AI85339); the Centers for Disease Control and Prevention (grant U64/CCU202219-13 [to P.P.]); and the Pediatric AIDS Foundation, of which J.P.M., K.L., and B.T.K. are Elizabeth Glaser Scientists and H.M.L.S. is a Scholar.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 3.Buseyne F, Janvier G, Teglas J P, Ivanoff S, Burgard M, Bui E, Mayaux M-J, Blanche S, Rouzioux C, Riviere Y. Impact of heterozygosity for the chemokine receptor CCR5 32-bp-deleted allele on plasma virus load and CD4 T lymphocytes in perinatally human immunodeficiency virus-infected children at 8 years of age. J Infect Dis. 1988;178:1019–1023. doi: 10.1086/515660. [DOI] [PubMed] [Google Scholar]
- 4.Cao Y, Krogstad P, Korber B T, Koup R A, Muldoon M, Macken C, Song J L, Jin Z, Zhao J Q, Clapp S, Chen I S, Ho D D, Ammann A J. Maternal HIV-1 viral load and vertical transmission of infection: the Ariel Project for the prevention of HIV transmission from mother to infant. Nat Med. 1997;3:549–552. doi: 10.1038/nm0597-549. [DOI] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Connor E M, Sperling R S, Gelber R, Kiselev P, Scott G, O’Sullivan M J, VanDyke R, Bey M, Shearer W, Jacobson R L, Jimenez E, O’Neill E, Basin B, Delfraissy J-F, Gulnane M, Coombs R, Elkins M, Moye J, Stratton P, Balsley J the Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. Reduction of mother-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. N Engl J Med. 1994;331:1173–1180. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- 7.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 8.Deng H, Unutmaz D, Kewalramari V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 9.Dickover R E, Garratty E M, Herman S A, Sim M S, Plaeger S, Boyer P J, Keller M, Deveikis A, Stiehm E R, Bryson Y J. Identification of levels of maternal HIV-1 RNA associated with risk of perinatal transmission. Effect of maternal zidovudine treatment on viral load. JAMA. 1996;275:599–605. [PubMed] [Google Scholar]
- 10.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 13.Ioannidis J P, O’Brien T R, Rosenberg P S, Contopoulos-Ioannidis D G, Goedert J J. Genetic effects on HIV disease progression. Nat Med. 1998;4:536. doi: 10.1038/nm0598-536. [DOI] [PubMed] [Google Scholar]
- 14.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 15.Kostrikis L G, Tyagi S, Mhlanga M M, Ho D D, Kramer F R. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 16.Lee B, Doranz B J, Rana S, Yi Y, Mellado M, Frade J M, Martinez A C, O’Brien S J, Dean M, Collman R G, Doms R W. Influence of the CCR2-V64I polymorphism on human immunodeficiency virus type 1 coreceptor activity and on chemokine receptor function of CCR2b, CCR3, CCR5, and CXCR4. J Virol. 1998;72:7450–7458. doi: 10.1128/jvi.72.9.7450-7458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 18.Lucotte G, Mercier G. Distribution of the CCR5 gene 32-bp deletion in Europe. J Acquired Immun Defic Syndr. 1998;19:174–177. doi: 10.1097/00042560-199810010-00011. [DOI] [PubMed] [Google Scholar]
- 19.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O’Brien T R, Hilgartner M W, Vlahov D, O’Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 20.Martinson J J, Chapman N H, Rees D C, Liu Y T, Clegg J B. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16:100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 21.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 22.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 23.Michael N L, Nelson J A, KewalRamani V N, Chang G, O’Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O’Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misrahi M, Teglas J P, N N G, Burgard M, Mayaux M J, Rouzioux C, Delfraissy J F, Blanche S. CCR5 chemokine receptor variant in HIV-1 mother-to-child transmission and disease progression in children. French Pediatric HIV Infection Study Group. JAMA. 1998;279:277–280. doi: 10.1001/jama.279.4.277. [DOI] [PubMed] [Google Scholar]
- 25.Moore J P. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276:51–52. doi: 10.1126/science.276.5309.51. [DOI] [PubMed] [Google Scholar]
- 26.Mummidi S, Ahuja S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O’Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 27.Mummidi S, Ahuja S S, McDaniel B L, Ahuja S K. The human CC chemokine receptor 5 (CCR5) Gene. J Biol Chem. 1997;272:30662–30671. doi: 10.1074/jbc.272.49.30662. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien T R, Winkler C, Dean M, Nelson J A, Carrington M, Michael N L, White G C., II HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 29.Patterson B K, Landay A, Andersson J, Brown C, Behbahani H, Jiyamapa D, Burki Z, Stanislawski D, Czerniewski M A, Garcia P. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153:481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paxton W A, Liu R, Kang S, Wu L, Gingeras T R, Landau N R, Mackay C R, Koup R A. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244:66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 31.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 32.Shearer W T, Kalish L A, Zimmerman P A. CCR5 HIV-1 vertical transmission. Women and Infants Transmission Study Group. J Acquired Immun Defic Syndr. 1998;17:180–181. doi: 10.1097/00042560-199802010-00014. [DOI] [PubMed] [Google Scholar]
- 33.Sheon A, Fox H, Rich K, Stratton P, Diaz C, Tuomala R, Mendez H, Carrington J, Alexander G for the Women and Infants Transmission Study Group. The women and infants transmission study (WITS) of mother-infant transmission: study design, methods, and baseline data. J Womens Health. 1996;5:69–78. [Google Scholar]
- 34.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 35.Stephens J C, Reich D E, Goldstein D B, Shin H D, Smith M W, Carrington M, Winkler C, Huttley G A, Allikmets R, Schriml L, Gerrard B, Malasky M, Ramos M D, Morlot S, Tzetis M, Oddoux C, di Giovine F S, Nasioulas G, Chandler D, Aseev M, Hanson M, Kalaydjieva L, Glavac D, Gasparini P, Dean M. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR5 delta 32. Seroco Study Group. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 37.Wade N A, Birkhead G S, Warren B L, Charbonneau T T, French P T, Wang L, Baum J B, Tesoriero J M, Savicki R. Abbreviated regimens of zidovudine prophylaxis and perinatal transmission of the human immunodeficiency virus. N Engl J Med. 1998;339:1409–1414. doi: 10.1056/NEJM199811123392001. [DOI] [PubMed] [Google Scholar]
- 38.The Working Group on Mother-To-Child Transmission of HIV. Rates of mother-to-child transmission of HIV-1 in Africa, America, and Europe: results from 13 perinatal studies. J Acquired Immun Defic Syndr. 1995;8:506–510. doi: 10.1097/00042560-199504120-00011. [DOI] [PubMed] [Google Scholar]
- 39.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, He T, Talal A, Wang G, Frankel S S, Ho D D. In vitro distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 42.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]