Abstract
Fermentative production of natural colorants using microbial strains has emerged as a cost-effective and sustainable alternative to chemical synthesis. Visual pigments are used as signal outputs in colorimetric bacterial biosensors, a promising method for monitoring environmental pollutants. In this study, we engineered four self-sufficient indigo-forming enzymes, including HbpAv, bFMO, cFMO, and rFPMO, in a model bacterium E. coli. TrxA-bFMO was chosen for its strong ability to produce indigo under T7 lac and mer promoters’ regulation. The choice of bacterial hosts, the supplementation of substrate l-tryptophan, and ventilation were crucial factors affecting indigo production. The indigo reporter validated the biosensors for Hg(II), Pb(II), As(III), and Cd(II). The biosensors reported Hg(II) as low as 14.1 nM, Pb(II) as low as 1.5 nM, and As(III) as low as 4.5 nM but increased to 25 μM for Cd(II). The detection ranges for Hg(II), Pb(II), As(III), and Cd(II) were quantified from 14.1 to 225 nM, 1.5 to 24.4 nM, 4.5 to 73.2 nM, and 25 to 200 μM, respectively. The sensitivity, responsive concentration range, and selectivity are comparable to β-galactosidase and luciferase reporter enzymes. This study suggests that engineered enzymes for indigo production have great potential for green chemical synthesis. Additionally, heterologous biosynthesis of indigo production can lead to the development of novel, low-cost, and mini-equipment bacterial biosensors with zero background noise for visual monitoring of pollutant heavy metals.
1. Introduction
Whole-cell biosensors have garnered increasing attention in recent decades, mainly within synthetic biology.1 These genetically engineered microorganisms, usually bacteria, can detect and report specific compounds or analytes by emitting a detectable signal through various reporter systems. These highly effective and economical bacterial biosensors provide qualitative and quantitative information about the environment they are introduced to in an in situ manner.2
A cellular biosensor is a two-stage processor that can be divided into two modules. The first is the sensory module, which recognizes and transduces external signals into intracellular transcriptional signals. The second is the actuating module, which executes physiological responses.3 Furthermore, various exquisitely designed genetic circuits were used to amplify the biosensing signal.4,5 Versatile bacterial biosensors have been developed successfully to detect and report reactive oxygen species, DNA damage, protein damage, and membrane damage caused by various chemicals.1,6 Outstanding from them, heavy metal biosensors are known for their high specificity.2
Microorganisms can adjust their intracellular concentrations of metal ions to adapt to environmental changes and maintain cellular homeostasis. It is achieved through the help of metal-responsive transcriptional regulators, which play essential roles in metal uptake, pumping out, sequestration, and transformation to a less toxic status.7 These regulators are responsible for controlling the expression of genes related to resistance. The metalloregulators have become popular biological parts in synthetic biology due to their sensory and regulatory functions. Two critical families, MerR and SmtB/ArsR, exhibit remarkable sensitivity and selectivity toward metal ions.7 These families can regulate heavy metals and metalloids, including the highly toxic ones such as Pb, Hg, Cd, and As. Heavy metal biosensors have been developed using these families to address the widespread problem of heavy metal contamination.8−10
Fluorescent proteins and enzymes like β-galactosidase and luciferase are widely used as reporters in heavy metal whole-cell biosensors that are fluorescent, colorimetric, or bioluminescent.11−13 Colorimetric biosensors have always been considered cost-effective biological devices with minimum requirements.14 It is now possible to create various natural colorants in chassis cells through synthetic biology and metabolic engineering.15,16 As visible reporters, natural pigments offer significant advantages, such as simple visual examination and spectrophotometric analysis, which enable qualitative and quantitative measurement.17,18 Researchers have developed bacterial biosensors using various pigments such as water-insoluble violacein, pyocyanin, β-carotene, and water-soluble indigoidine and anthocyanin. These pigments are generated from gene clusters like vioABCDE, phzMS, crtEBIY, bpsA-pcpS, and 3GT-ANS. Such biosensors detect heavy metal ions such as Hg(II), Pb(II), and Cd(II).19−22 Pigment-based bacterial biosensors are a promising solution for field tests due to their substrate-free nature, small equipment requirements, and signal amplification properties.1,23 Biotechnological advances possibly enable the production of more pigments in bacterial chassis cells as a novel, green chemical and visual indicator for bacterial biosensing.
Indigo, an ancient dye, remains highly demanded, especially in the denim industry.24 Indigo was initially sourced from plants, but it has been synthesized from petroleum using a chemical process (as shown in Figure 1a) and has replaced most plant-based production.25 Since the early 1980s, researchers have explored the production of indigo by fermentation after identifying bacterial strains capable of synthesizing indigo.26 Although various bacterial strains and enzymes have been identified as being able to produce indigo, a large-scale industrial biotechnological process still needs to be developed for producing indigo.27 The demand for a biotechnological indigo production process has become more urgent, and modern enzyme and strain engineering techniques make it more feasible.28
Figure 1.
Indigo production history and its metabolic engineering (Public domain images sourced from the Internet, not subject to copyright restrictions). (a) The timeline of the processes involved in the production of indigo. (b) Metabolic engineering of indigo could have potential applications in whole-cell visual biosensing.
Indigo-forming enzymes convert indole via an oxygenation reaction.29 The redox enzymes that have been identified can be classified into three different enzyme classes based on the cofactor used for performing the oxygenation of indole. These enzyme classes include nonheme iron oxygenases, heme-containing oxygenases, and flavin-dependent monooxygenases.30 In the current study, four self-sufficient indigo-forming enzymes were chosen from identified oxygenases and engineered under the control of Hg(II)-responsive MerR in the model bacterium E. coli. In Figure 1b, it is shown that E. coli breaks down l-tryptophan into indole using endogenous tryptophanase TnaA.31 The produced indole is then converted into indigo using recombinant redox enzymes. The redox enzyme that is most effective in Hg(II) whole-cell biosensing will be selected and further tested for its efficacy in biosensing Pb(II), As(III), and Cd(II).
Self-sufficient oxygenases were engineered in this study instead of indigo-forming oxygenases that are part of a multicomponent system. We somewhat compared the four candidate enzymes and observed significant variations in their indigo productivity, indicating their diverse capabilities for producing indigo in E. coli. Our research suggests that engineered oxygenases have great potential for the green chemical synthesis of indigo. Compared to previously developed visual signals, such as violacein and pyocyanin, indigo produces a charming, noncytotoxic sky-blue color. Additionally, the production of indigo is triggered by a single enzyme, making it an ideal pigment reporter for developing biosensors. Heavy metals-responsive indigo production allows the synthetic bacterium to function as a visual biosensor.
2. Materials and Methods
2.1. Bacterial Strains, Recombinant Constructs, and Reagents
Various indigo-producing enzymes have been identified in microorganisms with the rapid development of metagenome mining, mainly in bacteria. Four oxygenases are chosen from them, engineered in this study, and listed in Table 1. The amino acid and DNA sequences of indigo-forming enzymes after E. coli encoding optimization are listed in Table S1.
Table 1. Indigo-Forming Enzymes used in this Study.
| enzymes | name | origin | ref |
|---|---|---|---|
| 2-hydroxybiphenyl-3-monooxygenase variant | HbpAv | Pseudomonas azelaica HBP1 | (32) |
| flavin-containing monooxygenase | bFMO | Methylophaga sp. strain SK1 | (33) |
| flavin-containing monooxygenase | cFMO | Corynebacterium glutamicum | (34) |
| two-component flavoprotein monooxygenases | rFPMO | Rhodococcus strain ATCC 21145 | (35) |
The following Table 2 displays the bacterial hosts and recombinant constructs. The bacterial cultures were grown in Luria–Bertani (LB) broth containing 1% tryptone, 0.5% yeast extract, and 1% sodium chloride. If required, the cultures were supplemented with 50 μg/mL ampicillin. All metal salts, such as mercuric chloride, lead nitrate, sodium arsenite, and cadmium chloride, were of analytical grade and were dissolved in purified water at 1 mM as a stock solution. All of the reagents used were obtained from Sigma-Aldrich unless specified otherwise.
Table 2. Bacterial Strains and Plasmids Used in this Study.
| strains and plasmids | genotypes or description | reference |
|---|---|---|
| E. colistrains | ||
| TOP10 | F– Φ80lacZΔM15 ΔlacX74 recA1 | Tiangen |
| BL21(DE3) | F–ompT hsdSB (rB–mB–) gal dcm (DE3) | Tiangen |
| DH5α | F– φ80 lacZΔM15 Δ(lacZYA-argF) | Tiangen |
| Rosseta(DE3) | F–ompT hsdSB (rB–mB–) gal dcm (DE3) pRARE | Tiangen |
| Plasmids | ||
| pET-21a | AmpR, T7 promoter, lac operator | Merck |
| pET-32a | AmpR, T7 promoter, lac operator, TrxA tag | Merck |
| pPmer | pET-21a derivative containing natural merR and Pmer divergent promoter region cloned into BglII and XbaI sites | (11) |
| pPpbr | pET-21a derivative containing a redesigned lead sensory element from Klebsiella pneumoniae CG43 plasmid pLVPK | this study |
| pPars | pET-21a derivative containing a redesigned arsenic sensory element from Escherichia coli K12 chromosomal ars operon cloned into BglII and XbaI sites | this study |
| pCadR10-DV | The vioABCE gene cluster fused downstream of a redesigned cadmium sensory element from Halomonas caseinilytica JCM 14802 | (36) |
| pET-HbpAv | pET-21a derivative containing the HbpAv coding gene | this study |
| pET-bFMO | pET-21a derivative containing the bFMO coding gene | this study |
| pET-cFMO | pET-21a derivative containing the cFMO coding gene | this study |
| pET-rFPMO | pET-21a derivative containing the rFPMO coding gene | this study |
| pET-TrxA-bFMO | pET-32a derivative containing the fused TrxA-bFMO coding gene | this study |
| pET-TrxA-cFMO | pET-32a derivative containing the fused TrxA-cFMO coding gene | this study |
| pHg-bFMO | pPmer derivative containing the bFMO coding gene under transcriptional control of Hg(II) sensory element | this study |
| pPb-bFMO | pPb-vioABCE derivative containing the bFMO coding gene under transcriptional control of Pb(II) sensory element | this study |
| pCd-bFMO | pCadR10-DV derivative containing the bFMO coding gene under transcriptional control of Cd(II) sensory element | this study |
| pHg-TrxA-bFMO | pPmer derivative containing the fused TrxA-bFMO coding gene under transcriptional control of Hg(II) sensory element | this study |
| pHg-TrxA-cFMO | pPmer derivative containing the fused TrxA-cFMO coding gene under transcriptional control of Hg(II) sensory element | This study |
| pHg-rFPMO | pPmer derivative containing the rFPMO coding gene under transcriptional control of Hg(II) sensory element | this study |
| pPb-TrxA-bFMO | pPb-vioABCE derivative containing the fused TrxA-bFMO coding gene under transcriptional control of Pb(II) sensory element | this study |
| pAs-TrxA-bFMO | pPars derivative containing the fused TrxA-bFMO coding gene under transcriptional control of As(III) sensory element | this study |
| pCd-TrxA-bFMO | pCadR10-DV derivative containing the fused TrxA-bFMO coding gene under transcriptional control of Cd(II) sensory element | this study |
2.2. Assembly of Recombinant Constructs
The strategy used for the plasmid construction and bacterial transformation is shown in Figure S1. Encoding genes for four indigo-producing enzymes, including HbpAv, bFMO, cFMO, and rFPMO were artificially synthesized and cloned into pET-21a using NdeI-SacI sites, to generate pET-HbpAv, pET-bFMO, pET-cFMO, and pET-rFPMO, respectively. The NcoI-SacI fragments containing the bFMO and cFMO encoding sequences were amplified by PCR and inserted into pET-32a to generate pET-TrxA-bFMO and pET-TrxA-cFMO, respectively. The plasmids pPmer, pPpbr, pPars, and pCadR10-DV contain natural Hg(II), redesigned Pb(II), redesigned As(III), and redesigned Cd(II) sensory elements. These elements were amplified by PCR and inserted into pET-21a-derived vectors as BglII-XbaI fragments. The result is a variety of biosensing constructs, including pHg-bFMO, pPb-bFMO, pCd-bFMO, and pHg-rFPMO. All sensory elements of heavy metals and metalloids are listed in Table S2. The TrxA fused oxygenase-encoding genes were PCR amplified from pET-32a-derived vectors and inserted into pPmer, pPpbr, pPars, and pCadR10-DV using NdeI and SacI sites, generating five biosensing constructs: pHg-TrxA-bFMO, pHg-TrxA-cFMO, pPb-TrxA-bFMO, pAs-TrxA-bFMO, and pCd-TrxA-bFMO.
2.3. Recombination Expression and Validation of Indigo-Producing Enzymes in E. coli
The pET-derived plasmids were freshly transformed into E. coli BL21(DE3). Single colonies were selected and activated overnight at 37 °C. The activated cultures were then diluted 1:100 in fresh LB broth and cultured at 37 °C with shaking at 250 rpm for 3 h before being induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h. Then, the bacteria were collected by centrifugation, treated with ultrasonication as described previously,37 and subjected to 10% SDS-PAGE.
The heavy metal biosensing constructs were freshly transformed into E. coli TOP10. Recombinant bacteria in the early stage of logarithmic growth were exposed to target metals, such as Hg(II) at 450 nM, Cd(II) at 50 μM, and Pb(II) at 50 μM, with shaking at 250 rpm for 12 h.
After IPTG or heavy metals induction, the bacterial culture was measured for density at 600 nm in 100 μL. Then, 900 μL of the culture was centrifuged at 12000 rpm for 2 min. The bacterial pellets were dehydrated with ethanol, extracted with 200 μL DMSO, and vortexed extensively. The DMSO phases were scanned using visible light, and the absorbances at 620 nm were recorded in 100 μL using a microplate reader (BioTek Epoch, USA).
2.4. Validation of Indigo’s Visual Reporting Potential Using Hg(II) Whole-Cell Biosensing
The vector pHg-TrxA-bFMO was transformed into E. coli TOP10 to create a biosensor for Hg(II). We first selected single colonies for the time-dose–response assay and activated them overnight at 37 °C. Next, we diluted the overnight incubated cultures by a factor of 1:100 in fresh LB broth. We then induced the cultures with varying concentrations of Hg(II) in a gradient of 0, 56.3, and 450 nM. Finally, we cultured the induced cells at 37 °C with shaking at 250 rpm for 13 h. At 1-h intervals, 1 mL aliquots of cultures were sampled to determine bacterial density and indigo-derived signal.
Overnight activated TOP10/pHg-TrxA-bFMO was diluted at a 1:100 ratio into fresh LB medium. Induction groups with varying Hg(II) concentrations were obtained using a 2-fold dilution method in a dose–response assay. It resulted in exposure groups with 0, 14.1, 28.1, 56.3, 112.5, 225, 450, and 900 nM Hg(II) concentrations. Various metal ions were supplemented in induction groups at 5 μM in a selectivity assay. Following a 12-h incubation at 37 °C with shaking at 250 rpm, bacterial density and indigo-derived color signal were measured as described above.
2.5. Optimization of Hg(II) Whole-Cell Biosensing System
The Hg(II) biosensing construct pHg-TrxA-bFMO was transformed into four E. coli hosts, including TOP10, BL21(DE3), DH5α, and Rosseta(DE3). The resultant engineered bacteria were picked up, activated overnight, and exposed to increased Hg(II) concentrations in a 2-fold dilution method at 250 rpm. Bacterial density and indigo-derived color signal were measured as described above after a 12-h incubation at 37 °C.
After being activated overnight, TOP10/pHg-TrxA-bFMO was diluted 1:100 into a fresh LB medium. The culture was supplemented with excessive up to 1 mM l-tryptophan, which was noncytotoxic, and increased Hg(II) concentrations were induced in a 2-fold dilution. After a 12-h incubation at 37 °C with shaking at 250 rpm, bacterial density and indigo-derived color signal were measured as described above.
The TOP10/pHg-TrxA-bFMO was activated overnight. It was then diluted at a ratio of 1:100 into a fresh LB medium. The medium was supplemented with 1 mM l-tryptophan and induced with increased Hg(II) concentrations. The mixture was shaken at 150 rpm and incubated for 12 h at 37 °C. Finally, the bacterial density and indigo-derived color signal were measured as described previously.
2.6. Validation of Indigo’s Visual Reporting Potential Using Different Biosensing Models
The plasmids pPb-TrxA-bFMO, pAs-TrxA-bFMO, and pCd-TrxA-bFMO were transformed into E. coli TOP10 strains to create biosensors for Pb(II), As(III), and Cd(II), respectively. Overnight activated biosensor cells were diluted 1:100 in fresh LB medium with 1 mM l-tryptophan. In a time–dose–response assay, TOP10/pPb-TrxA-bFMO was induced with 0, 14.4, and 1550 nM Pb(II) for 8 h, TOP10/pAs-TrxA-bFMO was induced with 0, 18.3, and 4680 nM As(III) for 13 h, and TOP10/pCd-TrxA-bFMO was induced with 0, 25, and 100 μM Cd(II) for 13 h. All were shaken at 150 rpm. At 1-h intervals, 1 mL aliquots of cultures were sampled to determine bacterial density and indigo-derived signal as described above.
In a dose–response assay, TOP10/pPb-TrxA-bFMO was exposed to 0–100 μM Pb(II) for 6 h, TOP10/pAs-TrxA-bFMO was exposed to 0–150 μM As(III) for 12 h, and TOP10/pCd-TrxA-bFMO was exposed to 0–400 μM Cd(II) for 12 h. All the groups were added with 1 mM l-tryptophan and were agitated at a speed of 150 rpm.
In a selectivity assay, TOP10/pPb-TrxA-bFMO and TOP10/pAs-TrxA-bFMO were exposed to various metal ions at 5 μM for 6 and 12 h, respectively. All cultures were shaken at 150 rpm and supplemented with 1 mM l-tryptophan. After the induction, the bacterial density and the color signal derived from indigo were measured as described above.
3. Results and Discussion
3.1. Indigo Biosynthesis in E. coli Expressing Self-Sufficient Oxygenases
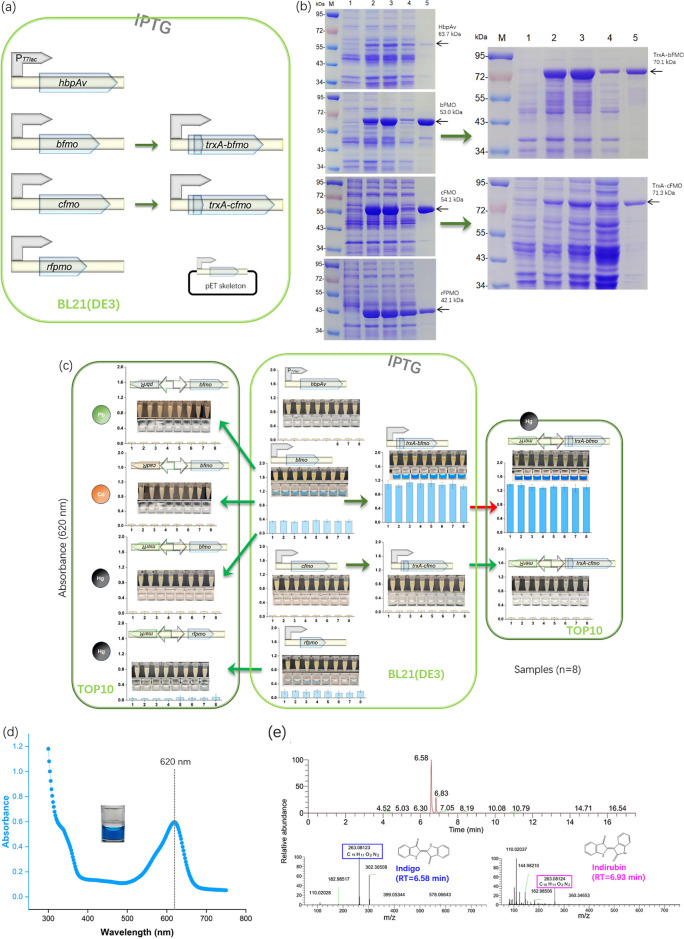
In recent years, scientists have discovered several enzymes that produce indigo in microorganisms, primarily bacteria.30 Some oxygenases that produce indigo are self-sufficient and may be expressed recombinantly in E. coli. Four enzymes, namely HbpAv from Pseudomonas azelaic HBP1, bFMO from Methylophaga sp. strain SK1, cFMO from Corynebacterium glutamicum, and rFPMO from Rhodococcus strain ATCC 21145, were selected for the study. The genes encoding these enzymes were artificially synthesized and optimized for bacterial coding. They were then cloned downstream of the T7 lac promoter (Figure 1a).
According to the data presented in Figure 1b, the HbpAv expression was deficient. On the other hand, rFPMO was highly expressed and was predominantly present in a soluble form. However, despite the high expression of bFMO and cFMO, most recombinant oxygenases were wrongly folded, insoluble, and lacked biological activity.
The TrxA partner, commonly used to increase the yield and solubility of recombinant proteins,38 was fused with the two enzymes. However, the enhancement of soluble expression was insignificant. Additionally, TrxA-cFMO expression decreased.
Various microbial origins have been shown to use l-tryptophan as a substrate to biosynthesize indigo via these four oxygenases.32−35 We first validated and compared their capacities in E. coli under the strong T7 lac promoter as a part of our study. Based on the findings depicted in Figure 1c, it was observed that pigment biosynthesis was not possible with recombinant HbpAv and cFMO. Even when TrxA was fused with cFMO, indigo production was not successful. However, recombinant bFMO and rFPMO successfully produced water-insoluble blue pigment. While the soluble expression of bFMO was lower than that of rFPMO, bFMO showed significantly more efficient pigment synthesis catalyzing than rFPMO. The bFMO is a homodimeric protein of 54 kDa.33 Recombinant E. coli expressing bFMO produced up to 160 mg/L of indigo in tryptophan medium after 12 h of cultivation.33 Its pigment-forming capacity was further increased when fused with the TrxA partner.
Previous research has demonstrated that augmenting the expression of enzymes alone is insufficient to boost pigment biosynthesis.21,39 Then, we engineered several promising oxygenases under weak, heavy metals-responsive promoters, including mer, cad, or pbr promoters. However, only the expression of TrxA-bFMO responding to the Hg(II)-responsive mer promoter synthesized blue pigment visible to the naked eye (Figure 1c). The TrxA-bFMO was then used as a novel biosensing module in the following studies.
Figure 2d displays the visible light scan of the blue pigment triggered by Hg(II). The highest absorbance was observed at around 620 nm, similar to the indigo standard from Sigma-Aldrich (Figure S2). The LC–MS test (Figure 2e) revealed that the blue pigment was indigo. Besides indigo, another peak appeared with a comparable retention time of 6.83 min, later identified as indirubin, a common byproduct generated during indole biotransformation.40 Furthermore, indigo was stable in acidic conditions under low temperatures (Figure S3).
Figure 2.
Heterologous expression of various oxygenases and their capacities in catalyzing the biosynthesis of indigo. (a) Recombinant production of four oxygenases in E. coli and (b) analysis of soluble expression. Lane M was loaded with protein molecular weight markers. Lanes 1–5 showed uninduced culture, IPTG-induced culture, induced culture broken by ultrasonication, the supernatant after ultrasonication, and the precipitation after ultrasonication. (c) Indigo biosynthesis in various engineered E. coli. The photos show the induced culture (above) and the DMSO extraction phase (below). Absorbance at 620 nm was measured in eight groups with at least three replicates. (d) Visible light scan of indigo biosynthesized in Hg(II)-induced recombinant bacteria. (e) The LC–MS chromatogram of DMSO containing indigo. It displayed primary (above) and secondary (below) mass spectra.
3.2. Hg(II)-Responsive Indigo Biosynthesis Enables Engineered E. coli Whole-Cell Biosensors
The TrxA-bFMO encoding element as an actuator module was genetically connected with a natural Hg(II) regulatory element41 as a sensory module to validate the visual biosensing potential of indigo. Dimer metalloregulator MerR can bind with Hg(II) and trigger the downstream reporter gene transcription (Figure 3a). The biosensor cells were slightly negatively affected by increased Hg(II) concentrations, as shown in Figure 3b. Figure 3c demonstrated an apparent time-dose–response relationship, and the signal amplification of enzymatic catalysis was apparent. No more indigo accumulation was observed after an 11-h induction in the high-dose group. No background noise was observed in the 0 nM Hg(II) group. However, high background fluorescent intensity was consistently observed in MerR-based fluorescent biosensors.5,11 No growth inhibition was observed after overnight incubation with Hg(II) exposure below 450 nm (Figure 3d). The maximum indigo output was also observed at that dose (Figure 3e). At 56.3 nM, a sudden appearance of indigo was visible to the naked eye (Figure 3e). However, a slow and gradual synthesis of pigments usually occurred in other pigment-based reporters, such as deoxyviolacein, indigoidine, and carotenoid.17,21,22 Thanks to the signal amplification of pigment biosynthesis, indigo-based Hg(II) biosensors have a lower detection limit than most fluorescent biosensors.9 The detection range for Hg(II) was quantified between 56.3 and 450 nM using indigo-based biosensors (Figure 3e) with an excellent selectivity (Figure 3f), which can potentially become a qualitative sensor that indicates whether Hg(II) exceeds the limit value.
Figure 3.
MerR-triggered indigo biosynthesis enables a visual biosensor toward Hg(II). (a) Molecular mechanism of TrxA-bFMO expression responsive to Hg(II). The time–dose–response relationship of TOP10/pHg-TrxA-bFMO was studied by determining bacterial density (b) and indigo absorbance (c) at 1-h intervals. The dose–response curve of TOP10/pHg-TrxA-bFMO was drawn by measuring bacterial density (d) and indigo absorbance (e) with increased concentrations of Hg(II) exposure. The asterisk indicates p < 0.05 against the background by t-Student analysis (n = 3). (f) Response of TOP10/pHg-TrxA-bFMO to various metal ions at 5 μM.
3.3. Optimization of Biosensing Conditions Improves the Performance of Hg(II) Bacterial Biosensors
The optimization of fermentation conditions, including microbial hosts, adequate substrates, and air exchange state, is crucial for indigo production.42 The different genetic backgrounds of bacterial hosts contribute to producing different amounts of pigments.17,22 Although the growth of TOP10 was the weakest among four E. coli hosts (Figure 4a), the indigo production was significant and comparable to that of wildly used protein expression host BL21(DE3). However, the indigo output of the latter fell immediately at above 225 nM Hg(II) (Figure 4b). It is worth noting that TOP10 gave the most sensitive and visual response to Hg(II) at a concentration of 56.3 nM. Surprisingly, the production of indigo in DH5α was significantly lower than in other bacterial strains and only slightly visible to the naked eye at a concentration of 225 nM. Therefore, it can be concluded that bacterial genetic makeup significantly impacts indigo biosynthesis.
Figure 4.
Optimizing biosensing conditions using an indigo-based Hg(II) biosensor. The host effect on bacterial growth (a) and indigo production (b) was investigated after induced with increased concentrations of Hg(II). The impact of extra substrate supplementation on bacterial growth (c) and indigo production (d) was investigated after exposing TOP10/pHg-TrxA-bFMO to increased concentrations of Hg(II). Bacterial growth (e) and indigo production (f) in TOP10/pHg-TrxA-bFMO were studied under varying ventilation conditions. The asterisk indicates p < 0.05 against the background by t-Student analysis (n = 3).
Extra substrate supplements positively affect pigment production during high-density fermentation but have little to no effect under conventional laboratory conditions.21,42 We observed that adding 1 mM l-tryptophan did not harm bacterial growth, as shown in Figure 4c. However, it increased the sensitivity to Hg(II) response to 28.1 nM. Interestingly, the maximum indigo absorbance at 620 nm was about 1.5 regardless of l-tryptophan addition. However, indigo output fell rapidly in the l-tryptophan supplement group upon exposure to above 450 nM Hg(II) (Figure 4d).
Overventilation had a detrimental impact on the production of colorants in genetically modified bacteria.16 Therefore, we decreased the rotation rate from 250 to 150 rpm. As the Hg(II) concentration increased, the cultures darkened due to intracellular indigo accumulation, increasing OD600 values (Figure 4e). As depicted in Figure 4f, the maximum absorbance of indigo at 620 nm increased to 1.7. The minimum concentration of Hg(II) that elicited a visual response decreased further to 14.1 nM. After optimizing the biosensing conditions, the quantitative range of Hg(II) detection extended from 14.1 to 225 nM.
3.4. Indigo Biosynthesis Triggered by Other Metalloregulators in a Dose-Dependent Manner
Microorganisms can adapt to changes in their environment and maintain cellular balance by regulating the concentrations of metal ions inside their cells. This regulation is usually achieved through transcriptional control by metal-responsive regulators such as PbrR, ArsR, and CadR.7 These regulators have been used to develop synthetic bacteria for the detection and bioremediation of Pb(II), As(III), and Cd(II).43,44 In the following study, we validated the indigo reporter module’s visual biosensing potential by engineering it under the control of the three sensory elements above.
The TrxA-bFMO genetic module was placed downstream of a redesigned PbrR-based sensory element (Figure 5a). The Pb(II) concentration had no noticeable effect on bacterial growth. There was a slight increase in the values of OD600 due to the darkening of the culture, as shown in Figure 5b. No production of indigo background was detected. However, fluorescent biosensors using the PbrR regulator observed a certain background level.45,46 The blue color signal remained stable following a 6-h Pb(II) induction in a time and dose-dependent manner (Figure 5c). No cytotoxicity was observed until 100 μM Pb(II) (Figure 5d). Still, the indigo output was stable since 24.4 nM (Figure 5e). The developed biosensors were capable of detecting Pb(II) at concentrations as low as 1.5 nM, which is more sensitive than fluorescent whole-cell biosensors.46,47 The biosensors could quantify Pb(II) detection, ranged from 1.5 to 24.4 nM, and showed excellent specificity toward Pb(II) (Figure 5f), meeting various environmental test requirements.10
Figure 5.
PbrR-triggered indigo biosynthesis enables a visual biosensor toward Pb(II). (a) Molecular mechanism of TrxA-bFMO expression responsive to Pb(II). The time–dose–response relationship of TOP10/pPb-TrxA-bFMO was investigated by determining bacterial density (b) and indigo absorbance (c) at 1-h intervals. The concentration-dependent response of TOP10/pPb-TrxA-bFMO was evaluated by measuring bacterial density (d) and indigo absorbance (e) upon increasing concentrations of Pb(II) exposure. The asterisk indicates p < 0.05 against the background by t-Student analysis (n = 3). (f) Response of TOP10/pPb-TrxA-bFMO to various metal ions at 5 μM.
The arsenic-responsive biosensor genetic circuit was constructed as Figure 6a. In addition to the accumulation of indigo, the concentration of As(III) did not affect bacterial growth (Figure 6b). No indigo background production was detected, outperforming reported biosensors.44,48 The blue color signal remained stable after 8 h of As(III) induction in a dose and time-dependent manner, as shown in Figure 6c. No cytotoxicity was observed up to 150 μM As(III) (Figure 6d). At the same time, the indigo output remained stable since 73.2 nM (Figure 6e). The biosensors developed could detect As(III) at concentrations as low as 4.5 nM and quantify As(III) from 4.5 to 73.2 nM, which is more sensitive than fluorescent biosensors.49 The response to group 15 antimony is a congenital deficiency of ArsR,50 also observed in this sensor (Figure 6f). Available whole-cell biosensors employing natural ArsR and its variants failed to distinguish As(III) from Sb(III) efficiently.51 However, the molecular evolution of ArsR and its congenital promoter had the potential to improve biosensing selectivity toward As(III).49,52
Figure 6.
Indigo biosynthesis regulated by ArsR has been developed as a visual biosensor for detecting As(III). (a) The molecular mechanism behind the expression of TrxA-bFMO in response to As(III). The time–dose–response relationship of TOP10/pAs-TrxA-bFMO was studied by measuring bacterial density (b) and indigo absorbance (c) every hour. The dose–response curve of TOP10/pAs-TrxA-bFMO was drawn by measuring bacterial density (d) and indigo absorbance (e) with increased concentrations of As(III) exposure. The asterisk indicates p < 0.05 against the background by t-Student analysis (n = 3). (f) Response of TOP10/pAs-TrxA-bFMO to different 5 μM metal ions.
A genetic circuit for the detection of cadmium was constructed, as shown in Figure 7a. No cytotoxic Cd(II) concentrations were used (Figure 7b). A high background output of biosensing signals was consistently observed in CadR-based biosensors.20 However, no leaked indigo synthesis was observed in the control group. The time–dose–response relationship was demonstrated (Figure 7c). A slight OD600 value increased due to intracellular indigo accumulation upon exposure to 50 μM Cd(II) above (Figure 7d). Compared to fluorescent and pigment-based biosensors,11,53 the dose–response relationship has shifted back at concentrations between 25 and 200 μM. It is speculated that promoters with different activities induce different levels of indigo synthase expression, but the optimization of indigo synthesis always occurs at an appropriate indigo synthase level.
Figure 7.
CadR-triggered indigo biosynthesis enables a visual biosensor toward a high concentration of Cd(II). (a) Molecular mechanism of TrxA-bFMO expression responsive to Cd(II). The time–dose–response relationship of TOP10/pCd-TrxA-bFMO was investigated by determining bacterial density (b) and indigo absorbance (c) at 1-h intervals. The dose–response curve of TOP10/pCd-TrxA-bFMO was plotted by measuring bacterial density (d) and indigo absorbance (e) under increased concentrations of Cd(II) induction. The asterisk indicates p < 0.05 against the background by t-Student analysis (n = 3).
Instrumental techniques are crucial for detecting heavy metal pollution, but for speciation, the high barriers to instrumental methods make it challenging to popularize them.54 Various cell-based biosensors emerging with the rapid development of synthetic biology specifically respond to bioavailable heavy metals, which are toxic to organisms.13 Many studies indicate that whole-cell biosensors are a powerful supplement to traditional instrumental ways.1,9,10 The emergence of colorimetric whole-cell biosensors further lowers their application threshold.23 The development of colorful reporters facilitates the design of versatile biological devices to monitor environmental contaminations using green analytical chemistry.
4. Conclusion
This study has successfully developed a novel green chemical, visual reporter for bacterial biosensing with zero background noise based on an engineered indigo biosynthesis module. We have created a visual monitoring method for metal pollutants by engineering four self-sufficient indigo-forming enzymes in E. coli. TrxA-bFMO was chosen for its robust ability to produce indigo under IPTG and Hg(II) induction.
We optimized indigo production by selecting better bacterial hosts, supplementing l-tryptophan, and optimizing ventilation conditions. These indigo biosensors have demonstrated susceptible detection of Hg(II), Pb(II), As(III), and Cd(II) with detection limits of 14.1 nM, 1.5 nM, 4.5 nM, and 25 μM, respectively. These biosensors’ sensitivity, responsive concentration range, and selectivity are superior to fluorescent reporter enzymes. However, indigo-based biosensors are a cost-effective and mini-equipment requirement.
Furthermore, this research investigated the potential of heterologous biosynthesis of indigo. This exploration opens new possibilities for environmentally friendly chemical synthesis and is a foundation for developing innovative bacterial biosensors to monitor pollutants visually. The biosensors’ ability to respond to different heavy metals highlights the potential of engineered enzymes for producing natural pigments like indigo.
This study contributes significant scientific knowledge to green chemistry synthesis and biosensing, providing a new biotechnological tool for environmental pollutant monitoring. Further optimization of biosensor performance and exploration of real-world applications in ecological tracking will be pursued in future work.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (82073517), the Natural Science Foundation of Guangdong Province (2023A1515011184), the Natural Science Foundation of Shenzhen Municipality (JCYJ20230807151400002), the Shenzhen Key Medical Discipline Construction Fund (SZXK068), and the Shenzhen Fund for Guangdong Provincial High-level Clinical Key Specialties (SZGSP015).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c03613.
Primary sequences and DNA sequences of indigo-forming enzymes (Table S1); DNA sequence of biosensing sensory elements (Table S2); the assembly of recombinant plasmids and engineered bacteria (Figure S1); the comparison of Hg(II)-induced blue pigment with indigo sample from Sigma (Figure S2); the stability of indigo (Figure S3) (PDF)
Author Contributions
# Y.G. and S.-Y.H. equally contributed to this work. C.-Y.H. and Y.G. contributed to conceptualization; C.-X.G. and Y.G. contributed to methodology; S.-Y. H. and C.W. contributed to investigation; C.-Y.H. contributed to writing–review and editing; C.-Y.H., and Y.G. contributed to funding acquisition. All authors have read and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Liu C.; Yu H.; Zhang B.; Liu S.; Liu C. G.; Li F.; Song H. Engineering whole-cell microbial biosensors: Design principles and applications in monitoring and treatment of heavy metals and organic pollutants. Biotechnol. Adv. 2022, 60, 108019. 10.1016/j.biotechadv.2022.108019. [DOI] [PubMed] [Google Scholar]
- Somayaji A.; Sarkar S.; Balasubramaniam S.; Raval R. Synthetic biology techniques to tackle heavy metal pollution and poisoning. Synth. Syst. Biotechnol. 2022, 7 (3), 841–846. 10.1016/j.synbio.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X.; Volpetti F.; Petrova E.; French C.; Maerkl S. J.; Wang B. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 2019, 15 (5), 540–548. 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Lim J. W.; Jeong H.; Lee S. J.; Lee D. W.; Kim T.; Lee S. J. Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry. Biosens. Bioelectron. 2016, 79, 701–708. 10.1016/j.bios.2015.12.101. [DOI] [PubMed] [Google Scholar]
- Du R.; Guo M.; He X.; Huang K.; Luo Y.; Xu W. Feedback regulation mode of gene circuits directly affects the detection range and sensitivity of lead and mercury microbial biosensors. Anal. Chim. Acta 2019, 1084, 85–92. 10.1016/j.aca.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Hu S. Y.; Yang X. Q.; Guo Y. A panel of visual bacterial biosensors for the rapid detection of genotoxic and oxidative damage: A proof of concept study. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2023, 888, 503639. 10.1016/j.mrgentox.2023.503639. [DOI] [PubMed] [Google Scholar]
- Jung J.; Lee S. J. Biochemical and biodiversity insights into heavy metal ion-responsive transcription regulators for synthetic biological heavy metal sensors. J. Microbiol. Biotechnol. 2019, 29 (10), 1522–1542. 10.4014/jmb.1908.08002. [DOI] [PubMed] [Google Scholar]
- a Zhang N. X.; Guo Y.; Li H.; Yang X. Q.; Gao C. X.; Hui C. Y. Versatile artificial mer operons in Escherichia coli towards whole cell biosensing and adsorption of mercury. PLoS One 2021, 16 (5), e0252190 10.1371/journal.pone.0252190. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hui C. Y.; Guo Y.; Liu L.; Yi J. Recent advances in bacterial biosensing and bioremediation of cadmium pollution: a mini-review. World J. Microbiol. Biotechnol. 2022, 38 (1), 9. 10.1007/s11274-021-03198-w. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Ma B. C.; Hu S. Y.; Wu C. Tailored bacteria tackling with environmental mercury: Inspired by natural mercuric detoxification operons. Environ. Pollut. 2024, 341, 123016. 10.1016/j.envpol.2023.123016. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Ma B. C.; Wang Y. Q.; Yang X. Q.; Cai J. M. Designed bacteria based on natural pbr operons for detecting and detoxifying environmental lead: A mini-review. Ecotoxicol. Environ. Saf. 2023, 267, 115662. 10.1016/j.ecoenv.2023.115662. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Guo Y.; Li H.; Chen Y. T.; Yi J. Differential detection of bioavailable mercury and cadmium based on a robust dual-sensing bacterial biosensor. Front. Microbiol. 2022, 13, 846524. 10.3389/fmicb.2022.846524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. Y.; Lin Y. J.; Kao Y. L.; Kuo P.; Grauffel C.; Lim C.; Cheng Y. S.; Chou H. D. Sensitive and specific cadmium biosensor developed by reconfiguring metal transport and leveraging natural gene repositories. ACS Sens. 2021, 6 (3), 995–1002. 10.1021/acssensors.0c02204. [DOI] [PubMed] [Google Scholar]
- Thai T. D.; Lim W.; Na D. Synthetic bacteria for the detection and bioremediation of heavy metals. Front. Bioeng. Biotechnol. 2023, 11, 1178680. 10.3389/fbioe.2023.1178680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Yu D.; Li R.; Sun X.; Zhang H.; Yu H.; Dong S. Colorimetric and electrochemical dual-signal method for water toxicity detection based on Escherichia coli and p-Benzoquinone. ACS Sens. 2021, 6 (7), 2674–2681. 10.1021/acssensors.1c00651. [DOI] [PubMed] [Google Scholar]; b Yu D.; Li R.; Rong K.; Fang Y.; Liu L.; Yu H.; Dong S. A novel, environmentally friendly dual-signal water toxicity biosensor developed through the continuous release of Fe(3). Biosens. Bioelectron. 2023, 220 (220), 114864. 10.1016/j.bios.2022.114864. [DOI] [PubMed] [Google Scholar]
- Yang D.; Park S. Y.; Lee S. Y. Production of rainbow colorants by metabolically engineered Escherichia coli. Adv. Sci. 2021, 8 (13), e2100743 10.1002/advs.202100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.; Park S. Y.; Park Y. S.; Eun H.; Lee S. Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020, 38 (7), 745–765. 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Ma B. C.; Guo Y.; Lin Y. R.; Zhang J.; Wang X. Q.; Zhang W. Q.; Luo J. G.; Chen Y. T.; Zhang N. X.; Lu Q.; et al. High-throughput screening of human mercury exposure based on a low-cost naked eye-recognized biosensing platform. Biosens. Bioelectron. 2024, 248, 115961. 10.1016/j.bios.2023.115961. [DOI] [PubMed] [Google Scholar]
- Hui C.; Guo Y.; Gao C.; Li H.; Lin Y.; Yun J.; Chen Y.; Yi J. A tailored indigoidine-based whole-cell biosensor for detecting toxic cadmium in environmental water samples. Environ. Technol. Innovation 2022, 27, 102511. 10.1016/j.eti.2022.102511. [DOI] [Google Scholar]
- a Hui C. Y.; Guo Y.; Zhu D. L.; Li L. M.; Yi J.; Zhang N. X. Metabolic engineering of the violacein biosynthetic pathway toward a low-cost, minimal-equipment lead biosensor. Biosens. Bioelectron. 2022, 214, 114531. 10.1016/j.bios.2022.114531. [DOI] [PubMed] [Google Scholar]; b Guo Y.; Huang Z. L.; Zhu D. L.; Hu S. Y.; Li H.; Hui C. Y. Anthocyanin biosynthetic pathway switched by metalloregulator PbrR to enable a biosensor for the detection of lead toxicity. Front. Microbiol. 2022, 13 (13), 975421. 10.3389/fmicb.2022.975421. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wang D.; Zheng Y.; Fan X.; Xu L.; Pang T.; Liu T.; Liang L.; Huang S.; Xiao Q. Visual detection of Hg(2+) by manipulation of pyocyanin biosynthesis through the Hg(2+)-dependent transcriptional activator MerR in microbial cells. J. Biosci. Bioeng. 2020, 129 (2), 223–228. 10.1016/j.jbiosc.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Guo Y.; Li H.; Gao C. X.; Yi J. Detection of environmental pollutant cadmium in water using a visual bacterial biosensor. Sci. Rep. 2022, 12 (1), 6898. 10.1038/s41598-022-11051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C. Y.; Guo Y.; Li L. M.; Liu L.; Chen Y. T.; Yi J.; Zhang N. X. Indigoidine biosynthesis triggered by the heavy metal-responsive transcription regulator: a visual whole-cell biosensor. Appl. Microbiol. Biotechnol. 2021, 105 (14–15), 6087–6102. 10.1007/s00253-021-11441-5. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Hu S. Y.; Li L. M.; Yun J. P.; Zhang Y. F.; Yi J.; Zhang N. X.; Guo Y. Metabolic engineering of the carotenoid biosynthetic pathway toward a specific and sensitive inorganic mercury biosensor. RSC Adv. 2022, 12 (55), 36142–36148. 10.1039/D2RA06764A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer G.; Koubaa M.; El Chamy L.; Maroun R. G.; Louka N. Seeing colors: A literature review on colorimetric whole-cell biosensors. Fermentation 2024, 10 (2), 79. 10.3390/fermentation10020079. [DOI] [Google Scholar]
- Ruxandra Leontieş A.; Răducan A.; Cristina Culiţă D.; Alexandrescu E.; Moroşan A.; Eduard Mihaiescu D.; Aricov L. Laccase immobilized on chitosan-polyacrylic acid microspheres as highly efficient biocatalyst for naphthol green B and indigo carmine degradation. Chem. Eng. J. 2022, 439, 135654. 10.1016/j.cej.2022.135654. [DOI] [Google Scholar]
- Kaplan G.; Seferoglu Z. The synthetic approaches for preparation of indigo and applications in denim industry. Curr. Org. Synth. 2023, 20 (4), 361–364. 10.2174/1570179419666220830091956. [DOI] [PubMed] [Google Scholar]
- Aino K.; Hirota K.; Okamoto T.; Tu Z.; Matsuyama H.; Yumoto I. Microbial communities associated with indigo fermentation that thrive in anaerobic alkaline environments. Front. Microbiol. 2018, 9, 2196. 10.3389/fmicb.2018.02196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Shoji O.; Fujishiro T.; Ohki T.; Ueno T.; Watanabe Y. Construction of biocatalysts using the myoglobin scaffold for the synthesis of indigo from indole. Catal. Sci. Technol. 2012, 2 (4), 739–744. 10.1039/C2CY00427E. [DOI] [Google Scholar]
- Chen L.; Xu J.-K.; Li L.; Gao S.-Q.; Wen G.-B.; Lin Y.-W. Design and engineering of neuroglobin to catalyze the synthesis of indigo and derivatives for textile dyeing. Mol. Syst. Design Eng. 2022, 7 (3), 239–247. 10.1039/D1ME00123J. [DOI] [Google Scholar]
- Qu Y.; Ma Q.; Liu Z.; Wang W.; Tang H.; Zhou J.; Xu P. Unveiling the biotransformation mechanism of indole in a Cupriavidus sp. strain. Mol. Microbiol. 2017, 106 (6), 905–918. 10.1111/mmi.13852. [DOI] [PubMed] [Google Scholar]
- Fabara A. N.; Fraaije M. W. An overview of microbial indigo-forming enzymes. Appl. Microbiol. Biotechnol. 2020, 104 (3), 925–933. 10.1007/s00253-019-10292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Chen Q.; Deng H.; Ye S.; Chen R.; Keasling J. D.; Luo X. One-pot selective biosynthesis of Tyrian purple in Escherichia coli. Metab. Eng. 2024, 81, 100–109. 10.1016/j.ymben.2023.11.003. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Schmid A.; Held M.; Westphal A. H.; Rothlisberger M.; Kohler H. P.; van Berkel W. J.; Witholt B. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 2002, 277 (7), 5575–5582. 10.1074/jbc.M110018200. [DOI] [PubMed] [Google Scholar]
- Choi H. S.; Kim J. K.; Cho E. H.; Kim Y. C.; Kim J. I.; Kim S. W. A novel flavin-containing monooxygenase from Methylophaga sp strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 2003, 306 (4), 930–936. 10.1016/S0006-291X(03)01087-8. [DOI] [PubMed] [Google Scholar]
- Ameria S. P.; Jung H. S.; Kim H. S.; Han S. S.; Kim H. S.; Lee J. H. Characterization of a flavin-containing monooxygenase from Corynebacterium glutamicum and its application to production of indigo and indirubin. Biotechnol. Lett. 2015, 37 (8), 1637–1644. 10.1007/s10529-015-1824-2. [DOI] [PubMed] [Google Scholar]
- Hart S.; Kirby R.; Woods D. R. Structure of a Rhodococcus gene encoding pigment production in Escherichia coli. J. Gen. Microbiol. 1990, 136 (7), 1357–1363. 10.1099/00221287-136-7-1357. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Guo Y.; Lin Y. R.; Ma B. C.; Ge X. R.; Zhang W. Q.; Zhang N. X.; Yang S. M.; Hui C. Y. Detection of cadmium in human biospecimens by a cadmium-selective whole-cell biosensor based on deoxyviolacein. ACS Biomater. Sci. Eng. 2024, 10, 4046. 10.1021/acsbiomaterials.3c01814. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Guo Y.; Liu L.; Zheng H. Q.; Wu H. M.; Zhang L. Z.; Zhang W. Development of a novel bacterial surface display system using truncated OmpT as an anchoring motif. Biotechnol. Lett. 2019, 41 (6–7), 763–777. 10.1007/s10529-019-02676-4. [DOI] [PubMed] [Google Scholar]
- Ren J.; Hwang S.; Shen J.; Kim H.; Kim H.; Kim J.; Ahn S.; Kim M. G.; Lee S. H.; Na D. Enhancement of the solubility of recombinant proteins by fusion with a short-disordered peptide. J. Microbiol. 2022, 60 (9), 960–967. 10.1007/s12275-022-2122-z. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Guo Y.; Liu L.; Zhang N. X.; Gao C. X.; Yang X. Q.; Yi J. Genetic control of violacein biosynthesis to enable a pigment-based whole-cell lead biosensor. RSC Adv. 2020, 10 (47), 28106–28113. 10.1039/D0RA04815A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C.; Ma Q.; Li Y.; Zhou D.; Yang B.; Qu Y. Application of an efficient indole oxygenase system from Cupriavidus sp. SHE for indigo production. Bioprocess Biosyst. Eng. 2019, 42 (12), 1963–1971. 10.1007/s00449-019-02189-4. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Hui C. Y.; Liu L.; Chen M. P.; Huang H. Y. Development of a bioavailable Hg(II) sensing system based on MerR-regulated visual pigment biosynthesis. Sci. Rep. 2021, 11 (1), 13516. 10.1038/s41598-021-92878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel N.; Singh B. B.; Dureja C.; Yang Y. H.; Bhatia S. K. Indigo production goes green: a review on opportunities and challenges of fermentative production. World J. Microbiol. Biotechnol. 2024, 40 (2), 62. 10.1007/s11274-023-03871-2. [DOI] [PubMed] [Google Scholar]
- a Guo Y.; Hui C. Y.; Zhang N. X.; Liu L.; Li H.; Zheng H. J. Development of cadmium multiple-signal biosensing and bioadsorption systems based on artificial cad operons. Front. Bioeng. Biotechnol. 2021, 9, 585617. 10.3389/fbioe.2021.585617. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hui C. Y.; Guo Y.; Yang X. Q.; Zhang W.; Huang X. Q. Surface display of metal binding domain derived from PbrR on Escherichia coli specifically increases lead(II) adsorption. Biotechnol. Lett. 2018, 40 (5), 837–845. 10.1007/s10529-018-2533-4. [DOI] [PubMed] [Google Scholar]; c Hui C.; Guo Y.; Zhang W.; Gao C.; Yang X.; Chen Y.; Li L.; Huang X. Surface display of PbrR on Escherichia coli and evaluation of the bioavailability of lead associated with engineered cells in mice. Sci. Rep. 2018, 8 (1), 5685. 10.1038/s41598-018-24134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y.; Zhang Y.; Li R.; Wang B.; Ye B. C. De Novo design of the ArsR regulated Pars promoter enables a highly sensitive whole-cell biosensor for arsenic contamination. Anal. Chem. 2022, 94 (20), 7210–7218. 10.1021/acs.analchem.2c00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.; Liu X.; Sun P.; Wang X.; Zhu H.; Hong M.; Mao Z. W.; Zhao J. Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ. Sci. Technol. 2014, 48 (6), 3363–3371. 10.1021/es4046567. [DOI] [PubMed] [Google Scholar]
- Bereza-Malcolm L.; Aracic S.; Franks A. E. Development and application of a synthetically-derived lead biosensor construct for use in Gram-negative bacteria. Sensors 2016, 16, 2174. 10.3390/s16122174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chen H.; Shao S.; Yu Y.; Huang Y.; Zhu X.; Zhang S.; Fan J.; Yin G. Y.; Chi B.; Wan M.; et al. A dual-responsive biosensor for blood lead detection. Anal. Chim. Acta 2020, 1093, 131–141. 10.1016/j.aca.2019.09.062. [DOI] [PubMed] [Google Scholar]; b Hui C. Y.; Guo Y.; Liu L.; Zheng H. Q.; Gao C. X.; Zhang W. Construction of a RFP-lacZalpha bicistronic reporter system and its application in lead biosensing. PLoS One 2020, 15 (1), e0228456 10.1371/journal.pone.0228456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Cui M.; Zhao J.; Wang J.; Fang X. A self-amplifying plasmid based ultrasensitive biosensor for the detection of As(III) in water. Biosens. Bioelectron. 2023, 221, 114937. 10.1016/j.bios.2022.114937. [DOI] [PubMed] [Google Scholar]
- Chen S. Y.; Wei W.; Yin B. C.; Tong Y.; Lu J.; Ye B. C. Development of a highly sensitive whole-cell biosensor for arsenite detection through engineered promoter modifications. ACS Synth. Biol. 2019, 8 (10), 2295–2302. 10.1021/acssynbio.9b00093. [DOI] [PubMed] [Google Scholar]
- Chen X.; Yao H.; Song D.; Lin J.; Zhou H.; Yuan W.; Song P.; Sun G.; Xu M. A novel antimony-selective ArsR transcriptional repressor and its specific detection of antimony trioxide in environmental samples via bacterial biosensor. Biosens. Bioelectron. 2023, 220, 114838. 10.1016/j.bios.2022.114838. [DOI] [PubMed] [Google Scholar]
- Hui C. Y.; Liu M. Q.; Guo Y. Synthetic bacteria designed using ars operons: a promising solution for arsenic biosensing and bioremediation. World J. Microbiol. Biotechnol. 2024, 40 (6), 192. 10.1007/s11274-024-04001-2. [DOI] [PubMed] [Google Scholar]
- Li L.; Liang J.; Hong W.; Zhao Y.; Sun S.; Yang X.; Xu A.; Hang H.; Wu L.; Chen S. Evolved bacterial biosensor for arsenite detection in environmental water. Environ. Sci. Technol. 2015, 49 (10), 6149–6155. 10.1021/acs.est.5b00832. [DOI] [PubMed] [Google Scholar]
- Hui C.-Y.; Guo Y.; Gao C.-X.; Li H.; Lin Y.-R.; Yun J.-P.; Chen Y.-T.; Yi J. A tailored indigoidine-based whole-cell biosensor for detecting toxic cadmium in environmental water samples. Environ. Technol. Innovation 2022, 27, 102511. 10.1016/j.eti.2022.102511. [DOI] [Google Scholar]
- Marcinkowska M.; Baralkiewicz D. Multielemental speciation analysis by advanced hyphenated technique - HPLC/ICP-MS: A review. Talanta 2016, 161, 177–204. 10.1016/j.talanta.2016.08.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.