Abstract
Successful trans-complementation of the defective Kunjin virus (KUN) RNA FLdGDD with a deletion of the RNA polymerase motif GDD in the NS5 gene by using a BHK cell line, repBHK, that continuously produced a functionally active KUN replication complex (RC) from replicon RNA was recently reported (A. A. Khromykh, M. T. Kenney, and E. G. Westaway, J. Virol. 72:7270–7279, 1998). In order to identify whether this complementation of FLdGDD RNA was provided by the wild-type NS5 protein alone or with the help of other nonstructural (NS) proteins also expressed in repBHK cells, we generated BHK cell lines stably producing the individual NS5 protein (SRns5BHK) or the NS1-NS5 polyprotein (SRns1-5BHK) by using a heterologous expression vector based on a modified noncytopathic Sindbis replicon. Western blot analysis with anti-NS5 antibodies showed that the level of production of NS5 was significantly higher in SRns5BHK cells than in SRns1-5BHK cells. Despite the higher level of expressed NS5, trans-complementation of FLdGDD RNA was much less efficient in SRns5BHK cells than in SRns1-5BHK cells and produced at least 100-fold less of the secreted complemented virus. In contrast, efficient complementation of KUN RNA with lethal cysteine-to-alanine mutations in the NS1 gene was achieved both in BHK cells producing the individual KUN NS1 protein from the Sindbis replicon vector and in repBHK cells, with both cell lines expressing similar amounts of NS1 protein. These results clearly demonstrate that flavivirus NS5 coexpressed with other components of the viral replicase possesses much higher functional (trans-complementing) activity than individually expressed NS5 and that efficient trans-complementation of mutated flavivirus NS1 and NS5 proteins occurs by different mechanisms. The results are interpreted and discussed in relation to our proposed model of formation of the flavivirus RC largely based on previous ultrastructural and biochemical analyses of KUN replication.
The flavivirus Kunjin (KUN) genome consists of single-stranded RNA of positive polarity comprising one long open reading frame coding for three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1 to NS5) (8). RNA replication occurs in cytoplasm and is associated with a range of induced membrane structures. Previously, we postulated that flavivirus double-stranded RNA or replicative form functions as the recycling template for synthesis of genomic RNA late in infection (5, 6). Further characterization of the membrane-associated KUN replication complex (RC) by immunogold labelling and a variety of biochemical analyses showed that the KUN RC apparently comprises NS1, NS2A, NS3, NS4A, and NS5 proteins as the viral replicase plus the RNA template (7, 22, 32, 33). Others also showed possible involvement of NS1 in flavivirus RNA replication (18, 19, 21, 23), helicase activity of NS3 (17), and in vitro RNA-dependent RNA polymerase (RdRp) activity of NS5 (29). Specific cell proteins, such as elongation factor-1 alpha, which interacts with the terminal stem-loop of the 3′ untranslated region (3′UTR) of several flaviviruses (3), may also be involved.
Although RdRp activity was demonstrated for the purified dengue virus type 1 NS5 protein, the activity was shown to be nonspecific and inefficient (29), indicating requirements for other virus-specific and/or cell-specific factors. In an attempt to assess the virus-specific factors required for formation of the flavivirus RC, trans-complementation analysis in repBHK cells of a mutated KUN RNA genome for NS5 with a deletion of the RNA polymerase motif GDD in the NS5 gene (FLdGDD) was previously employed (14). It was shown that the defective RNA polymerase function in FLdGDD RNA could be complemented by the functional KUN RC produced from the persistently replicating KUN replicon RNA (14). Since repBHK cells were continuously producing not only individual KUN NS proteins but also a fully operational RC comprising NS proteins and replicating RNA, it was difficult to identify whether defective NS5, alone or in combination with other components of the RC, was involved in trans-complementation of the defective RC in these experiments. Lindenbach and Rice (18) recently described successful complementation of yellow fever virus (YF) RNA with a large deletion in the NS1 gene in BHK cells stably expressing YF NS1 protein from noncytopathic Sindbis virus replicon vectors. It was therefore reasonable to assume that this approach would be applicable to complementation of individual KUN NS proteins.
In this article, we report trans-complementation of the defective KUN NS1 and NS5 proteins by the wild-type KUN NS1 and NS5 proteins, expressed individually from the Sindbis virus replicon vector. We also show that efficient complementation of the defective NS5 protein, but not of the defective NS1 protein, requires expression of the complementing protein from the NS1-NS5 gene cassette. Interpretation of the presented results is based on our proposed model for formation of flavivirus RC.
MATERIALS AND METHODS
Construction of plasmids.
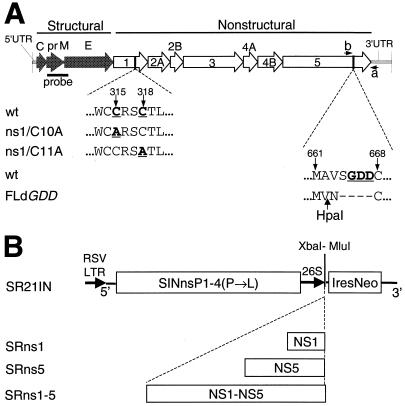
Cysteine-to-alanine mutations in the KUN NS1 gene (Fig. 1A) were prepared by overlapping PCR mutagenesis of the SphI2468-SphI3628 cDNA fragment from the FLSD clone (14) by using Pfu DNA polymerase and appropriate primers. The first two nucleotides, UG, in both the conserved 10th and 11th cysteine codons of the KUN NS1 gene (Fig. 1A) (8) were mutated to GC to produce alanine codons GCU and GCC, respectively. The resulting mutated SphI fragments were then used to substitute the corresponding SphI fragment in the FLSD clone to obtain ns1/C10A and ns1/C11A, respectively (Fig. 1A). Preparation of the FLdGDD plasmid (Fig. 1A) was described previously (14).
FIG. 1.
(A) Schematic representation of mutated KUN cDNA constructs ns1/C10A and ns1/C11A for NS1 and FLdGDD for NS5. The wild type (wt) represents the native KUN sequence flanking the mutated cysteine (C) residue numbers 10 and 11 (indicated by arrows) in NS1 and flanking the GDD motif in NS5. Cysteine residues were mutated to alanine (A) by PCR mutagenesis of the NS1 gene as described in Materials and Methods. Construction of FLdGDD cDNA was described previously (14). Numbers show amino acid positions coded in the KUN NS1 and NS5 genes (8). probe, the cDNA fragment in the prM-E region that was used for Northern blot analysis (see Materials and Methods); a and b, the positions of the primers used in RT-PCR (Fig. 4A); HpaI, the location of the HpaI restriction site in the coding region introduced into FLdGDD cDNA during construction (see reference 14). (B) Sindbis virus replicon constructs expressing KUN NS genes. The SR21IN vector was constructed from the noncytopathic DNA-based Sindbis virus replicon vector SINRep21 (2) as described in Materials and Methods. The KUN NS1 and NS5 genes and the KUN NS1-NS5 gene cassette were each cloned into the SR21IN vector as described in Materials and Methods to obtain the indicated SRns1, SRns5, and SRns1-5 constructs, respectively. XbaI-MluI, unique cloning sites; IresNeo, IRES of encephalomyelocarditis virus RNA followed by the NEO gene; RSV LTR, left terminal repeat of Rous sarcoma virus; 26S, Sindbis virus subgenomic promoter.
Sindbis virus replicon vector SR21IN (Fig. 1B) was constructed from the SINRep21 vector (kindly provided by C. M. Rice and colleagues) (2) by replacing the 26S promoter-puromycin N-acetyltransferase gene (pac) cassette with the encephalomyocarditis virus internal ribosome entry site-neomycin transferase (IRES-NEO) gene cassette derived from the plasmid pCIN4 (derivative of pCIN1 [27], obtained from S. Rees).
Plasmids SRns1 and SRns5 were prepared by cloning the KUN NS1 and NS5 genes (KUN nucleotides 2651 to 3525 and 7681 to 10398, respectively) (8, 11), PCR amplified with Pfu DNA polymerase from the FLSDX clone (14) with appropriate primers with incorporated translation initiation and termination codons, into the SR21IN vector. Primers for amplification of the NS1 gene were ns1MluF (forward, 5′-ggcacgcgtaccATGGCTCGAGATAGATCCA-3′) and pAcYM1ns1R (reverse, 5′-gctggatcctaGGCATTCACCTGTGA-3′). The resulting PCR fragment was cloned into the SR21IN vector digested with XbaI and blunt ended with the Klenow fragment of DNA polymerase. Primers for amplification of the NS5 gene were ns5MluF (5′-gaacgcgtaccATGGGTGGGGCAAAAGGA-3′) and ns5MluR (5′-ggaacgcgTTACAATACTGTATCCTCAA-3′). The underlined nucleotides in the above primer sequences represent MluI restriction sites, bold nucleotides show translation initiation and termination codons, and lowercase letters represent nonviral nucleotides. The resulting NS5 PCR fragment was digested with MluI and cloned into the MluI-digested SR21IN vector. The added methionine codon ATG is the only change to the N-terminal sequence of NS5. The plasmid SRns1-5 (Fig. 1B) containing the KUN NS1 to NS5 genes was constructed as follows. A 268-bp fragment containing the NS1 signal sequence and the beginning of the NS1 gene extending as far as the XbaI2651 site (KUN nucleotides 2392 to 2651) was purified from an MluI- and XbaI-digested PCR fragment amplified with Pfu DNA polymerase from the FLSDX clone (14), using the ns1MluF primer and a primer complementary to a sequence at the carboxy terminus of the NS1 gene. The resulting MluI-XbaI2651 fragment and an ∼8-kb fragment, extending from an XbaI2651 site to an MluI site 25 nucleotides downstream of the NS5 termination codon, excised from the FLSD cDNA clone (14) were then ligated into the SR21IN vector digested with MluI to obtain the SRns1-5 plasmid.
Antibodies.
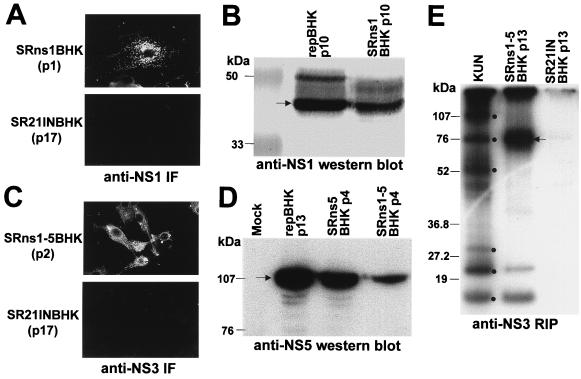
Anti-NS5 antibody was prepared by immunizing an adult rabbit subcutaneously several times with ∼100 μg of partially purified recombinant baculovirus-expressed glutathione S-transferase–NS5 fusion protein (12). The specificity of the antibody was confirmed by radioimmunoprecipitation (RIP) and Western blot (WB) analyses of NS5 in KUN-infected cells (data not shown) and by WB in KUN replicon-expressing cells (Fig. 2D, repBHK lane). Preparation of monoclonal antibodies to the KUN E and NS1 proteins has been described previously (1), as has preparation of rabbit polyclonal antibodies to KUN NS3 (33).
FIG. 2.
Characterization of BHK cell lines stably expressing KUN NS genes from a Sindbis virus replicon vector. BHK cell lines expressing KUN NS genes were generated by transfection with corresponding recombinant Sindbis virus replicon plasmid DNAs followed by G418 selection as described in Materials and Methods. (A and C) IF analysis of SRns1BHK (A) and SRns1-5BHK (C) cells with anti-NS1 and anti-NS3 antibodies at passages 1 and 2, respectively. (B) WB analysis of SRns1BHK cells (passage 10) and repBHK cells (passage 10) with anti-NS1 antibodies. The SR21INBHK cells in panels A and C show results of IF analysis of the control cells expressing vector (SR21IN) RNA by using anti-NS1 and anti-NS3 antibodies, respectively. (D) WB analysis of SRns5BHK (passage 4), SRns1-5BHK (passage 4), and repBHK (passage 13) cells with anti-NS5 antibodies. Approximately 5 × 104 cells were boiled in the SDS-PAGE sample buffer without (B) or with (D) β-mercaptoethanol, and samples were electrophoresed in an SDS–12.5% (B) or –10% (D) polyacrylamide gel. Blotting and detection of expressed proteins were performed as described in Materials and Methods. (E) RIP analysis of SRns1-5BHK and vector control SR21INBHK cell lysates with anti-NS3 antibodies. The control KUN lane represents radiolabelled KUN-infected BHK cells, with dots indicating positions of labelled KUN proteins (from top to bottom) NS5, NS3, E, prM, NS2A, and NS2B/NS4A. SRns1-5BHKp13 and SR21INBHKp13 lanes represent results of RIP analysis of cells that were transfected with SRns1-5 and SR21IN (vector only) DNAs, respectively, and given 13 cell passages in medium containing 0.5 to 1 mg of G418 per ml. Confluent monolayers of cells in 60-mm culture dishes were preincubated with 6 μg of actinomycin D per ml in methionine-cysteine-free medium for 1 h at 37°C and then labelled for 6 h in the same medium supplemented with 50 μCi of [35S]methionine-cysteine for 6 h. The lysate was solubilized in RIP assay buffer (1% deoxycholate, 1% NP-40, 0.1% SDS, 0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl) and centrifuged at 15,000 × g for 5 min to remove insoluble material, and a sample was radioimmunoprecipitated with anti-NS3 antibodies as described previously (33). The arrow shows the position of the NS3 protein.
IF, WB, and Northern blot analyses.
Immunofluorescence (IF) analysis of acetone-fixed cells with appropriate antibodies was performed as described previously (13, 33). For WB analysis with anti-NS5 antibodies, ∼5 × 104 mock BHK, repBHK, SRns5BHK, and SRns1-5BHK cells were boiled in the reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and the proteins were separated in an SDS–10% polyacrylamide gel and were electrophoretically transferred to a Hybond-P membrane (Amersham). The membrane was treated overnight in blocking buffer (5% skim milk powder in phosphate-buffered saline) at 4°C, washed in phosphate-buffered saline, and incubated in blocking buffer with rabbit anti-NS5 antibody diluted 1:50,000. Bound antibody was detected by using the ECL Plus chemiluminescence kit (Amersham) as described by the manufacturer. For anti-NS1 WB analysis, ∼105 cells were resuspended in RIP buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid sodium salt, 0.1% SDS) and clarified by centrifugation. Supernatants were then mixed with nonreducing SDS-PAGE sample buffer in a 1:1 ratio and boiled for 5 min, and the proteins were separated by electrophoresis in an SDS–10% PAGE minigel. Protein blotting and detection with KUN monoclonal anti-NS1 antibodies were performed as described previously (1).
Northern blot analysis of RNA purified from transfected cells was performed as described previously (14) by using a 32P-labelled AatII-ClaI cDNA fragment representing 567 nucleotides of the KUN prM-E region (KUN nucleotides 522 to 1089).
Generation of BHK cell lines expressing KUN NS proteins.
BHK21 cells were transfected with SR21IN, SRns1, SRns5, and SRns1-5 plasmid DNAs by electroporation under conditions described previously for electroporation of RNA (13), with the exception that only 1 to 2 μg of DNA was used for electroporation. Three days after electroporation, medium containing 0.5 to 1 mg of G418 (Geneticin; Gibco BRL) per ml was added (14) and cells were maintained in the medium with G418 during further passages to select for cells expressing KUN NS proteins. The expression of KUN proteins during passaging of cells was monitored by IF and/or WB analyses with appropriate antibodies (Fig. 2).
RT-PCR of RNA recovered from complemented secreted viruses.
Culture fluids (CFs; 630 μl) collected at day 5 after transfection of FLdGDD RNA into SRns5BHK and SRns1-5BHK cells were treated with 50 μg of RNase A (Sigma) per ml and 5 U of RQ1 DNase (Promega) per ml for 30 min at 37°C in order to ensure the absence of any possible DNA and RNA contaminations from transfected in vitro transcription mixtures. CFs still containing RNase A and DNase were then incubated overnight at 4°C with 70 μl of anti-E monoclonal antibodies to allow binding of secreted KUN particles followed by a further 2 h of incubation with 100 μl of a slurry of 10% protein A-Sepharose (Pharmacia). The precipitates on the washed protein A-Sepharose beads were treated with proteinase K in the presence of 0.5% SDS, followed by phenol-chloroform extraction and ethanol precipitation of the RNA. Precipitated RNA was dissolved in 6 μl of diethyl pyrocarbonate-treated H2O, and 1 to 3 μl of this RNA was used in a 10-μl reverse transcription (RT)-PCR with the SuperScript One-Step RT-PCR System (Gibco BRL) essentially as described by the manufacturer and with primers a and b (Fig. 1A). Primers were as follows: primer a, 5′-CACACTAAACACTATTATAAAGCTAAA-3′, minus sense, complementary to nucleotides 10443 to 10469 of KUN RNA in the 3′UTR region (8, 11, 14); and primer b, 5′-CGGCCCAGATGATGTGGAGAAA-5′, plus sense, representing nucleotides 9576 to 9597 of KUN RNA, approximately 100 nucleotides upstream of the GDD deletion (see Fig. 7 in reference 14).
RESULTS
Generation and characterization of stable BHK cell lines expressing KUN NS proteins from a modified DNA-based Sindbis virus replicon expression vector.
BHK cell lines stably expressing individual KUN NS genes NS1 and NS5, as well as all the KUN NS genes (NS1-NS5 cassette), were established by using DNA-based Sindbis virus replicon expression vector pSR21IN (Fig. 1B), which we constructed from the vector SINRep21 (2). In an attempt to increase the percentage of cells expressing higher levels of desired genes, we replaced the 26S promoter-puromycin resistance gene cassette in the SINRep21 vector with the IRES-NEO cassette from the pCIN4 vector (see Materials and Methods and Fig. 1B). The expression of low levels of the neomycin resistance gene NEO (obtained by specific mutations in the IRES sequence in the IRES-NEO cassette) (27), combined with simultaneous expression of a heterologous gene(s) from the same bicistronic mRNA (subgenomic RNA in the case of SINRep vectors), should allow for selection of cells producing only high levels of desired proteins during incubation in medium with a high concentration of the antibiotic G418 (27). Thus, using this modified vector SR21IN and selection with high concentrations of G418 (0.5 to 1 mg/ml), we established three BHK cell lines stably expressing individually the KUN NS1 and NS5 genes, as well as the NS1-NS5 gene cassette (see Materials and Methods). IF analysis showed that after 1 to 2 passages most of the SRns1BHK and SRns1-5BHK cells were producing KUN NS1 (Fig. 2A) and KUN NS3 (Fig. 2C) proteins, respectively. Production of the NS1 protein in SRns1BHK and repBHK cells and of the NS5 protein in repBHK, SRns5BHK, and SRns1-5BHK cells was then analyzed by WB analysis with the corresponding antibodies (Fig. 2B and D). Similar amounts of NS1 protein were produced in SRns1BHK and repBHK cells (Fig. 2B). In contrast, NS5 expression was significantly higher in repBHK and SRns5BHK cells than in SRns1-5BHK cells (Fig. 2D). It is possible that the lower level of expression of NS5 in SRns1-5BHK cells than in SRns5BHK cells was due to the much larger size of the inserted KUN sequence (∼8 kb versus ∼2.7 kb), which may influence the rate and efficiency of replication of the resulting Sindbis virus replicon RNA. Expression and correct processing of other KUN NS genes in SRns1-5BHK cells were demonstrated by detection of coprecipitated KUN NS3, NS2A, and NS2B/NS4A proteins in RIP analysis with anti-NS3 antibodies (Fig. 2E). We concluded from these results that KUN NS proteins were produced and correctly processed in cells selected for their stable expression by using the noncytopathic DNA-based Sindbis virus replicon vector SR21IN expressing the NEO gene under control of the encephalomyocarditis virus IRES.
Complementation of FLdGDD RNA in SRns1-5BHK and SRns5BHK cells and characterization of complemented viruses.
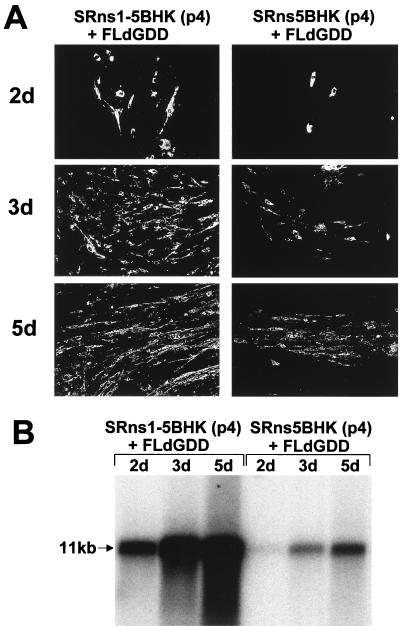
In order to evaluate the functional activity of KUN NS5 and NS1-NS5 proteins expressed from the Sindbis virus replicon in SRns5BHK and SRns1-5BHK cells, we used these cells in complementation experiments with NS5-defective FLdGDD RNA previously shown to be efficiently complemented in repBHK cells (14). Passage 4 of both Sindbis replicon cell lines (Fig. 2D) was used in complementation experiments. Transfection of FLdGDD RNA into both cell lines resulted in successful complementation of its replication as detected by IF and Northern blot analyses (Fig. 3). Interestingly, despite a significantly lower level of expressed NS5 in SRns1-5BHK cells than in SRns5BHK cells (Fig. 2D), complementation was much more efficient in SRns1-5BHK cells than in SRns5BHK cells (compare results in Fig. 3). While only rare foci of anti-E-positive cells were observed in transfected SRns5BHK cells by day 5 after transfection, most of the SRns1-5BHK cells were already positive by day 3 after transfection (Fig. 3A). These IF observations were confirmed by Northern blot analysis, showing a very slow accumulation of FLdGDD RNA in SRns5BHK cells by day 5 after transfection compared to a rapid accumulation of complemented FLdGDD RNA in SRns1-5BHK cells as early as day 2 posttransfection (Fig. 3B). No anti-E-positive cells or prM-E-specific RNA was detected after transfection of FLdGDD RNA into the control SR21INBHK cells (data not shown), as was shown previously for transfection of FLdGDD RNA into BHK cells (14). In a separate experiment, we observed similar efficiencies of complementation of FLdGDD RNA replication in repBHK and in SRns1-5BHK cells (data not shown).
FIG. 3.
Complementation of FLdGDD RNA in SRns1-5BHK and SRns5BHK cells. (A) IF analysis of SRns5BHK (passage 4) and SRns1-5BHK (passage 4) cells at 2, 3, and 5 days after transfection with FLdGDD RNA (14), using KUN anti-E antibodies. (B) Northern blot analysis of total RNA isolated from SRns5BHK (passage 4) and SRns1-5BHK (passage 4) cells at 2, 3, and 5 days after transfection with FLdGDD RNA. The cDNA probe for the prM-E region was described in the legend for Fig. 1A. The arrow indicates the position of RNA at about 11 kb, determined relative to migration in the same gel of ethidium bromide-stained 1-kb Plus DNA ladder (Gibco BRL).
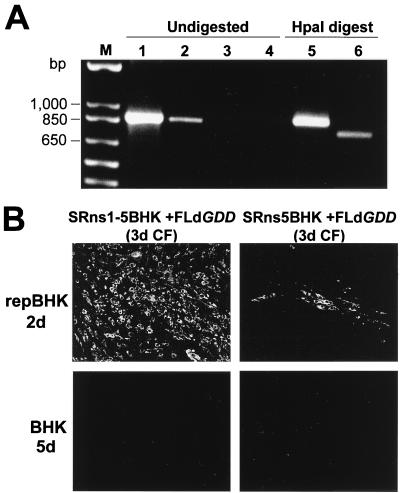
It was previously shown in the complementation experiments with FLdGDD RNA in repBHK cells that RNA of the secreted complemented virus retained the introduced deletion and that complemented virus was able to replicate in repBHK but not in normal BHK cells (14). Similar results were obtained with the viruses recovered in the CF after transfection of FLdGDD RNA into SRns5BHK and SRns1-5BHK cells. RT-PCR of RNAs isolated from virus particles precipitated with anti-E antibodies from 5-day CFs of FLdGDD-transfected SRns1-5BHK cells resulted in amplification of a DNA fragment with the predicted size of 817 bp (Fig. 4A, lane 2). An RT-PCR fragment of the same size was amplified in the control reaction with KUN virion RNA (Fig. 4A, lane 1). No RT-PCR amplification was observed from RNA recovered from CFs of FLdGDD-transfected SRns5BHK and normal BHK cells (Fig. 4A, lanes 3 and 4, respectively). Our failure to detect KUN RNA in the virus particles from the 5-day CF of FLdGDD-transfected SRns5BHK cells is in accord with the observed very low efficiency of complementation by the individually expressed NS5 protein (Fig. 3B). Digestion with HpaI restrictase resulted in a decrease by ∼100 nucleotides in the size of the fragment amplified from RNA isolated from SRns1-5BHK-transfected CFs, as expected (Fig. 4A, compare lanes 2 and 6), but not in the fragment amplified from the control KUN virion RNA (Fig. 4A, compare lanes 1 and 5). Hence the presence in the RT-PCR fragment of the HpaI site introduced in place of the GDD deletion during construction of the FLdGDD plasmid (Fig. 1A) (14) demonstrated retention of the GDD deletion in the recovered complemented viral RNA.
FIG. 4.
Characterization of secreted complemented FLdGDD viruses. (A) RT-PCR of complemented virus RNAs. KUN particles secreted from cells after complementation of the defective genomic RNA were treated with RNase A and DNase and immunoprecipitated with anti-E antibodies, and the virion RNA was extracted and used in RT-PCR analysis as described in Materials and Methods. Lane 1 represents an RT-PCR with KUN virion RNA purified as described previously (11). Lanes 2 to 4 represent the RT-PCRs of the RNA recovered from CFs after transfection of FLdGDD RNA into SRns1-5BHK cells (lane 2), SRns5BHK cells (lane 3), and BHK cells (lane 4). Lanes 5 and 6 show an HpaI digest of RT-PCR-amplified fragments from lanes 1 and 2, respectively. Lane M, molecular size markers. (B) IF analysis, using anti-E antibodies, of repBHK cells at 2 days and BHK cells at 5 days after infection with defective viruses recovered in 3-day CFs of FLdGDD-transfected SRns1-5BHK and SRns5BHK cells.
Recovered viruses were further characterized by their ability to replicate in repBHK cells (14) by using IF analysis of infected cells with anti-E antibodies. Initial IF analysis with anti-E antibodies showed that ∼100% of repBHK cells were positive by 2 days after infection with undiluted 3-day CF collected from FLdGDD-transfected SRns1-5BHK cells, while only a small number of anti-E-positive foci were detected at 2 days after infection of repBHK cells with undiluted 3-day CF collected from FLdGDD-transfected SRns5BHK cells (Fig. 4B). Thus, detection of a small number of anti-E-positive foci in infected repBHK cells demonstrated the presence of defective virus in very low concentrations in the CF collected from FLdGDD-transfected SRns5BHK cells, despite the negative results of RT-PCR analysis (Fig. 4A). We next determined infectious titers of viruses recovered from 3-day CFs by counting anti-E-positive foci in repBHK cells at 2 days after infection with serial dilutions of the corresponding CFs. The titers were ∼8 × 104 and ∼5 × 102 infectious units per ml for transfections in SRns1-5BHK and SRns5BHK cells, respectively. More than a 100-fold difference in the amounts of recovered secreted viruses confirmed the significantly higher efficiency of complementation of FLdGDD RNA in transfected SRns1-5BHK cells than in SRns5BHK cells.
We next examined recovered viruses for the presence of possible recombinants. It was previously shown that recombination between the defective NS5 gene in FLdGDD RNA and the wild-type NS5 gene in replicon RNA, which could lead to the appearance of virus able to replicate in normal BHK cells, did not occur in complementation experiments performed in repBHK cells (14). In agreement with these results, we were also not able to detect any anti-E-positive cells by day 5 after infection of normal BHK cells with 3-day CFs from FLdGDD-transfected SRns5BHK and SRns1-5BHK cells (Fig. 4B), clearly demonstrating the absence of recombinant virus in the recovered defective virus stocks. Thus, we concluded that KUN NS5 protein produced from a noncytopathic Sindbis virus replicon vector expressing either an NS1-NS5 polyprotein cassette or the NS5 gene alone is capable of trans-complementing replication of defective KUN RNA carrying a deletion of the RNA polymerase motif and that NS5 protein produced as a part of the polyprotein cassette is much more efficient in trans-complementation than individually expressed NS5.
Complementation of RNAs with mutations in the NS1 gene and characterization of complemented viruses.
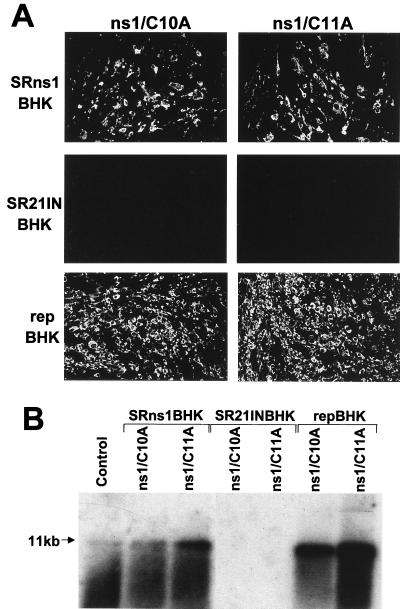
We were intrigued by the differences between inefficient trans-complementation of KUN NS5 observed in our experiments and the efficient trans-complementation of YF NS1 shown by Lindenbach and Rice (18, 19) when providing individually expressed NS5 and NS1 proteins, respectively. In order to show that the results obtained with complementation of YF NS1 were applicable to KUN NS1, we examined complementation of defective KUN RNAs with lethal mutations in the KUN NS1 gene by transfection into SRns1BHK cells expressing the wild-type KUN NS1 gene (see first section of Results). Two full-length KUN RNAs, ns1/C10A and ns1/C11A, in which conserved flavivirus codons for the 10th and 11th cysteines in the C-terminal part of the KUN NS1 gene, respectively, had been mutated to alanine were generated (Fig. 1A). Note that the corresponding cysteine residues remained in the coding sequence of YF RNA with a deletion in NS1 (18). We assumed that the cysteine-to-alanine mutations would be lethal due to irreversible conformational changes induced by disruption of disulfide bonds. Mutations of the third and fourth cysteines in dengue virus type 2 NS1 were shown to abort replication of full-length dengue virus type 2 RNA (26). It was also shown that mutations of the 10th and 11th cysteines did not affect the stability of dengue virus type 2 NS1 protein but completely inhibited its dimerization and secretion when expressed individually from a mammalian expression vector (25). In our experiments, transfection of KUN ns1/C10A and ns1/C11A RNAs into control BHK cells stably transfected with SR21IN vector alone (SR21INBHK) (Fig. 1B and 2E) or into normal BHK cells did not result in replication of these mutated RNAs by 2 days after transfection (Fig. 5A, SR21INBHK panels, and Fig. 5B, SR21INBHK lanes; data not shown for normal BHK cells), indicating that the mutations were lethal. In contrast, efficient replication of both cysteine mutant RNAs mediated by trans-complementation was detected at day 2 after their transfection in SRns1BHK cells and in repBHK cells by IF analysis with anti-E antibodies (Fig. 5A) and by Northern blot analysis with a prM-E-specific probe (Fig. 5B). In some experiments, replication of KUN ns1/C11A RNA was detected also in SR21INBHK or normal BHK cells late after transfection (days 4 to 6) (data not shown), suggesting that reversion of the point mutation to a wild-type sequence may have occurred during initially undetectable replication of mutated RNA in transfected cells (i.e., prior to day 4). Alternatively, a very low number of RNA molecules containing a wild-type sequence may be present in the in vitro-transcribed RNA stock due to the relatively low fidelity of SP6 RNA polymerase. Both these events would lead to the detectably delayed replication of these RNA molecules in SR21INBHK and normal BHK cells. Similar results were previously observed in detection of replication of defective KUN RNA FLGVD containing a point mutation in the NS5 gene after prolonged incubation of transfected normal BHK cells (14). Nevertheless, these observations did not compromise the results observed earlier of efficient complementation of defective KUN RNAs in SRns1BHK and repBHK cells (2 days) after transfection.
FIG. 5.
Complementation of KUN NS1 mutant RNAs ns1/C10A and ns1/C11A in SRns1BHK and repBHK cells. (A) IF analysis of SRns1BHK, SR21INBHK, and repBHK cells with KUN anti-E antibodies at day 2 after transfection with ns1/C10A and ns1/C11A RNAs. (B) Northern blot analysis of total RNA isolated from SR21INBHK, SRns1BHK, and repBHK cells at 2 days after transfection with ns1/C10A and ns1/C11A RNAs. The probe (prM-E region) was the same as that in Fig. 3B. The arrow indicates the position of RNA at about 11 kb, determined as described in the legend for Fig. 3B.
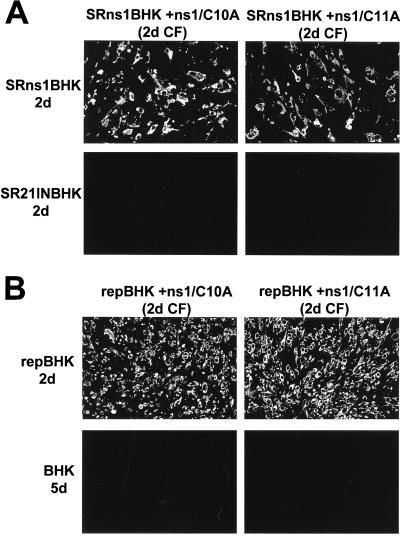
We next examined whether replication of defective viruses collected in the CFs of ns1/C10A- and ns1/C11A-transfected SRns1BHK cells could occur in SRns1BHK or SR21INBHK cells. No anti-E-positive SR21INBHK cells were detected at 2 days (Fig. 6A) or 5 days (data not shown) after infection with defective viruses recovered from 2-day CF of SRns1BHK cells transfected with either cysteine mutant RNA, confirming the defective genotype of complemented viruses and the absence of recombination in these complementation experiments. In contrast, when these recovered defective viruses were used to infect SRns1BHK cells, complementation occurred and all the cells were anti-E positive by 2 days after infection (Fig. 6A). Similarly, complementation of defective viruses recovered after transfection of ns1/C10A and ns1/C11A RNAs into repBHK cells occurred in repBHK cells but not in normal BHK cells (Fig. 6B). Viruses recovered from 2-day CFs of transfected SRns1BHK and repBHK cells were titrated by infection of repBHK cells with serial dilutions and counting of anti-E-positive foci at 2 days after infection. This titration showed that all four complemented viruses had similar titers in the range of 1 × 105 to 4 × 105 infectious units per ml in repBHK cells. Overall, the results presented in this section clearly demonstrate that defective full-length KUN RNAs with lethal mutations in the NS1 gene were complemented with similar efficiency in cells expressing KUN NS1 protein either as an individual protein from the Sindbis virus replicon vector or as part of a KUN polyprotein cassette from KUN replicon RNA. Clearly, a milieu of other KUN NS proteins being coexpressed had no inhibitory or enhancing effect on complementation of defective NS1.
FIG. 6.
Characterization of complemented NS1 Cys− mutant viruses. (A) IF analysis, with anti-E antibodies, of SRns1BHK and SR21INBHK cells at 2 days after infection with 2-day CFs collected from SRns1BHK cells transfected with ns1/C10A or ns1/C11A RNAs. (B) IF analysis with anti-E antibodies of repBHK and BHK cells at 2 days (repBHK) and 5 days (BHK) after infection with 2-day CFs collected from repBHK cells transfected with ns1/C10A or ns1/C11A RNAs.
DISCUSSION
Difference in complementation requirements for NS5 and NS1 proteins.
The main objective of this study was to determine whether RNAs with replication defects caused by a deletion or mutations in the flavivirus KUN genes NS5 and NS1 could be rescued by providing wild-type proteins in trans either as individual proteins or as a part of the flavivirus polyprotein cassette. Although it was recently shown that replication of FLdGDD RNA containing a deletion of the RNA polymerase motif GDD in the NS5 gene could be complemented in repBHK cells persistently expressing KUN replicon RNA (14), these results did not identify whether complementation was achieved by rescue of the defective function of mutated NS5 protein via the individual wild-type NS5 protein expressed from replicon RNA or by rescue via the RC preformed in repBHK cells. We have now established that although wild-type NS5 protein produced individually in SRns5BHK cells (using a noncytopathic Sindbis virus replicon vector expressing the KUN NS5 gene) could complement replication of FLdGDD RNA, the efficiency of complementation was low. However, this efficiency was dramatically enhanced when NS5 was expressed as a part of the NS1-NS5 polyprotein gene cassette in SRns1-5BHK cells (using the same Sindbis virus replicon vector).
In contrast to the results with NS5 complementations, we observed similar efficiency of complementation of replication of KUN RNAs with lethal cysteine-to-alanine mutations in NS1 both in SRns1BHK cells expressing NS1 individually from the Sindbis virus replicon vector and in repBHK cells expressing all the NS proteins from KUN replicon RNA. Efficient complementation of YF RNA with a large deletion in the NS1 gene was reported in BHK cells stably expressing YF NS1 from a noncytopathic Sindbis virus replicon (18). Dengue virus type 2 NS1 could also complement the same YF NS1 deletion but only when Asn at position 42 of the YF NS4A gene was mutated (19). In preliminary experiments, we were able to rescue replication of full-length dengue virus type 2 RNA with a lethal mutation of both glycosylation sites in the NS1 gene (310NS1-G1,2−, kindly provided by M. J. Pryor) (23) by complementation in SRns1BHK cells expressing KUN NS1 (10). Taken together, our complementation results with NS1-defective KUN and dengue virus type 2 RNAs in KUN NS1-expressing cells and the cited results with NS1-defective YF RNA in YF NS1- and dengue virus type 2 NS1-expressing cells (18, 19) demonstrate that the replicative function(s) of defective flavivirus NS1 protein could be complemented by individually expressed wild-type NS1.
The observed difference in the efficiencies of complementation of defective NS5 suggests that other NS proteins and/or possibly polyprotein intermediates were required for efficient complementation of defective RNA polymerase function. Interestingly, inefficient complementation of poliovirus RNA replication defects was also observed when individual poliovirus gene products were supplied in trans to rescue some of the mutated NS proteins (30, 31). In contrast, site-specific lesions in several poliovirus NS proteins were efficiently complemented when corresponding wild-type proteins were provided by a helper virus infection (reviewed in reference 31) by expression from cotransfected replicon RNA (24) or by expression from a polyprotein precursor during in vitro HeLa cell-free translation/replication reactions (31). This last report demonstrated that the function of a defective 3AB protein was complemented in an in vitro translation/replication reaction only when the cleavable 3AB polyprotein precursor P3, and not 3AB alone, was provided in trans by translation from another RNA molecule. Hence, Towner et al. (31) concluded that for successful complementation of poliovirus RC to occur, the defective function must be rescued by an interchangeable protein unit such as P3. No data on trans-complementation of defective poliovirus RNA polymerase by an individually expressed 3Dpol protein are available. Although we showed that the KUN RC with defective NS5 could be complemented (inefficiently) by wild-type NS5 alone, our other results favor the notion that rescue in trans of RNA polymerases occurs more readily by exchange with a partially or fully assembled replicase rather then with an individually expressed protein component. Further experiments with complementation of mutated NS5 and possibly other NS proteins in cells expressing truncated versions of the NS1-NS5 polyprotein cassette should define the minimal exchangeable replicase subunits required for efficient trans-complementation.
Relevance of complementation results to modelling the flavivirus RC.
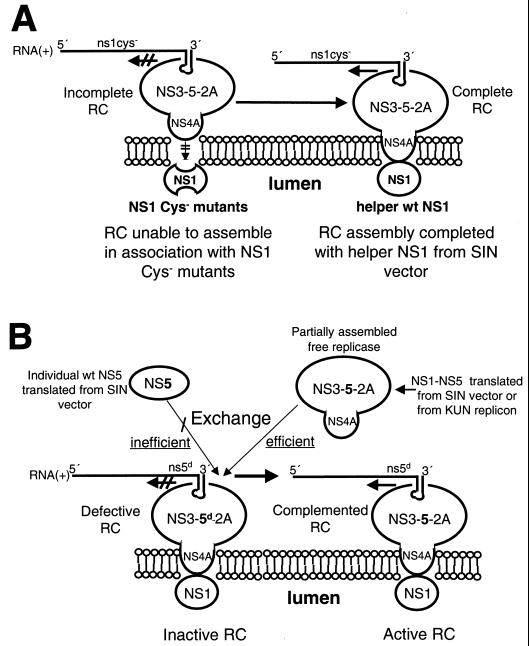
The consensus composition of the active flavivirus RC associated with the RNA template is NS1-NS3-NS5-NS2A-NS4A, based on electron microscopy of immunogold-labelled cryosections of KUN-infected cells, glutathione S-transferase-fusion protein binding assays, RIP of radiolabelled infected cell lysates by antibodies reactive with double-stranded RNA or replicative form functioning as the putative recycling template (22, 33), and purification of radiolabelled cell membrane fractions with retained RdRp activity (7). The association of the cytosolic components of the RC (NS3-NS5-NS2A-NS4A; incomplete RC in Fig. 7A) with NS1 presumably located in the lumen of the endoplasmic reticulum (ER) is intriguing. Recent genetic analyses of YF replication established an essential interaction between NS1 and NS4A for replicase activity (19). Based on the effect of specific mutations in the N-terminal (cytosolic) region of NS4A, it was suggested that this region affects the structure of an NS4A luminal peptide (located between predicted transmembrane spanning domains of NS4A) which might interact with NS1 (19). It was also proposed that such an interaction might induce a conformational effect on this region of NS4A, allowing it to recruit cytoplasmic components of the replicase. The ultrastructural definition of the RC in our immunogold labelling experiments late in KUN infection was unable to determine whether NS1 in the RC was located on the same side of the ER as the other components. However, the flavivirus translation model of Coia et al. (8) and the arguments advanced in complementation experiments with YF (19) favor the view that NS1 is confined within the lumen. We have incorporated this notion in our current model of formation of the flavivirus RC (15), where we have proposed that luminal NS1 interacts with NS4A via an exposed region between transmembrane spanning domains of NS4A and that NS4A is also associated with replicase components of the RC which have assembled on the 3′UTR adjacent to the ER. The inability of the NS1 cysteine mutants to interact with NS4A may result from incorrect folding of mutated NS1. If the proposed physical separation of NS1 from the remainder of the RC is correct, then it is reasonable that complementation in trans of defective NS1 can readily be achieved because of the availability of helper NS1, also in the lumen and independently expressed, without any requirement for a milieu of other NS proteins within the lumen. In other words, any source of wild-type NS1 could independently associate in the manner proposed with the remainder of the RC. A simplified model of such an association is shown in Fig. 7A.
FIG. 7.
Model showing how defective NS1 (NS1 Cys−) and NS5 with a deletion of the GDD polymerase motif (NS5d) in the RC can be complemented in trans by the wild-type (wt) NS1 and individual NS5 or NS1 to NS5, respectively, expressed from the Sindbis virus (SIN) replicon vector. (A) The RC assembles on the 3′UTR but is unable to associate correctly with the improperly folded NS1 Cys− mutant protein located in the lumen of the ER. However, this incomplete RC is able to associate with luminal wt NS1 supplied in trans from the Sindbis virus vector in SRns1BHK cells, thus completing the assembly of now functional RC. (B) A defective RC assembled with NS5d can be activated by an inefficient exchange with wt NS5 supplied independently in trans from the Sindbis virus vector in SRns5BHK cells. In contrast, a partially assembled wt KUN replicase supplied in trans by the Sindbis virus vector in SRns1-5BHK cells or by the KUN replicon in repBHK cells can readily exchange with the defective RC to form an active RC.
As noted above, complementation in trans of defective NS5 was much less efficient when helper NS5 was expressed independently rather than in association with the other NS proteins, in contrast to the observations with NS1. It is possible that the additional methionine which may remain (if not cleaved by methionine aminopeptidase) at the N terminus of the individually expressed NS5 relative to wild-type NS5 (see Materials and Methods) adversely affected the efficiency of complementation, as was reported for complementation by similarly modified Sindbis virus nsP4 RNA polymerase (16). However, we consider this unlikely based on our results demonstrating in vitro RNA polymerase activity of purified KUN NS5 proteins either with the additional N-terminal methionine or with an additional 26 N-terminal amino acids (9). In addition, RNA polymerase activity of NS5B protein of hepatitis C virus (another member of the Flaviviridae family) was also not apparently affected by the presence of additional N-terminal methionine (20).
Figure 7B illustrates how different mechanisms of trans-complementation by helper NS5 expressed independently or from a polyprotein cassette might occur. The requirement for the association of NS5 with other NS proteins accords with the proposal in our present model of replication that during translation in cis the RC commences to assemble by binding of NS3 (at least) to the N-terminal half of NS5, attaches to the adjacent 3′UTR on completion of translation, and finally moves to the proposed membrane attachment site involving NS4A and NS1, as discussed above (15). NS3 and NS5 of Japanese encephalitis virus have been shown to bind to each other and to the terminal conserved stem-loop in the 3′UTR (4). It is possible that the replicase can begin to assemble during translation in cis of KUN NS1 to NS5 encoded by the Sindbis virus vector that contains no KUN 3′UTR site for binding; hence, the partially assembled replicase (NS3-NS5-NS2A-NS4A) remains free and is able to readily exchange with the defective RC forming on FLdGDD RNA (Fig. 7B). Similarly, partially assembled replicase formed during or after completion of translation of the KUN replicon in repBHK cells may be briefly unattached to the proposed binding region of the RNA template and hence able to readily exchange with defective RC (Fig. 7B). Furthermore, during recycling of the native viral replicase and template (5, 6, 34) they will be transiently dissociated and hence free to exchange. In contrast, independently expressed NS5 is at a disadvantage in attempted trans-complementation because it must exchange with the defective NS5 in the assembled defective RC (Fig. 7B) and this exchange may be inefficient, possibly due to steric hindrance by defective NS5 protein for binding of helper NS5 to the defective RNA template or other components of defective replicase. While it is possible that assembly of NS proteins in the RC involves precursor polyprotein, we consider this scenario unlikely, at least late in infection. For example, in translation mapping experiments at 24 h postinfection which defined the correct order of translation in KUN-infected cells synchronized in initiation, all NS proteins were translated in about 17 min and all were apparently correctly cleaved within a chase period of 30 min (28). Furthermore, a complete block in translation by cycloheximide inhibition for several hours late in infection failed to stop KUN RNA synthesis (34). The interactions within the viral replicase shown in Fig. 7 are based on published KUN results discussed above and on other cited flavivirus data. We believe that the model represents a logical interpretation of accumulated past and present observations.
Although elusive details of flavivirus RNA replication are beginning to emerge, clearly more data including protein-RNA and protein-protein interactions are required to understand how the RC is assembled and how complementation in trans is achieved. The results and the model presented should contribute to such studies on flavivirus replication.
ACKNOWLEDGMENTS
We are grateful to Tatiana Khromykh and Jacqueline Scherret for technical assistance in preparation of NS1 mutants.
This work was supported by grant N981442 from the National Health and Medical Research Council of Australia.
Footnotes
This is publication no. 96 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Adams S C, Brown A K, Sammels L M, Hartnett A C, Howard M J, Coelen R J, Mackenzie J S, Hall R A. Glycosylation and antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. doi: 10.1016/s0042-6822(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 2.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C-J, Kulo M-D, Chien L-J, Hssu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 6.Chu P W, Westaway E G. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology. 1987;157:330–337. doi: 10.1016/0042-6822(87)90275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu P W, Westaway E G. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 8.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. Unpublished data.
- 10.Khromykh, A. A. Unpublished data.
- 11.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khromykh A A, Harvey T J, Abedinia M, Westaway E G. Expression and purification of the seven nonstructural proteins of the flavivirus Kunjin in the E. coli and the baculovirus expression systems. J Virol Methods. 1996;61:47–58. doi: 10.1016/0166-0934(96)02068-x. [DOI] [PubMed] [Google Scholar]
- 13.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh A A, Sedlak P L, Westaway E G. trans-complementation analysis of the flavivirus Kunjin NS5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol. 1999;73:9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm J A, Rumenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Clum S, You S, Ebner K E, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindenbach B D, Rice C M. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbach B D, Rice C M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie J M, Jones M K, Young P R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 23.Muylaert I R, Galler R, Rice C M. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 25.Pryor M J, Wright P J. The effects of site-directed mutagenesis on the dimerization and secretion of the NS1 protein specified by dengue virus. Virology. 1993;194:769–780. doi: 10.1006/viro.1993.1318. [DOI] [PubMed] [Google Scholar]
- 26.Pryor M J, Gualano R C, Lin B, Davidson A D, Wright P J. Growth restriction of dengue virus type 2 by site-specific mutagenesis of virus-encoded glycoproteins. J Gen Virol. 1998;79:2631–2639. doi: 10.1099/0022-1317-79-11-2631. [DOI] [PubMed] [Google Scholar]
- 27.Rees S, Coote J, Stables J, Goodson G, Harris S, Lee M G. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. BioTechniques. 1996;20:102–110. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- 28.Schrader A P, Westaway E G. Translation mapping with the flavivirus Kunjin: gene order and anomalities in translation of NS5. Virus Res. 1988;9:323–334. doi: 10.1016/0168-1702(88)90091-3. [DOI] [PubMed] [Google Scholar]
- 29.Tan B-H, Fu J, Sugrue R J, Yap E-H, Chan Y-C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 30.Teterina N L, Zhou W D, Cho M W, Ehrenfeld E. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J Virol. 1995;69:4245–4254. doi: 10.1128/jvi.69.7.4245-4254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towner J S, Mazanet M M, Semler B L. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol. 1998;72:7191–7200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westaway E G, Khromykh A A, Kenney M T, Mackenzie J M, Jones M K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 33.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westaway E G, Khromykh A A, Mackenzie J M. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]