Abstract
Head and neck squamous cell carcinomas (HNSCC) are associated with significant treatment-related morbidity and poor disease-free and disease-specific survival, especially in the recurrent and metastatic (R/M HNSCC) setting. Inhibition of the programmed death-1/ligand-1 (PD-1/PD-L1) immune checkpoint is accepted as a first-line treatment strategy for R/M HNSCC and has expanded into the neoadjuvant, definitive, and adjuvant settings. To understand cellular signals modulating the PD-L1 in HNSCC, we profiled a HNSCC cell-line with a genome-wide open reading frame (ORF) library of 17,000 individual constructs (14,000 unique genes). We identified 335 ORFs enriched in PD-L1high cells and independently validated five of these ORFs (FGF6, IL17A, CD300C, KLR1C and NFKBIA) as drivers of PD-L1 upregulation. We showed that exogenous FGF ligand is sufficient to induce PD-L1 expression in multiple HNSCC cell lines and human immature dendritic cells. Accordingly, overexpression of FGFR1, FGFR3 or the FGFR3 S249C and D786N mutants common to HNSCC tumors also induced PD-L1 overexpression on tumor cells. Small molecule inhibition of FGF signaling abrogated PD-L1 upregulation in these models and also blocked “classical” IFNγ-regulated PD-L1 expression in a STAT1-independent manner. Finally, we found that FGF specifically upregulated a glycosylated form of PD-L1 in our study, and exogenous FGF led to concomitant upregulation of glycosyltransferases that may stabilize PD-L1 on the surface of HNSCC cells. Taken together, our study supports a potential role for FGF/FGFR pathway signaling as a mechanism driving immune escape and rationalizes further exploration of novel combination therapies to improve clinical responses to PD-1/PD-L1 axis inhibition in HNSCC.
Introduction
Head and neck squamous cell carcinomas (HNSCC) are aggressive cancers with poor disease-free and disease-specific survival.1,2 In the recurrent and metastatic (R/M) setting in particular, treatments are limited, often impart significant toxicities, and typically yield only modest survival benefit.3 Thus, novel treatment strategies with acceptable safety profiles that tangibly improve outcomes are desperately needed. Immune checkpoint blockade targeting the PD-1/PD-L1 axis has become a first line strategy for R/M HNSCC.4 Increasingly, unique combinatorial regimens targeting the PD-1/PD-L1 axis and separate inhibitory immune pathways have shown impressive response rates and survival outcomes in select patients.5,6
Despite these advances, and the now widespread use of PD-1/PD-L1 inhibitors across many cancer types, the complex molecular mechanisms regulating PD-L1 expression and function on tumor and other antigen-presenting cells (APCs) in the HNSCC microenvironment remain only partially clarified.7,8 A better understanding of these mechanisms may facilitate discovery of new targetable pathways to overcome innate and acquired resistance to PD-1/PD-L1 axis blockade in HNSCC. While interferon gamma (IFNγ), secreted by tumor infiltrating lymphocytes, has long been known to induce cell surface expression of PD-L1 on tumor and other APCs, additional cell-intrinsic signals have been shown to potentiate PD-L1 expression and function independent of this “classical” pathway. For example, treatment of HNSCC cell lines with exogenous epidermal growth factor (EGF) upregulated PD-L1 expression in a JAK2/STAT1-dependent manner.9 Additionally, the EGFR effector STAT3 was shown to be a potent driver of PD-L1 expression in a Tgfbr1/Pten 2cKO HNSCC murine model.10 These studies suggest that EGF pathway activation, a hallmark of HNSCC biology, may serve as an IFNγ-independent mechanism for PD-L1 upregulation and tumor immune evasion. However, as tumor PD-L1 staining did not correlate with EGFR expression in a large clinical cohort of primary HNSCC specimens,11 it is likely that multiple distinct cellular mechanisms may drive PD-L1 expression and function in the HNSCC tumor microenvironment.
Recently, whole genome screening techniques, such as pooled CRISPR-Cas9 screens, have been adapted for a variety of applications to identify mediators of particular cellular phenotypes.12 These strategies entail lentiviral delivery of pooled short hairpin RNA (shRNA) or RNA-guided Cas9 libraries to induce genome wide knock-out of targeted gene expression. The transduced cell population is then monitored for dropout of specific constructs, indicating the essentiality of their genomic targets for cell viability.13 Here, we have modified this scheme to utilize a genome-scale open reading frame (ORF) library for overexpression of 17,000 genes in a pooled format, from which we can select individual cells in which PD-L1 expression has been upregulated. Through this high-throughput profiling approach, we sought to identify and validate novel regulators of PD-L1 expression in HNSCC cells with potential translational implications for improved patient selection and combinatorial immunotherapies.
Materials and Methods
Ethics Statement
All experiments were completed under a protocol reviewed and approved by the University of Michigan Institutional Review Board.
Cell Lines and Reagents
All UM-SCC cell lines used herein were derived and characterized within the Michigan Otolaryngology and Translational Oncology Laboratory at the University of Michigan with informed consent of the patient donors.14 The oral cavity and larynx cell lines used in this report were selected from models with comprehensive integrated SNP array, exome and transcriptome sequencing data recently published by our team.15,16 All UM-SCC cell lines were maintained in exponential growth at 37° C in a humidified atmosphere of 5% CO2 in DMEM supplemented with 10% fetal bovine serum (FBS), 7 μg/mL penicillin/streptomycin and 1% nonessential amino acids. Cal-33 cells (a kind gift from Dr. Anthony Nichols) were cultured similarly. HSC-2, HSC-4 (Japanese Collection of Research Bioresources, Sekisui XenoTech, Kansas City, KS) and Detroit 562 (ATCC, Manassas, VA) cells were cultured similarly in Eagle’s Minimum Essential Medium (EMEM) with FBS and penicillin/streptomycin. Human dendritic cells were purchased from Lonza and cultured in similar environment in LGM-3 media (Lonza) supplemented with IL-4 (50 ng/mL) and GM-CSF (50 ng/mL) to maintain cells as immature dendritic cells (iDCs). All cell lines were genotyped periodically throughout the study to ensure authenticity and tested for mycoplasma contamination using the MycoAlert detection kit (Lonza).15,16
Small molecule inhibitors (BGJ398, PD173074, Fedratinib, gefitinib, tunicamycin and MK-2206), were purchased from Selleck Chemicals and stored in DMSO at −80° C for no longer than one year. PNGase F was purchased in solution from New England BioLabs and stored at −20° C for no longer than two years. Recombinant IFN-γ was purchased from R&D Systems and stored in PBS at −20° C for no longer than two months. FGF ligands were purchased from Thermo Fisher Scientific and stored in PBS containing 0.5% BSA at −20° C for no longer than two months.
ORF Library Transduction
The genome-wide ORF library was purchased from Sigma Aldrich as a lentiviral transduction-ready pool (MISSION® TRC3 human whole genome lentiviral ORF pool). We first determined the appropriate multiplicity of infection (MOI) by transduction of UM-SCC-49 cells followed by puromycin selection and cell count assays, as described.17 Using an MOI of 0.3 – 0.5, we then transduced UM-SCC-49 and expanded the population of puromycin resistant clones after seven days of antibiotic selection. UM-SCC-49 ORF library transduced cells were maintained in culture and treated in pools of no less than 6 million cells. From the transduced pool, cell subsets were cloned out from control and treated populations to ensure enrichment of PD-L1 expression in individual clones, as described below.
Flow Cytometry
Suspensions of one million cells/mL per condition were incubated with anti-PD-L1 antibody #14–5983-82 or IgG1 kappa isotype control antibody #14–4714-82 (Thermo Fisher Scientific) at 0.5 μg/mL antibody dilution in PBS containing 1% FBS for 15 minutes, followed by PBS wash. Control and PD-L1 stained cells were then incubated with PE-conjugated rat anti-mouse secondary antibody (#12–4015-82, Thermo Fisher Scientific) at 0.2 μg/mL antibody dilution in PBS containing 1 % FBS for 15 minutes. Cell suspensions were then gated on live cells and sorted on a MoFlo Astrios #2 cell sorter (Beckman Coulter) within the University of Michigan Flow Cytometry Core. Following ORF library transduction of UM-SCC-49 cells as above, flow cytometry was used to select the top 2.0 % of PD-L1 expressing cells in the transduced pool. These cells were expanded in culture and sorted again to select the top 11.0 %.
Sequencing Library Preparation
Genomic DNA (gDNA) was harvested from ORF library transduced UM-SCC-49 unsorted and PD-L1 serially sorted cell populations using the Gentra Puregene kit (Qiagen), according to the manufacturer’s protocol. For each sample, triplicate reactions with 10 μg gDNA input each were run to amplify ORF sequences using Herculase II Fusion DNA polymerase (Agilent) and primers listed in S. Table 1. These PCR products were pooled and used as template for a second PCR amplification to create sequencing-ready libraries (S. Table 1). Amplicons were purified using the QIAquick PCR purification kit (Qiagen) and submitted to the University of Michigan Advanced Genomics Core for sequencing on the Illumina MiSeq V3 platform.
Analysis of ORF Libraries
After sequencing, adapter contamination in the samples was removed using Trim_galore (v 0.4.4) and reads were mapped to the ORF reference library using SeqMap (v 1.0.13). Because SeqMap requires FASTA files as input, the read FASTQ files were converted to FASTA by extracting only the sequence information from the former. Additionally, all the FASTA files were mapped to the reverse complement of the ORF library barcodes. The barcode counts obtained by mapping the reads to the ORF library were normalized by the total read counts for each sample and the log2 fold change was calculated and compared between the unsorted and PD-L1 sorted populations. Then, the log-rank gene set lists were uploaded into GSEA (v14, Broad Institute) to identify significant overlap with Hallmark, KEGG and GO biological process pathways with false discovery rate (FDR) q-value ≤ 0.05 considered statistically significant.
Validation of Candidate Drivers of PD-L1 Expression
cDNA for the following top differentially enriched ORF constructs in the PD-L1 sorted populations were obtained (GeneCopoeia): CD300C, KLRC1, FGF6, IL17A, and NFKBIA. These did not produce transducible lentivirus. Therefore, we cloned each gene into the pCR8 vector, confirmed the correct orientation and sequence by Sanger sequencing, and subsequently transferred the expression cassettes to pLenti6/V5-DEST vector using Clonase according to the manufacturer’s protocol (Thermo Fisher Scientific). Sanger sequencing was again used to confirm correct orientation and sequence of the inserts, all of which were cloned without a stop codon such that the ORF of each construct would contain a C-terminal V5-tag. The wild type FGFR1, FGFR3 and mutant FGFR3 (S249C and D786N) constructs were acquired from Addgene.18 Transduction-ready virus was made for each candidate ORF construct and FGFR constructs within the University of Michigan Vector Core. UM-SCC-49 cells were then transduced and stably expressing populations were developed under blasticidin selection.
Western Blotting
Western blot analysis was performed as described.19 Briefly, UM-SCC cell lines in log-phase growth were treated as indicated, rinsed twice with PBS, and then lysed in mild detergent buffer (150 mM NaCl, 10% Glycerol, 1% NP40, 0.1% Triton X-100, 1 mM PIPES, 1 mM MgCl, 50 mM Tris pH 8.0) containing protease and phosphatase inhibitors (#78430, #78420, Thermo Fisher Scientific). Separation by SDS-PAGE was then performed, and primary antibodies were used to visualize target proteins, as described (S. Table 2).20
Quantitative PCR
For qPCR analysis, cells were lysed in QIAzol and RNA was isolated using the QIAgen RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. cDNA synthesis was performed with the Superscript™ VILO kit (Thermo Fisher Scientific). Amplification by qPCR was performed with the QuantiTect SYBR Green RT-qPCR kit (Qiagen) on a QuantStudio™ 5 System (Applied Biosystems) under cycling conditions recommended by the manufacturer. All qPCR primer sequences are provided in S. Table 3.
TCGA Transcriptome Analysis
Log2(RSEM+1) values for CD274 (PD-L1), candidate PD-L1 drivers CD300C, KLRC1, FGF6, IL17A, and NFKBIA, and FGF pathway members from primary tumors in the head and neck squamous cell carcinoma TCGA cohort (n = 566) were retrieved from the UCSC Genome Browser (www.xenabrowser.net). Statistical correlations between CD274 and CD300C, KLRC1, FGF6, IL17A, and NFKBIA gene expression were examined with Pearson’s correlation coefficient. Linear regression and box-and-whisker plots were created with GraphPad Prism 8 software.
Transcriptome Sequencing and Analysis
RNA sequencing was previously performed on 43 UM-SCC cell lines using Illumina stranded transcriptome library kits, as described in detail in Mann et al.16 For RNA sequencing and analysis of IFNγ- or FGF2-treated UM-SCC-14a, total RNA was isolated with the QIAgen RNeasy Mini Kit (Qiagen) and submitted to the University of Michigan Advanced Genomics Core for library generation with an Illumina stranded total RNA kit according to the manufacturer’s protocol. Samples were then paired end sequenced to >100x depth on the Illumina NovaSeq 6000 with a 300-cycle run as previously described.16 Quality of the sequencing reads was then assessed with FastQC v.0.11.5 and reads were mapped with a two-step alignment workflow of STAR v2.7.3a in which the hg19 reference genome and annotated transcriptome files were used to generate genome indexes. Then, STAR was used to guide read mapping in the second step for these genome index files.21 Samtools v1.9 was then used to extract uniquely mapped reads, which were then used to compute FPKMs with Cufflinks v2.2.1. Due to the high depth of coverage of RNA-seq data, the --max-bundle-frags argument of cufflinks was changed to 100000000 from its default value of 1000000. This modification allowed us to compute FPKMs at loci with high depth of coverage. Log2 fold change was subsequently calculated for all genes and used to order rank lists. Heatmaps were generated using MeV software version 4.9 based on log2(FPKM+1) values.22
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed on all differential rank lists with publicly available GSEA v4.03 software from the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp). Accordingly, the pre-ranked gene lists were entered based on the log2-fold change in FPKM and the gene sets selected from the Molecular Signatures Database v7.0 for analysis including the Hallmark gene sets, gene ontology (GO) gene sets and oncogenic signatures gene sets.
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Additional data are available upon request from the corresponding author.
Results
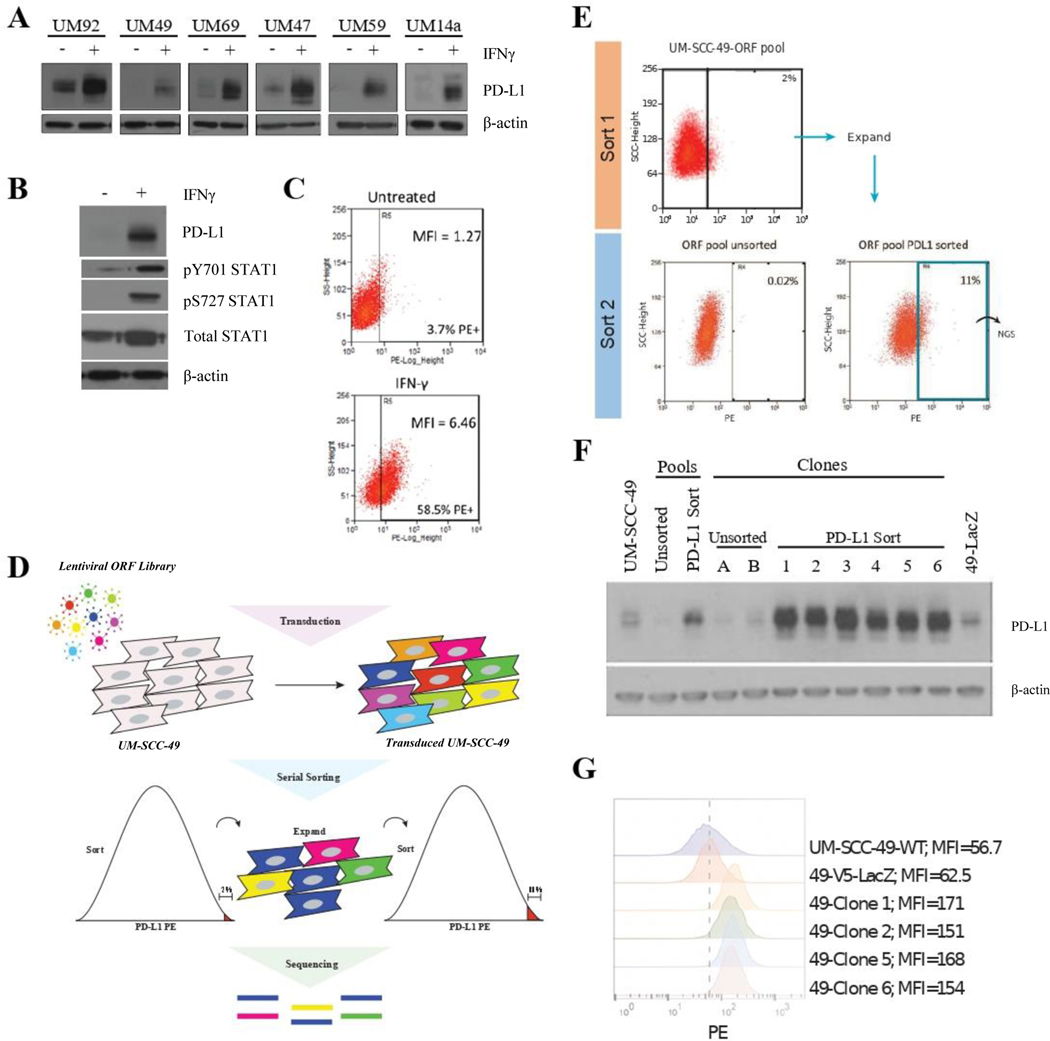
We first analyzed a panel of six UM-SCC cell lines to determine their baseline and inducible PD-L1 expression after treatment with IFNγ (Figure 1A). ImageJ intensity analysis of the bands demonstrated a mean PD-L1 induction of six-fold across the six cell lines, with UM-SCC-92 exhibiting the most modest induction of PD-L1 expression (two-fold), and UM-SCC-49 and −59 exhibiting the most significant increase (approximately 14-fold) (S. Figure 1). For our ORF screen, we chose to use the UM-SCC-49 cell line due to its degree of inducible PD-L1 expression. Previously, several groups have characterized a JAK2- and STAT1-dependent mechanism by which IFNγ drives increased CD274 transcription and PD-L1 expression.10 We confirmed that IFNγ drives increased STAT1 activation by phosphorylation in UM-SCC-49 as well (Figure 1B). We further confirmed IFNγ-inducible cell surface expression of PD-L1 in UM-SCC-49 cells by flow cytometry (Figure 1C). Thus, we concluded that detection of cell surface PD-L1 expression by flow cytometry could be used to sort individual cells to select for specific phenotypic alterations.
Figure 1. Genome-Wide ORF Screen to Identify Drivers of PD-L1 Expression in UM-SCC-49.
A, Individual cell lines were treated with 10 ng/mL IFNγ (or vehicle control) for 72 hours, followed by protein harvest and immunoblot for PD-L1. B, UM-SCC-49 cells were treated with 10 ng/mL IFNγ for 72 hours, followed by protein harvest and immunoblot for PD-L1, phosphorylated STAT1, and total STAT1. C, UM-SCC-49 cells were treated with 10 ng/mL IFNγ (or vehicle control) for 72 hours, trypsinized, and stained for flow cytometric analysis of cell surface PD-L1; MFI, mean fluorescence intensity. D, Schematic of genome-wide ORF library transduction. UM-SCC-49 cells were transduced with the 17,000 gene ORF library at an MOI of 0.3 followed by puromycin selection. Cells were then serially sorted for the top 2.0 % of PD-L1 expressing cells, followed by expansion and a subsequent sort for the top 11.0 %, of PD-L1 expressing cells in the population. PCR-amplified barcodes from the genomic DNA of PD-L1 sorted and unsorted cell populations were then sequenced on an Illumina MiSEQ platform. E, UM-SCC-49 ORF library cells were stained for cell surface expression of PD-L1 and the top 2.0 % of cells with highest PE positivity were selected and expanded in culture (top). This selected population was subjected to a second sort, this time with the top 11.0 % collected for next-generation sequencing (NGS) (bottom right). The initial ORF pool was also analyzed for PD-L1 expression for comparison (bottom left). F, PD-L1 western blot confirmed differential expression in all sorted clones relative to unsorted pools and clones. G, Subset of clones from (E) were stained for PD-L1 and analyzed by flow cytometry. Dashed line represents MFI for UM-SCC-49-LacZ-V5.
Genome-Wide ORF Screen to Identify Drivers of PD-L1 Expression
We then created stable pools of UM-SCC-49 cells transduced with a genome-wide ORF library containing 17,000 unique ORF constructs (each with in-frame C-terminal V5-tag) by lentiviral transduction and puromycin selection (Figure 1D). The population was then sorted by flow cytometry to select the top 2.0 % of cells with highest cell surface PD-L1 expression. This subpopulation was then expanded in culture and a subsequent sort for the highest 11.0 % was used to further enrich the population for ORFs that drove increased cell surface expression of PD-L1 (PD-L1high) (Figure 1E). To confirm that this approach was successful, we selected and expanded two clones from the unsorted population and seven clones from the serially sorted population. Comparison of total and cell surface PD-L1 expression between the unsorted, pooled ORF cells and clones and PD-L1 sorted clones demonstrated a substantial increase in total PD-L1 expression in all sorted clones, confirming that our strategy of enriching for this phenotype was successful (Figure 1F, G). Sanger sequencing of these clones revealed the ORF constructs listed in S. Table 4. We then isolated DNA and created Illumina MiSEQ compatible libraries from the unsorted ORF pool and PD-L1high population and sequenced libraries to a depth of > 1.5 million reads per library. Mapping barcodes to the reference identified a significant enrichment in several genes (S. Table 5) including IL17A, which has previously been shown to regulate PDL1 expression in a murine model of breast cancer,23 supporting that our overall strategy was successful.
Detailed analysis of sequencing results using gene family annotation of the overrepresented genes demonstrated a statistically significant increase in cytokines, transcription factors, and kinases in the gene set (S. Table 6), suggesting that the enrichment process identified ORFs that regulate PD-L1 expression on multiple cellular levels from transcription to autocrine stimulation. GSEA demonstrated a statistically significant 34-gene overlap with genes in the GO pathway “regulation of immune response” (FDR q-value = 1.55 × 10−7) demonstrating that the ORFs driving increased PD-L1 expression in our model were also strongly associated with cellular immune response in previously defined gene sets. Similarly, we observed enrichment within gene sets associated with “response to external stimulus,” “regulation of response to stress,” and “TNF alpha signaling driven by NFκB” (S. Table 7). Collectively, GSEA supported the role of ORFs identified in our screen in the regulation of immune response but also suggested that the ORFs drive PD-L1 expression through mechanistically diverse pathways, some of which may be NFκB-dependent.
Validation of Candidate PD-L1 Drivers
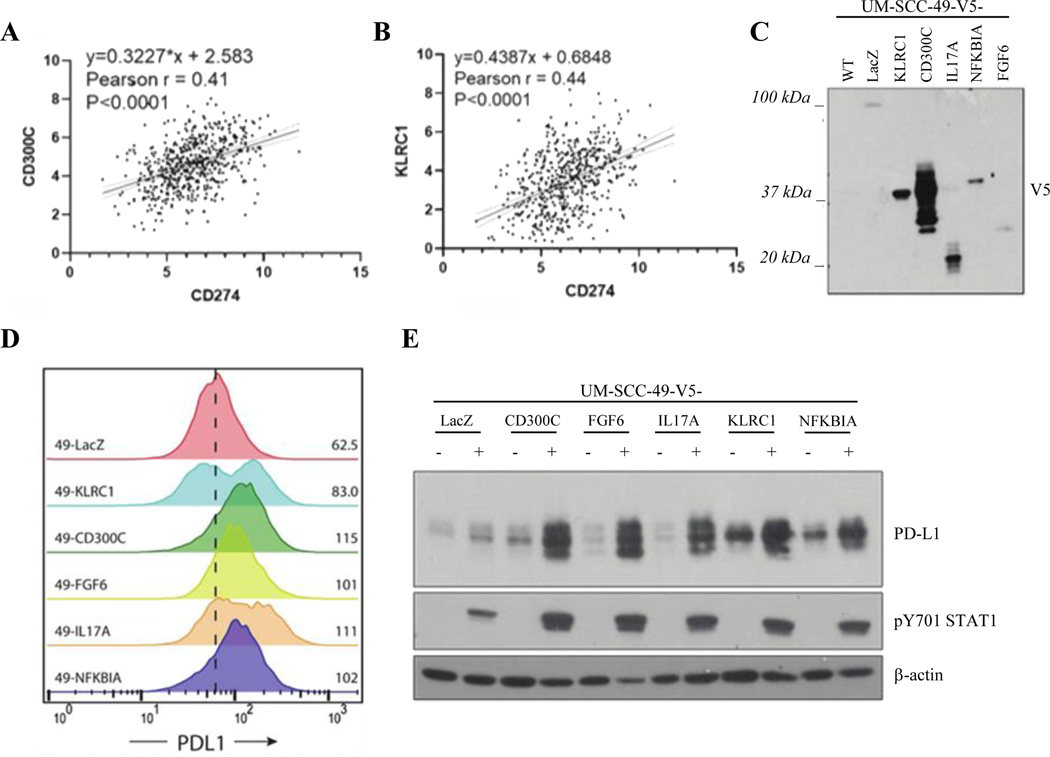
From the top overall screen hits, we selected a diverse set of ORFs for subsequent validation experiments, including CD300C, FGF6, IL17A, KLRC1, and NFKBIA. We opted to validate IL17A as it has recently been shown to modulate PD-L1 expression in murine breast and lung cancers but has not been studied in the context of HNSCC.23,24 We also interrogated HNSCC TCGA transcriptome data to examine expression of genes of interest nominated by the screen in relation to PD-L1 (Figure 2A, B; S. Figure 2). The expression of several genes nominated by our screen positively correlated with CD274 mRNA, including CD300C (Pearson’s r = 0.41, p < 0.0001) and KLRC1 (Pearson’s r = 0.44, p < 0.0001). CD300LB, an uncharacterized gene related to CD300C, also weakly correlated with PD-L1 expression in TCGA (Pearson’s r = 0.22, p < 0.0001; S. Figure 2), and was enriched in the PD-L1 sorted pool to a similar degree to CD300C. However, we focused our analysis on CD300C due to its stronger correlation with CD274 in TCGA. We also proceeded with validation of two additional genes of interest that did not correlate with PD-L1 expression in TCGA, NFKBIA and FGF6. Notably, however, FGF6 expression was undetectable in the majority of HNSCC TCGA specimens. We were intrigued by the finding of FGF6 in our dataset as a driver of PD-L1 expression due to the known tumorigenic role for FGFR signaling in HNSCC and other cancers,25 and because EGFR, which acts through similar downstream mechanisms to FGFR, was previously shown to upregulate PD-L1 expression in HNSCC.9
Figure 2. Validation of Candidate Drivers of PD-L1 Expression in UM-SCC-49.
A and B, Linear regression analysis showing positive correlation between CD300C and CD274 (A) and KLRC1 and CD274 (B) gene expression in HNSCC TCGA (Bonferroni-adjusted p values). C, Lentiviral ORF-V5 tagged expression vectors were cloned and used to overexpress each of five candidate genes and one control (LacZ) in UM-SCC-49 cells. Expression of V5 tag was confirmed in each cell line by anti-V5 western blot. Expected molecular weights of LacZ, 121 kD; KLRC1, 31 kD; CD300C, 29 kD; IL17A, 22 kD; NFKBIA, 40 kD; FGF6, 28 kD. D, Flow cytometry confirmed increase in PD-L1 MFI in each UM-SCC-49-ORF cell line. Dashed line represents MFI for UM-SCC-49-LacZ-V5. E, UM-SCC-49-ORF cells were treated with 10 ng/mL IFNγ for 72 hours (+/− notation in figure), followed by protein harvest and western blot for PD-L1 and phosphorylated STAT1 protein expression.
The ORFs of interest were each cloned into a lentiviral vector with a C-terminal V5-tag and expressed in wild-type UM-SCC-49 cells following lentiviral transduction. Western blot confirmed overexpression of the V5-tagged construct in each case (Figure 2C). Flow cytometry confirmed that each gene drove an increase in cell surface expression of PD-L1, which validated these genes as PD-L1 drivers (Figure 2D). To then determine the effect of these constructs on PD-L1 expression, we cultured cells with or without IFNγ and characterized the changes to total PD-L1 expression by western blot (Figure 2E). All constructs led to increases in IFNγ-regulated total PD-L1 expression while only some increased baseline PD-L1, suggesting that differences exist in the mechanism by which each gene regulates PD-L1 expression. For example, individual genes may modulate cell surface presentation of PD-L1, while others modulate total PD-L1 protein expression.
JAK2/STAT1 and FGF Pathway Inhibition in HNSCC Cell Lines
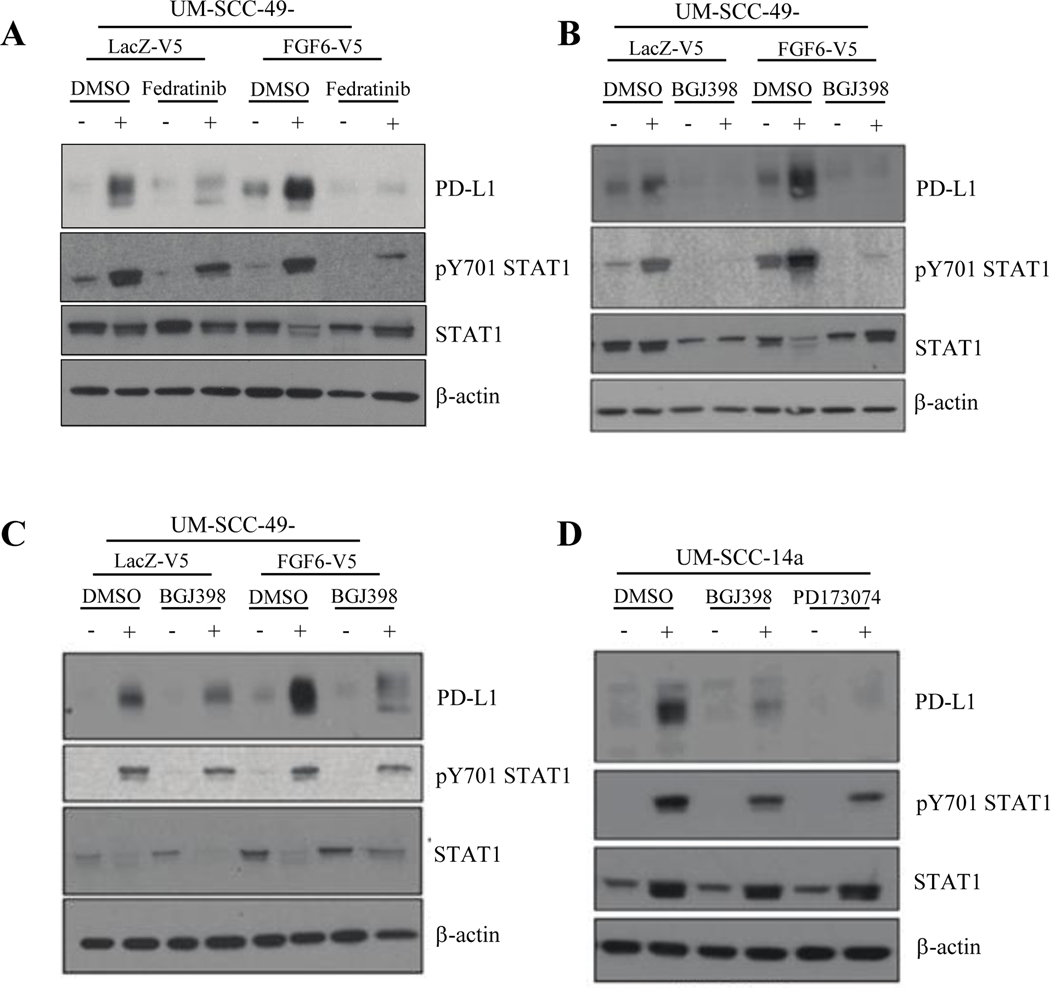
Given the immediate potential to advance FGF pathway inhibitors clinically, especially with the defined pathogenetic role of FGFR1 and FGFR3 in HNSCC,19,26 we next focused our study on the relationship of FGF pathway activity and PD-L1 expression in HNSCC models. As shown in Figure 3A, phosphorylated STAT1 in UM-SCC-49-FGF6-V5 is comparable to that in UM-SCC-49-LacZ-V5 in both untreated- and IFNγ-treated conditions. JAK2 inhibition with Fedratinib, a selective ATP-competitive inhibitor,27 blocked IFNγ-induced PD-L1 expression in FGF6-overexpressing cells and control cells, but also reduced baseline PD-L1 in UM-SCC-49-FGF6-V5, suggesting that the JAK2/STAT1 pathway may modulate FGF6-V5-induced PD-L1 expression as well. We then questioned whether inhibition of FGF receptors would alter constitutive or induced PD-L1 expression in UM-SCC-49-FGF6-V5. We noted a modest reduction in constitutively expressed total PD-L1 in these cells following treatment with either BGJ398, a small molecule inhibitor of FGFR1–328, or PD173074, an FGFR1-specific inhibitor29 (Figure 3B, C). Interestingly, we also observed that in both LacZ-expressing control cells and UM-SCC-49-FGF6-V5, inhibition of FGFRs abrogated IFNγ-induced PD-L1 expression, supporting a mechanistic crosstalk between FGFR and IFNγ pathways in HNSCC cells. Thus, we asked whether inducible PD-L1 expression could be modulated by FGFR inhibition in other models. Indeed, we observed loss of IFNγ-inducible PD-L1 expression in UM-SCC-14-a, an additional wild-type HNSCC cell line (Figure 3D).
Figure 3. JAK2/STAT1 and FGF Pathway Inhibition Abrogates PD-L1 Expression in HNSCC Cell Lines.
A, UM-SCC-49-V5-LacZ and -V5-FGF6 cells were pre-treated with JAK2 inhibitor Fedratinib (2 μM) or vehicle control (DMSO) for 6 hours, followed by addition of IFNγ (10 ng/mL) as indicated. Cells were harvested 72 hours after IFNγ treatment and PD-L1 and STAT1 expression were assessed by western blot. B, UM-SCC-49-V5-LacZ and -V5-FGF6 cells were pre-treated with FGFR1–3 inhibitor BGJ398 (3μM), FGFR1 inhibitor PD173074 (3μM) or vehicle control (DMSO) for 6 hours, followed by addition of IFNγ (10ng/mL) as indicated. Cells were harvested 72 hours after IFNγ treatment and total PD-L1 and STAT1 expression were assessed by western blot. C, UM-SCC-14a cells were pre-treated with BGJ398 (3μM), PD173074 (3μM) or vehicle control (DMSO) for 6 hours, followed by addition of IFNγ (10ng/mL) as indicated. Cells were harvested 72 hours after IFNγ treatment and total PD-L1 and STAT1 expression were assessed by western blot.
Expression and Activity of FGF/FGFR Family Proteins in HNSCC
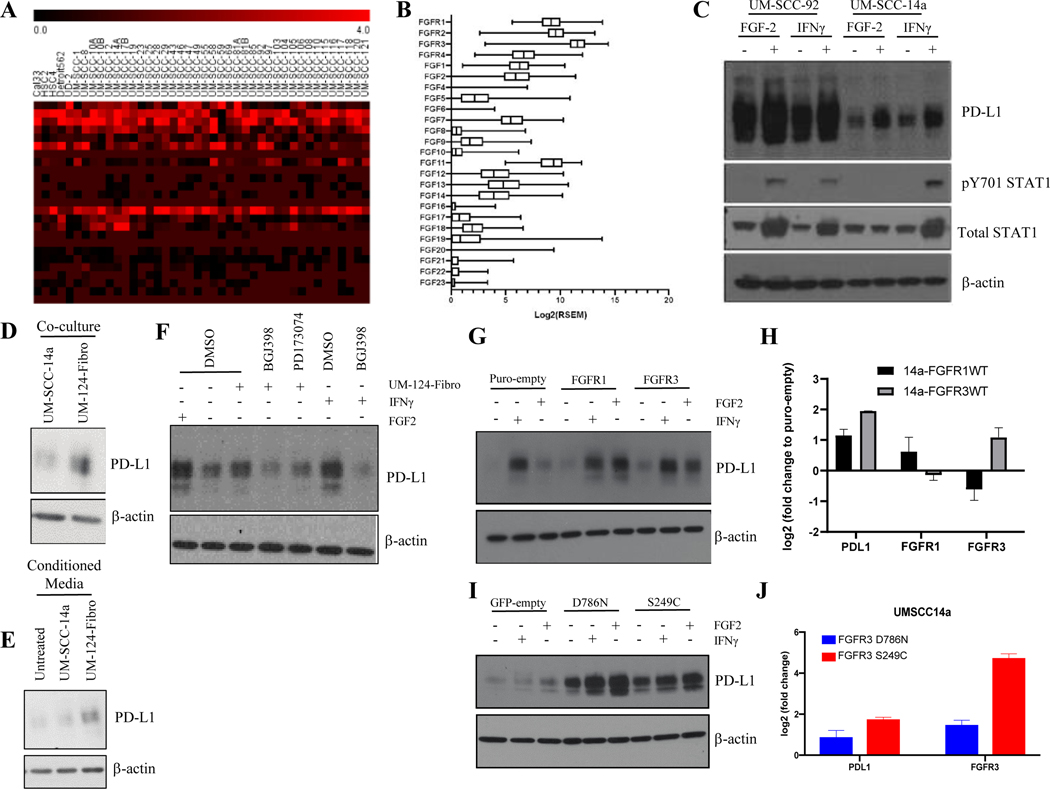
To then begin to understand the role of activated FGF receptors in PDL1 regulation, we first characterized expression of the FGF/FGFR pathway components using publicly available data. RNAseq analysis of 40 HNSCC cell lines revealed high expression of FGFR2, FGFR3, and FGF11 across nearly all cell lines, with many also exhibiting high levels of FGF2, FGFR1, and FGFR4 (Figure 4A). We next sought to compare our observations with HNSCC TCGA expression data and found that expression of FGF/FGFR family genes in our cell lines were similar to that observed in the TCGA HNSCC cohort (Figure 4B). This analysis demonstrated that FGF receptors FGFR1, FGFR2, and FGFR3 were highly expressed in HNSCC tumors, which is consistent with previous data30, and that the FGF ligands FGF2 and FGF11 were the most highly expressed FGF family genes in both our HNSCC cell lines and the TCGA cohort. We next asked whether expression of any FGF ligands or receptors correlated with PD-L1 expression and surprisingly, found no positive correlations and weak negative correlations for FGFR1 and FGFR2 (S. Figure 3). Thus, it was important to understand the role of activated FGF receptors in our models. Notably, FGF6 was not highly expressed in any of the 40 cell lines, nor in the TCGA cohort. While the highly-expressed FGF11 is not known to be secreted or interact with any FGFRs,31 FGF2 has been reported to promote growth, angiogenesis, survival, motility, and cell proliferation in numerous in vitro and in vivo HNSCC models and other cancers, and because it was highly expressed in some models and tissues it represented a clinically relevant secreted ligand to advance for functional studies.32 We treated UM-SCC cells with recombinant FGF2 to assess impact on PD-L1 expression and to understand whether a role for FGFRs in regulating PD-L1 could be recapitulated across multiple models. In UM-SCC-14a and UM-SCC-92, FGF2 upregulated PD-L1 expression even in the absence of IFNγ stimulation, further supporting the ability of FGF pathway signaling to promote PD-L1 expression in HNSCC (Figure 4C). Notably, exogenous FGF-2 also upregulated PD-L1 expression in the absence of IFNγ in cultured human immature dendritic cells (iDCs), supporting a broader role for FGF-2 signaling in mediating immunosuppressive HNSCC tumor microenvironments (S. Figure 4).
Figure 4. FGF/FGFR Family Genes are Highly Expressed and FGF Pathway Stimulation Enhances PD-L1 Expression in HNSCC.
A, Expression heat map (Log2[FPKM+1]) for FGF/FGFR family genes in 40 HNSCC cell lines. B, Box and whisker plot (Log2[RSEM]) for expression of FGF/FGFR family genes in HNSCC TCGA dataset. C, UM-SCC-14a and UM-SCC-92 cells were treated +/− FGF2 (30 ng/mL), +/− IFNγ (10 ng/mL), or vehicle control (DMSO) for 72 hours. PD-L1 expression was assessed by western blot. D, UM-SCC-14a cells were seeded in a 6-well dish with a permeable membrane insert (0.4 μm pore size) upon which either additional UM-SCC-14a cells or UM-124-Fibroblasts were seeded as indicated. Cells were cultured for 72 hours followed by protein harvest and western blot for PD-L1 expression. E, UM-SCC-14a cells were seeded in a 6-well dish. Eighteen hours after seeding, media was left unchanged (untreated) or decanted and replaced with media collected from separate UM-SCC-14a cells or UM-124-Fibroblasts. Protein was harvested 24 hours after media change and PD-L1 expression was assessed by western blot. F, UM-SCC-14a cells were pre-treated with BGJ398 (5 μM), PD173074 (5 μM), or vehicle control (DMSO) in 1 mL of media for six hours. An additional 1 mL media was then added, either UM-124-Fibro conditioned media as in (E) or standard media +/− FGF2 (30 ng/mL), +/− IFNγ (10 ng/mL), or vehicle control (DMSO). Cells were then cultured for an additional 24 hours, followed by protein harvest and western blot for PD-L1. G, UM-SCC-14a cells overexpressing empty vector control (Puro-empty), FGFR1 wild-type (WT), or FGFR3 WT were treated +/− IFNγ (10 ng/mL), FGF2 (30 ng/mL), or vehicle control (DMSO) as indicated for 24 hours, followed by protein and RNA harvest. PD-L1 expression was assessed by western blot (G) and qPCR (H). RNA expression was assessed by qPCR in untreated samples only. I, UM-SCC-14a cells overexpressing empty vector control (GFP-empty), GFP-FGFR3-S249C mutant, or GFP-FGFR3-D786N mutant were treated +/− IFNγ (10 ng/mL), FGF2 (30 ng/mL), or vehicle control (DMSO) as indicated for 24 hours, followed by protein and RNA harvest. PD-L1 expression was assessed by western blot (I) and qPCR (J). RNA expression was assessed by qPCR in untreated samples only.
Given the ability of UM-SCC cell lines to upregulate PD-L1 in response to FGF2 alone, we sought to recapitulate unique phenotypes with accentuated FGF pathway activity. We co-cultured UM-SCC-14a cells in a dish with either more UM-SCC-14a cells or UM-124-Fibroblasts (UM-124-Fibro) seeded onto a 0.4 μm membrane and discovered a modest increase in UM-SCC-14a PD-L1 protein when cells were grown in the presence of fibroblasts (Figure 4D). We further found that UM-124-Fibro conditioned media was sufficient to upregulate PD-L1 expression in UM-SCC-14a (Figure 4E). Pretreatment of UM-SCC-14a with FGFR inhibitors attenuated PD-L1 upregulation in the presence of UM-124-Fibro-conditioned media (Figure 4F).
Next, we examined whether FGFR overexpression or expression of recurrent FGFR hotspot mutations identified in the HNSCC TCGA project that are known to drive FGFR activation could alter PD-L1 expression. We overexpressed FGFR1 or FGFR3 in UM-SCC-14a (Figure 4G, H) and UM-SCC-59 (S. Fig 5) and observed higher constitutive, IFNγ-induced, and FGF2-induced PD-L1 expression. We then overexpressed FGFR3 constructs bearing either of two activating mutations: S249C, observed in 4/6 tumors from TCGA with FGFR3 point mutations,30 and D786N, a predicted gain-of-function mutation.33 Similarly, dramatic increases in constitutive and inducible PD-L1 expression were observed (Figure 4I, J), although the specific phenotypic impact of these mutations has yet to be determined.
IFNγ and FGF2 Activate Distinct but Overlapping Signaling Pathways to Upregulate PD-L1
To better understand the interplay between IFNγ and FGF signaling pathways, we examined expression of various downstream targets of FGFR and IFNGR activation by qPCR in UM-SCC-14a. We observed that RNA expression of genes regulated by IFNγ in other model systems, such as STAT1, IRF9, SOCS1, and SOCS333 were indeed induced by IFNγ but not FGF2 (Figure 5A, B). Instead, FGF2 induced a separate transcriptional program that included upregulation of CXCL8, IRF7, IL6, and SPRY. JAK2 inhibition with Fedratinib reduced RNA expression of both the IFNγ- and FGF2-regulated gene sets, while FGFR1–3 inhibition with BGJ398 reduced RNA expression of only the FGF2-regulated gene set. CD274 gene expression, however, is induced by IFNγ and this induction is reduced in cells pre-treated with BGJ398, consistent with protein data from above. Also of interest, despite the known ability of FGFRs to activate JAK/STAT signaling,25 only IFNγ (not FGF2) induced STAT1 expression in this setting (Figure 5A, B). To further explore proteins downstream of FGF-2 that mediate PD-L1 upregulation, we treated UM-SCC-14a cells with MK-2206, a selective inhibitor of Akt 1/2/3.34 MK-2206 did not appreciably alter PD-L1 expression in the presence of IFNγ or FGF2, suggesting an alternative downstream mechanism predominates (S. Figure 6).
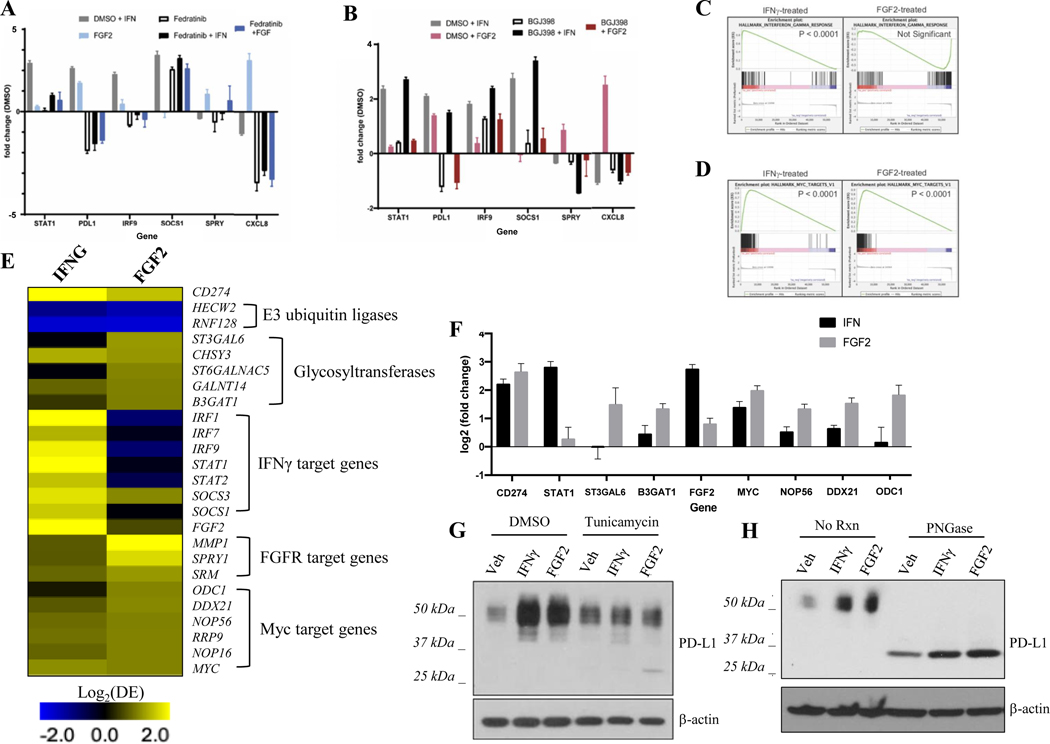
Figure 5. Analysis of Mediators Downstream of IFNγ and FGF2 that Upregulate PD-L1 in HNSCC.
UM-SCC-14a cells were pre-treated with Fedratinib (5 μM) (A), BGJ398 (5 μM) (B) or vehicle control (DMSO) for three hours as indicated. Then, IFNγ (10 ng/mL) or FGF2 (30 ng/mL) was added for an additional three-hour treatment. RNA was harvested and expression of genes of interest analyzed by qPCR. C, GSEA of RNAseq data showed significant upregulation of IFNγ response gene set in UM-SCC-14a cells after 24-hour treatment with IFNγ but not FGF2. D, GSEA of RNAseq data showed significant upregulation of MYC family genes in UM-SCC-14a cells after 24-hour treatment with IFNγ or FGF2. E, Differential expression (Log2[DE]) of target genes in IFNγ- or FGF2-treated (24 hours), relative to untreated, UM-SCC-14a cells shows distinct but overlapping transcriptional programs. F, qPCR validation of MYC gene family upregulation in IFNγ- and FGF2-treated (24 hours), relative to untreated, UM-SCC-14a cells. G, UM-SCC-14a cells were pre-treated with tunicamycin (50 ng/mL) or vehicle control (DMSO) for six hours. Then, IFNγ (10 ng/mL) or FGF2 (30 ng/mL) was added for an additional three-hour treatment. Protein was harvested and PD-L1 expression was assessed by western blot. H, UM-SCC-14a cells were treated with IFNγ (10 ng/mL), FGF2 (30 ng/mL), or vehicle control (DMSO) for 72 hours. Protein was harvested and lysate was treated with PNGase or control (RIPA buffer). PD-L1 expression was assessed by western blot.
We next performed RNA-seq on UM-SCC-14a cells stimulated with either IFNγ or FGF2 for 24 hours. GSEA of the rank lists for INFγ-treated and FGF2-treated UM-SCC-14a cells identified 168 and 121 positively correlated gene sets, respectively, and 1 and 3 negatively correlated genes sets, respectively (FDR q-value < 0.01, S. Table 8.). Importantly, the top upregulated gene set in the IFNγ-treated cells was the Hallmark_Interferon_gamma_response gene set, while there was no significant enrichment of this signature in the FGF2-treated cells (Figure 5C), confirming that FGF2-mediated PD-L1 regulation is distinct from classical IFNγ signaling pathways. Unexpectedly, we observed that the top upregulated gene set in the FGF2-treated cells, Hallmark_MYC_targets_V1, was also significantly enriched in IFNγ-treated cells (FDR q-value < 0.0001, enrichment rank 5th; Figure 5D), suggesting that IFNγ regulated both a unique signature and the most significant components of the FGF2-regulated signature. Consistent with this observation, analysis of the differentially expressed genes demonstrated that IFNγ induced a 2.77-Log2 fold upregulation of FGF2 expression (Figure 5E), while FGF2 did not significantly alter IFNG expression. This suggests that IFNγ may drive a feed-forward loop through FGF2 that acts to maintain PD-L1 cell surface expression. Upregulation of MYC and its target genes ODC1, NOP56, and DDX21 in response to IFNγ and FGF2 was validated by qPCR (Figure 5F).
FGF Signaling May Promote Glycosylation and Stabilization of Cell Surface PD-L1 by Concomitant Upregulation of Glycosyltransferases in HNSCC
Because cell surface PD-L1 is stabilized by glycosylation in other model systems, we assessed whether PD-L1 is similarly glycosylated in UM-SCC cell lines.35,36 We observed PD-L1 protein as a 40–50 kD smear on western blots. When we pre-treated UM-SCC-14a cells with tunicamycin, an inhibitor of N-linked glycosylation,37 we observed a dramatic decrease in IFNγ- and FGF2-stimulated PD-L1 expression at 40–50 kD along with a concomitant accumulation of non-glycosylated PD-L1 at 33 kD.35 (Figure 5G). Furthermore, treatment of protein lysates from IFNγ- or FGF2-treated UM-SCC-14a cells with PNGase, an enzyme catalyzing N-linked de-glycosylation, abolished detectable PD-L1 at 40–50 kD with all PD-L1 protein migrating to 33 kD (Figure 5H). Recent studies have described a role for EGFR in stabilizing PD-L1 via regulation of glycosylation and degradation processes in breast cancer.35,36 EGFR signaling can induce upregulation of B3GNT3, an enzyme that catalyzes PD-L1 glycosylation and may also promote inactivation of GSK3β, a kinase responsible for targeting non-glycosylated PDL1 for E3 ligase mediated degradation.35,36 We therefore speculated that FGFR signaling may act through analogous mechanisms to stabilize PD-L1 in HNSCC. We first confirmed that EGFR inhibition by the small molecule inhibitor gefitinib resulted in loss of the inhibitory serine 9 phosphorylation of GSK3β, but FGFR inhibition had no effect (S. Fig 7). We therefore hypothesized that FGF signaling may activate glycosylation machinery, thereby protecting PD-L1 from targeting by GSK3β, and interrogated our RNA-seq dataset to identify differentially regulated glycosyltransferases. We found that a small subset of glycosyltransferases was indeed modestly upregulated in response to FGF2 and confirmed these findings by qPCR (Figure 5E, F). Thus, one hypothesis for future research focused on the mechanism of action is that FGF signaling may concomitantly upregulate glycosyltransferases to stabilize cell surface PD-L1 expression in HNSCC.
Discussion
HNSCC is a strongly immunosuppressive malignancy; understanding the molecular mechanisms that drive immune escape and innate and acquired resistance to immunotherapies is critical to improving outcomes for afflicted patients. Here, we are the first to perform a genome-wide ORF screen to identify novel and potentially targetable PD-L1 regulatory mechanisms in HNSCC. Overall, our approach identified 335 enriched genes as candidate drivers of PD-L1 expression in HNSCC, several of which we independently validated. Supporting the validity of our screening approach, several of our candidate drivers have been shown to regulate PD-L1 expression in other cancers. For example, inhibition of IL17A in estrogen receptor (ER)-negative breast cancer models has been shown to abrogate PD-L1 expression.23 Many HNSCC tumors are characterized by high tumor-infiltrating lymphocyte content.38–40 Several reports have reported significant enrichment of Th17-cells specifically, which express high levels of IL17A, in both pre-malignant oral lesions41 and invasive tumors,39,40 supporting a critical role for IL17A-mediated PD-L1 upregulation in these settings.
Our screen also identified several genes closely linked to pathways known to regulate PD-L1 expression in other cancers, including the NFKB (NFKBIA) and FGF/FGFR signaling pathways (FGF6). In fact, while NFKB signaling is one of the most well characterized regulators of PD-L1 expression thus far,42,43 NFKBIA itself has not been previously implicated in PD-L1 regulation.42,43 In the HNSCC TCGA project, NFKBIA is amplified in 8/517 (1.5%) cases,30 and a recent independent report also demonstrated a significant enrichment of NFKBIA alterations in human papillomavirus (HPV)-positive oropharyngeal cancers (12/149, 8% of tumors).44 Thus, NFKBIA activity is a potentially useful biomarker of tumor immunosuppressive phenotypes and response to PD-1/PD-L1 axis blockade in HNSCC requiring further study.
We and others have previously shown FGFR signaling to be a driver of certain HNSCC tumors.19, 45–47 Further, FGFR1 is overexpressed in > 80% of HPV-positive HNSCCs and 75% of HPV- HNSCCs, supporting a critical role for FGF signaling in a substantial subset of these cancers.46 Similarly, FGFR signaling has been shown to be upregulated in HNSCC cancer stem cell (CSC) populations in response to platinum-based therapy in vitro.47 When considering our data showing the strong upregulation of PD-L1 expression by activated FGF signaling, these findings support the hypothesis that cisplatin-mediated adaptive FGFR1 upregulation may activate the PD-L1 checkpoint in the CSC population in some tumors, thereby preventing their recognition and control by tumor-infiltrating lymphocytes. This mechanism may have important implications for patients with R/M HNSCC treated with immune checkpoint blockade after failing previous platinum-based therapy.48,49
The observations herein are of particular interest given recent focus on the role of the HNSCC tumor microenvironment in promoting tumorigenesis, immune evasion, and metastasis. Specifically, our findings that recombinant FGF, as well as secreted signals from patient-derived fibroblasts, can significantly upregulate PD-L1 expression on malignant cells support a novel role for cancer-associated fibroblasts (CAFs) in directly facilitating tumor immune escape. Additionally, we show that FGF-2 is capable of potent upregulation of PD-L1 on iDCs, independent of IFNγ. In the HNSCC tumor microenvironment, upregulation of inhibitory immune checkpoints in APCs such as iDCs and myeloid-derived suppressor cells (MDSCs) leads to particularly potent immunosuppression and poor response to immune checkpoint inhibition.50,51 Thus, our data supports a more global role for intercellular FGF-2 signaling in the HNSCC tumor microenvironment.
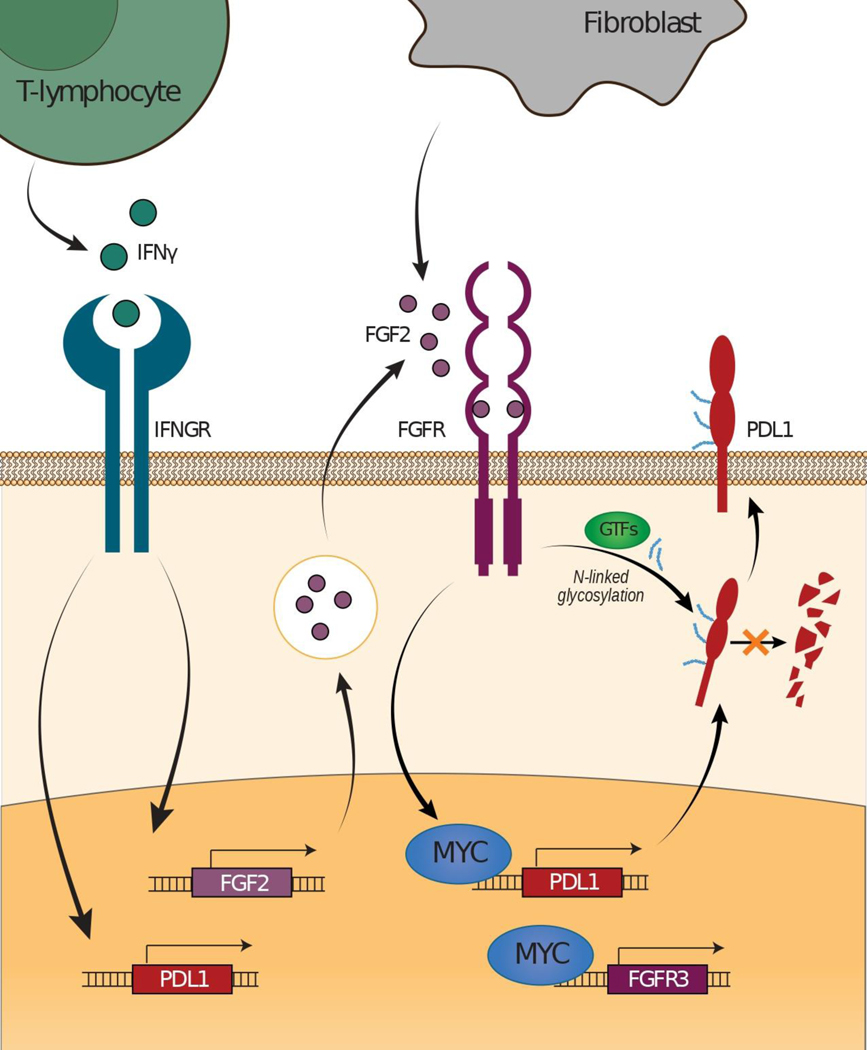
We also nominate multiple downstream effectors that may mediate PD-L1 regulation in response to FGF signaling, including MYC signaling. MYC is a known transcriptional regulator of PD-L1,52 correlates with PD-L1 protein expression in lung cancers,53 and regulates PD-L1 protein translation in murine liver cancers.54 Furthermore, several reports describe a role for aberrant FGFR1 and FGFR3 activation in upregulating MYC25,55 and suggest that MYC may be a key regulator of FGFR-targeted therapeutic response in FGFR-altered cancers.56 In fact, Mahe et al57 observed a positive feedback loop in bladder cancers in which MYC mRNA was upregulated and MYC protein stabilized downstream of altered FGFR3. In turn, the accumulated MYC bound to enhancers upstream of FGFR3, directly leading to its upregulation. Given these data and our observation that PD-L1, MYC, and other MYC targets are upregulated in FGF2-stimulated HNSCC cells, we propose a novel mechanism whereby FGFR signaling, activated by either genetic aberrations or exogenous signals from the tumor microenvironment, upregulates PD-L1 protein in a MYC-dependent manner (Figure 6). Furthermore, as we also observed that IFNγ stimulation leads to upregulation of FGF2 mRNA, we postulate that IFNγ may induce PD-L1 expression rapidly via JAK/STAT but may also promote a paracrine signaling mechanism that serves to sustain PD-L1 expression via FGFR activation (Figure 6). Thus, our data support multiple novel mechanisms involving both extracellular signals and genetic aberrations through which PDL1-mediated immune escape could occur in HNSCC.
Figure 6.
Hypothesized PD-L1 Regulatory Mechanisms Mediated by FGF/FGFR Pathway Signaling in HNSCC.
Further studies will be needed to fully understand the impact of FGF signaling on upregulation of glycosylation machinery that may stabilize PD-L1 expression on the cell surface and render PD-L1 immune checkpoint blockade less effective.35,36 Our preliminary data is only hypothesis-generating in this regard. Ultimately, our study suggests a novel role for FGF/FGFR signaling beyond its known function in promoting growth and proliferation in HNSCC.25 Thus, we believe this work could have important translational impact, particularly for tumors harboring activating FGFR alterations. Further, our findings rationalize exploration of a role for FGFRs and FGF signaling in the tumor microenvironment as a biomarker of response to PD-L1 blockade, as well as assessing FGFR-targeted therapies in conjunction with traditional immunotherapies.
Supplementary Material
S. Figure 1. ImageJ intensity analysis of relative PD-L1 induction in panel of UM-SCC cell lines.
S. Figure 2. Linear regression analysis showing positive correlation between FGF2 and CD274 (left), NFKBIA and CD274 (middle), and IL17A and CD274 (right) gene expression in HNSCC TCGA (Bonferroni-adjusted p values shown).
S. Figure 3. Linear regression analysis showing correlations between FGF/FGFR and CD274 gene expression in HNSCC TCGA (Bonferroni-adjusted p values shown).
S. Figure 4. FGF-2 upregulates PD-L1 expression in human immature dendritic cells (iDCs). iDCs were treated +/− FGF2 (30 ng/mL), +/− IFNγ (10 ng/mL), or vehicle control (DMSO), as indicated. Cells were harvested at indicated timepoints, lysed, and PD-L1 expression was assessed by western blot.
S. Figure 5. PD-L1 expression is upregulated in UM-SCC-59 cells overexpressing mutant FGFR3- and treated +/− IFNγ and +/− FGF2. Cells overexpressing empty vector control (Puro-empty or GFP empty), FGFR1 wild-type (WT), FGFR3 WT, FGFR3 D786N or FGFR3 S249C were treated +/− IFNγ (10 ng/mL), FGF2 (30 ng/mL), or vehicle control (DMSO) as indicated for 24 hours, followed by protein harvest. PD-L1 expression was assessed by western blot.
S. Figure 6. The Akt 1/2/3 inhibitor MK-2206 fails to reverse PD-L1 upregulation stimulated by FGF2 in UM-SCC-14a. UM-SCC-14a cells were treated +/− MK-2206 (1 μM), FGF2 (30 ng/mL), or vehicle control (DMSO) for 24, 48, or 72 hours as indicated. Then, protein was harvested and total AKT, pS473 AKT, and PD-L1 expression was assessed by western blot.
S. Figure 7. Inhibition of EGFR, but not FGFR, leads to loss of inhibitory serine 9 phosphorylation of GSK3ꞵ in UM-SCC-14a. UM-SCC-14a cells were treated +/− vehicle control (DMSO), Fedratinib (JAK2i; 5 μM), BGJ398 (FGFR1/2/3i; 5 μM), gefitinib (EGFRi; 5 μM), EGF (10 ng/mL), IFNγ (10 ng/mL), or FGF2 (30 ng/mL) for 18h. Protein was then harvested and protein targets of interest were assessed by western blot.
Contributor Information
Jacqueline E Mann, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA; Department of Pathology, University of Michigan, Ann Arbor, MI 41809, USA..
Joshua D. Smith, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Aditi Kulkarni, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA..
Susan K. Foltin, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Erin B. Scheftz, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Isabel R. Murray, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Elizabeth Genterblum-Miller, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA; Program in Cellular and Molecular Biology, University of Michigan, Ann Arbor, MI 41809, USA..
Collin V. Brummel, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Apurva Bhangale, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA..
Rebecca C. Holesli, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
J. Chad Brenner, Department of Otolaryngology - Head and Neck Surgery, University of Michigan, Ann Arbor, MI 48109, USA; Program in Cellular and Molecular Biology, University of Michigan, Ann Arbor, MI 41809, USA; Department of Pharmacology, University of Michigan, Ann Arbor, MI 48109, USA; Rogel Cancer Center, University of Michigan, Ann Arbor, MI 48109, USA..
References
- 1.Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncotarget 2010;15:994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2023;9(1):4. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Li Y, Forastiere A. Prognostic factors and long-term survivorship in patients with recurrent or metastatic carcinoma of the head and neck. Cancer 2004;101(10):2222–29. [DOI] [PubMed] [Google Scholar]
- 4.Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, de Castro G Jr, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;10212:23–9. [DOI] [PubMed] [Google Scholar]
- 5.Sacco AG, Chen R, Worden FP, Wong DJL, Adkins D, Swiecicki, et al. Pembrolizumab plus cetuximab in patients with recurrent or metastatic head and neck squamous cell carcinoma: an open-label, multi-arm, non-randomised, multicentre, phase 2 trial. Lancet Oncol 2021;22(6):883–92. [DOI] [PubMed] [Google Scholar]
- 6.Cohen EEW, Nabell L, Wong DJ, Day T, Daniels GA, Milhem M, et al. Intralesional SD-101 in combination with pembrolizumab in anti-PD-1 treatment-naïve head and neck squamous cell carcinoma: Results from a multicenter, phase II trial. Clin Cancer Res 2022;28(6):1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 8.Cortes J, Rugo HS, Cescon DW, Im S, Yusof MM, Gallardo C, et al. Pembrolizumab plus chemotherapy in advanced triple-negative breast cancer. N Engl J Med 2022;387:217–26. [DOI] [PubMed] [Google Scholar]
- 9.Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, et al. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res 2016;76(5):1031–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bu LL, Yu GT, Wu L, Mao L, Deng WW, Liu JF, et al. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J Dent Res 2017;96(9):1027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steuer CE, Griffith CC, Nannapaneni S, Patel MR, Liu Y, Magliocca KR, et al. A correlative analysis of PD-L1, PD-1, PD-L2, EGFR, HER2, and HER3 expression in oropharyngeal squamous cell carcinoma. Mol Cancer Ther 2018;17(3):710–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katti A, Diaz BJ, Caragine CM, Sanjana NE, Dow LE. CRISPR in cancer biology and therapy. Nat Rev Cancer 2022;22:259–79. [DOI] [PubMed] [Google Scholar]
- 13.Chan Y, Lu Y, Wu J, Zhang C, Tan H, Bian Z, et al. CRISPR-Cas9 library screening approach for anti-cancer drug discover: overview and perspectives. Theranostics 2022;12(7):3329–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2010;32(4):417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig ML, Kulkarni A, Birkeland AC, Michmerhuizen NL, Foltin SK, Mann JE, et al. The genomic landscape of UM-SCC oral cavity squamous cell carcinoma cell lines. Oral Oncol 2018;87:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann JE, Kulkarni A, Birkeland AC, Kafelghazal J, Eisenberg J, Jewell BM, et al. The molecular landscape of the University of Michigan laryngeal squamous cell carcinoma cell line panel. Head Neck 2019;41(9):3114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343(6166):84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 2018;33(3):450–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillman BN, Yanik M, Birkeland AC, Liu C, Hovelson DH, Cani AK, et al. Fibroblast growth factor family aberrations as a putative driver of head and neck squamous cell carcinoma in an epidemiologically low-risk patient as defined by targeted sequencing. Head Neck 2016;38Suppl1:E1646–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michmerhuizen NL, Leonard E, Kulkarni A, Brenner JC. Differential compensation mechanisms define resistance to PI3K inhibitors in PIK3CA amplified HNSCC. Otorhinolaryngol Head Neck Surg 2016;1(2):44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003;34(2):374–8. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Chen C, Li D, Liu M, Lv Z, Ji Y, et al. Targeting of interleukin (IL)-17A inhibits PDL1 expression in tumor cells and induces anticancer immunity in an estrogen receptor-negative murine model of breast cancer. Oncotarget 2017;8(5):7614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbay EA, Koyoma S, Liu Y, Dries R, Bufe LE, Silkes M, et al. Interleukin-17A promotes lung tumor progression through neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J Thorac Oncol 2017;12(8):1268–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babina IS, Turner NC. Advances and challenges in targeting FGFR signaling in cancer. Nat Rev Cancer 2017;17(5):318–32. [DOI] [PubMed] [Google Scholar]
- 26.Koole K, Brunen D, van Kempen PMW, Noorlag R, de Bree R, et al. FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res 2016;22(15):3884–93. [DOI] [PubMed] [Google Scholar]
- 27.Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, et al. Efficacy of Fedratinib, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell 2008;13(4):311–20. [DOI] [PubMed] [Google Scholar]
- 28.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}−1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem 2011;54(20):7066–83. [DOI] [PubMed] [Google Scholar]
- 29.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J 1998;17(20):5896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, et al. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem 2003;278(36):34226–36. [DOI] [PubMed] [Google Scholar]
- 32.Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res 2011;17(15):5016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Bio Chem 1998;273(52):35056–62. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y, Ren X, Zhang Y, Patel R, Sharma A, Wu H, et al. eEF-2 kinase dictates cross-talk between autophagy and apoptosis induced by Akt inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res 71(7):2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016;7:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell 2018;33(2):187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer R, Weber TK, Arceci R, Ramchurren N, Kastrinakis WV, et al. Inhibition of N-linked glycosylation of P-glycoprotein by tunicamycin results in a reduced multidrug resistance phenotype. Br J Cancer 1995;71(4):670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann JE, Smith JD, Birkeland AC, Bellile E, Swiecicki P, Mierzwa M, et al. Analysis of CD103 resident memory T-cell content in recurrent laryngeal squamous cell carcinoma. Cancer Immunol Immunother 2019;68(2):213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoesli R, Birkeland AC, Rosko AJ, Issa M, Chow KL, Michmerhuizen NL, et al. Proportion of CD4 and CD8 tumor infiltrating lymphocytes predicts survival in persistent/recurrent laryngeal squamous cell carcinoma. Oral Oncol 2018;77:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann J, Hoesli R, Michmerhuizen N, Devenport SN, Ludwig ML, Vandenberg TR, et al. Surveilling the potential for precision medicine-driven PD-1/PD-L1 targeted therapy in HNSCC. J Cancer 2017;8(3):332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Costa AA, Schuyler CA, Walker DD, Young MRI. Characterization of the evolution of immune phenotype during the development and progression of squamous cell carcinoma of the head and neck. Cancer Immunol Immunother 2012;61(6):927–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim S, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquination and stabilization of PD-L1 by CSN5. Cancer Cell 2016;30(6):925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS One 2015;10(4):e0123410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillison ML, Akagi K, Xiao W, Jiang B, Pickard RKL, Swanson BJ, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res 2019;9(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res 2011;17(15):5016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubot C, Bernard V, Sablin MP, Vacher S, Chemlali W, Schnitzler A, et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur J Cancer 2018;91:47–55. [DOI] [PubMed] [Google Scholar]
- 47.McDermott SC, Rodriguez-Ramirez C, McDermott SP, Wicha MS, Nor JE. FGFR signaling regulates resistance of head and neck cancer stem cells to cisplatin. Oncotarget 2018;9(38):25148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen EEW, Soulieres D, Le Tourneau C, Dinis J, Licitra L, Ahn MJ, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head and neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 2019;393(10167):156–67. [DOI] [PubMed] [Google Scholar]
- 49.Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous cell carcinoma of the head and neck. N Engl J Med 2016;375(19):1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis RJ, Moore EC, Clavijo PE, Friedman J, Cash H, Chen Z, et al. Anti-PD-L1 efficacy can be enhanced by inhibition of myeloid-derived suppressor cells with a selective inhibitor of PI3Kδ/γ. Cancer Res 2017;77(10):607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang X, Fan H, Tang Y, Wang S, Cao M, Wang H, et al. Myeloid derived suppressor cells contribute to the malignant progression of oral squamous cell carcinoma. PLoS One 2020;15(2):e0229089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352(6282):227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim EY, Kim A, Kim SK, Chang YS. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer 2017;110:63–67. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Poggio M, Jin HY, Shi Z, Forester CM, Wang Y, et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med 2019;25(2):301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu T, Wu Q, Chong Y, Qin H, Poole CJ, van Riggelen J, et al. FGFR1 fusion kinase regulation of MYC expression drives development of stem cell leukemia/lymphoma syndrome. Leukemia 2018;32(11):2363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Ai J, Shen A, Chen Y, Wang X, Peng X, et al. c-Myc alteration determines the therapeutic response to FGFR inhibitors. Clin Cancer Res 2017;23(4):974–84. [DOI] [PubMed] [Google Scholar]
- 57.Mahe M, Dufour F, Neyret-Kahn H, Moreno-Vega A, Beraud C, Shi M, et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol Med 2018;10(4):e8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Figure 1. ImageJ intensity analysis of relative PD-L1 induction in panel of UM-SCC cell lines.
S. Figure 2. Linear regression analysis showing positive correlation between FGF2 and CD274 (left), NFKBIA and CD274 (middle), and IL17A and CD274 (right) gene expression in HNSCC TCGA (Bonferroni-adjusted p values shown).
S. Figure 3. Linear regression analysis showing correlations between FGF/FGFR and CD274 gene expression in HNSCC TCGA (Bonferroni-adjusted p values shown).
S. Figure 4. FGF-2 upregulates PD-L1 expression in human immature dendritic cells (iDCs). iDCs were treated +/− FGF2 (30 ng/mL), +/− IFNγ (10 ng/mL), or vehicle control (DMSO), as indicated. Cells were harvested at indicated timepoints, lysed, and PD-L1 expression was assessed by western blot.
S. Figure 5. PD-L1 expression is upregulated in UM-SCC-59 cells overexpressing mutant FGFR3- and treated +/− IFNγ and +/− FGF2. Cells overexpressing empty vector control (Puro-empty or GFP empty), FGFR1 wild-type (WT), FGFR3 WT, FGFR3 D786N or FGFR3 S249C were treated +/− IFNγ (10 ng/mL), FGF2 (30 ng/mL), or vehicle control (DMSO) as indicated for 24 hours, followed by protein harvest. PD-L1 expression was assessed by western blot.
S. Figure 6. The Akt 1/2/3 inhibitor MK-2206 fails to reverse PD-L1 upregulation stimulated by FGF2 in UM-SCC-14a. UM-SCC-14a cells were treated +/− MK-2206 (1 μM), FGF2 (30 ng/mL), or vehicle control (DMSO) for 24, 48, or 72 hours as indicated. Then, protein was harvested and total AKT, pS473 AKT, and PD-L1 expression was assessed by western blot.
S. Figure 7. Inhibition of EGFR, but not FGFR, leads to loss of inhibitory serine 9 phosphorylation of GSK3ꞵ in UM-SCC-14a. UM-SCC-14a cells were treated +/− vehicle control (DMSO), Fedratinib (JAK2i; 5 μM), BGJ398 (FGFR1/2/3i; 5 μM), gefitinib (EGFRi; 5 μM), EGF (10 ng/mL), IFNγ (10 ng/mL), or FGF2 (30 ng/mL) for 18h. Protein was then harvested and protein targets of interest were assessed by western blot.
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Additional data are available upon request from the corresponding author.