Abstract
Patients with human T-cell leukemia virus type 1 (HTLV-1)-associated myelopathy/tropical spastic paraparesis (HAM/TSP) typically have a high HTLV-1 proviral load in peripheral blood mononuclear cells and abundant, activated HTLV-1-specific cytotoxic T lymphocytes (CTLs). No effective treatment for HAM/TSP has been described so far. We report a 10-fold reduction in viral DNA for five patients with HAM/TSP during treatment with the reverse transcriptase inhibitor lamivudine. In one patient with recent-onset HAM/TSP, the reduction in viral DNA was associated with a fall in the frequency of CTLs specific to two peptides in the immunodominant viral antigen Tax. The half-life of peripheral blood mononuclear cell populations was estimated from changes in viral DNA copy number, CTL frequency, reduction in CD25 expression, and the loss of dicentric chromosomes following radiation-induced damage. Each of these four different techniques indicated a cellular half-life of approximately 3 days consistent with continuous lymphocyte replication and destruction. These results indicate that viral replication through reverse transcription significantly contributes to the maintenance of HTLV-1 viral DNA load. The relative contribution of proliferation versus replication may vary between infected people.
The human T-cell leukemia virus type 1 (HTLV-1), an endemic retrovirus, causes lifelong infection. The majority of persons infected are asymptomatic carriers. In a minority, HTLV-1 infection causes inflammatory diseases characterized by lymphocytic infiltration, of which HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) is the most notable and causes significant morbidity (7, 20). HTLV-1 also causes adult T-cell leukemia/lymphoma, an aggressive condition resistant to chemotherapy (9, 30). The proviral load of HTLV-1 in peripheral blood mononuclear cells (PBMCs) has been estimated by diverse techniques: mean proviral load is approximately 10 copies/100 PBMCs in patients with HAM/TSP and 10-fold less in asymptomatic carriers (16). HTLV-1 proviral load might be maintained either by lymphocyte proliferation, with duplication of the HTLV-1 genome at every cell division, or by classical retroviral replication via reverse transcription. The relative contribution of these two replication pathways to the total proviral load has not been determined. However, multiple clonal expansions of HTLV-1-infected lymphocytes have been demonstrated for patients with HAM/TSP and for asymptomatic carriers (2). Consequently, Wattel et al. have hypothesized that lymphocyte proliferation maintains HTLV-1 proviral load and that the absence of reverse transcription after a few initial rounds of replication might explain the relative conservation of the HTLV-1 genome compared to the other human retroviruses (32).
However, a number of observations suggest that viral antigen expression is continuous and that ongoing viral replication occurs in at least a subpopulation of infected cells. HTLV-1-infected subjects have high titers of antibody to structural gene products throughout the course of the infection (5). HTLV-1 mRNA is detected in peripheral blood lymphocytes especially in patients with a high proviral load (19). A persistent active cytotoxic T-cell response has been demonstrated for patients with HAM/TSP (11) and for asymptomatic carriers (4). Despite an interisolate diversity of only 4 to 8% between the major HTLV-1 subtypes (12), within-isolate sequence variation and a high dN/dS ratio in the tax gene have been demonstrated, particularly for asymptomatic carriers (17).
If proviral load were maintained by reverse transcription, sustained inhibition of reverse transcriptase (RT) may be expected to reduce proviral load, by analogy with human immunodeficiency virus (HIV) infection (1). In addition to its proven in vitro and in vivo efficacy against HIV, zidovudine has been shown previously to be effective against murine (21) (Rauscher murine leukemia virus) and feline (26) (feline leukemia virus) retroviruses in vitro and in vivo.
Matsushita et al. (14) found a profound suppression of HTLV-1 Gag protein production and a reduction in proviral DNA when primary CD4+ T lymphocytes were exposed to HTLV-1 and cultured in the presence of zidovudine. Nusinoff-Lehrman et al. (18) also found that at zidovudine concentrations of 2.4 μg/ml or above, HTLV-1-specific DNA could not be detected by Southern blot analysis when infected lymphocytes were cocultured with susceptible target cells. Macchi et al. (13) have shown that low concentrations (as low as 0.1 μM) of zidovudine inhibit transmission of HTLV-1 to adult PBMCs in vitro, inhibiting the production of viral DNA and RNA. Zidovudine decreased CD25 expression on T lymphocytes in culture exposed to HTLV-1 as well as in uninfected PBMCs, but down-regulation of HLA-DR expression was more marked in HTLV-1-infected cells. Zidovudine had no antiretroviral effect on PBMCs already infected with HTLV-1 (13). In rabbits, the administration of zidovudine inhibited HTLV-1 replication following inoculation of an HTLV-1-transformed cell line (10). The cytosine analogue 2′,3′-dideoxycytidine (zalcitabine) also inhibited the synthesis of HTLV-1 viral DNA in CD4+ lymphocytes in vitro (14). Two groups who had previously used zidovudine to treat patients with HAM/TSP did not report HTLV-1 proviral load (8, 24).
In order to elucidate the extent of HTLV-1 replication in vivo, we studied the effect of nucleoside analogue RT inhibitors on the quantity of HTLV-1 viral DNA in PBMCs, T-lymphocyte phenotype, anti-Tax cytotoxic T-lymphocyte (CTL) precursor frequency, and clinical status, first in a patient with early, progressing HAM/TSP and subsequently in four other patients with HAM/TSP and high HTLV-1 viral DNA copy number.
MATERIALS AND METHODS
Subjects.
The subjects were five patients with HAM/TSP, whose demographic and clinical characteristics are shown in Table 1; all cases satisfied the international criteria for diagnosis of HAM/TSP (34). Four patients were ambulant with aid, and one was wheelchair bound. Four had stable disease for more than 3 years, while one, patient TAN, developed HAM/TSP while participating in a prospective study of initially asymptomatic HTLV-1 carriers (28). Following a period of assessment with multiple baseline measurements, treatment with a nucleoside analogue RT inhibitor was initiated. Patient TAN was treated first with zidovudine (200 mg) three times daily for 3 months and subsequently with lamivudine (150 mg) twice daily; the other four patients were treated only with lamivudine (150 mg) twice daily. The mean period of treatment with lamivudine was 10.2 months (range, 5.6 to 17.2 months). No other treatment was taken concurrently, with the single exception of patient TAF, who was undergoing treatment with alpha interferon for hepatitis C virus-related chronic active hepatitis. The study was approved by the local ethical review board, and all study participants gave written informed consent.
TABLE 1.
Characteristics of subjectsa
| Patient ID | Age (yr) | Sex | Race | Duration of HAM | Western blot assay resultb | Anti-HTLV-1 antibody titerc
|
No. of HTLV-1 DNA copies/100 PBMCsd | |
|---|---|---|---|---|---|---|---|---|
| Serum | CSF | |||||||
| TAD | 35 | F | AC | 3 yr | HTLV-1 | 131,072 | NA | 2.2 |
| TAF | 47 | M | Cauc | 11 yr | HTLV-1 | 262,144 | NA | 1.6 |
| TAK | 53 | F | AC | 13 yr | HTLV-1 | 32,768 | NA | 7.7 |
| TAL | 60 | F | AC | 15 yr | HTLV-1 | 262,144 | NA | 1.4 |
| TAN | 39 | F | Cauc | 7 mo | HTLV-1 | 16,384 | 1,024 | 14 |
Abbreviations: ID, identifier; F, female; M, male; AC, Afro-Caribbean; Cauc, Caucasian; CSF, cerebrospinal fluid; NA, not available.
The assay used was Genelabs HTLV-I/II 2.3 (Genelabs, Singapore, Singapore).
The assay used was the Serodia HTLV-1 gel particle agglutination assay (Fujirebio, Tokyo, Japan).
Median number of HTLV-1 viral DNA copies prior to any antiviral treatment.
Quantification of HTLV-1 viral DNA in PBMCs.
To quantify HTLV-1 DNA in PBMCs, DNA was extracted from 2 × 106 PBMCs by the proteinase K method. Replicate serial dilutions of the DNA were amplified by a nested-PCR technique that reliably detected a single copy of HTLV-1 tax DNA in genomic DNA from 105 cells (29). The viral DNA copy number was calculated from the Poisson distribution of negative samples at the cutoff dilution. Interassay variability (0.3 log10) was determined by repeated testing of a random selection of patient samples (29).
Quantification of CTL frequency.
CTL effector frequency was determined in a limiting dilution assay (LDA) (4). CD8+ lymphocytes, separated from fresh PBMCs with M-450 anti-CD8-coated Dynabeads, were plated out at known cell numbers per well in 96-well plates. The CD8+ cells were stimulated with phytohemagglutinin (1 μg/ml) in culture medium (RPMI 1640 medium, 10% fetal calf serum, and 10% Lymphocult-T [Biotest UK Ltd., Solihull, United Kingdom]) on day 3. On day 7, the cells were divided into three duplicate 96-well plates and were made up to 200 μl/well with the same culture medium. LDAs were performed on day 10 with 3,000 51Cr-labelled, peptide-pulsed autologous Epstein-Barr virus-transformed B cells (target cells) per well. Target cells were pulsed for 1 h with 50 μM peptide X1 (MEPTLGQHLPTLSFPD) or X6 (VIFCHPGQLGAFLTN), which contained the immunodominant Tax epitopes recognized by patient TAN’s CTLs (data not shown). Target cells pulsed with medium alone were used as controls to measure nonspecific lysis. Lysis in each well was considered positive if the 51Cr release exceeded the mean + 3 standard deviations of the nonspecific lysis. The effector frequencies for both X1 and X6 were added at each time point and represent the minimum number of Tax-specific CTLs in each sample. The frequency of activated CTLs measured in the LDA correlated closely with the activity of CTLs in the bulk CTL assay (data not shown) and was independent of the background lysis.
Lymphocyte half-life.
To determine the lymphocyte half-life, 10 ml of peripheral blood at each time point was taken into preservative-free sodium heparin (20 IU/ml; Monoparin; CP Pharmaceuticals, Barnstable, United Kingdom) before and after computerized tomography of patient TAN’s thorax, which was performed (for a clinical indication) during the 6th week of therapy with lamivudine (week 186), which coincided with the nadir in HTLV-1 DNA copies and anti-HTLV-1 Tax CTL frequency. The lymphocytes were separated as described above and then cultured for 48 h in RPMI 1640 medium with 15% bovine serum (Serum Supreme; BioWhittaker, Walkersville, Md.) and 10 μg of lectin (Sigma Chemical Co., St. Louis, Mo.) per ml in a conical flask. After disaggregation by pipetting, the cells were incubated with 0.05 μg of N-deacetyl-N-methylcolchicine (Demecolcine; Sigma-Aldrich Co. Ltd., Poole, United Kingdom) per ml at 37°C for 30 to 180 min, pelleted (1,000 × g for 5 min), and resuspended in 10 ml of 0.075 M KCl for 0 to 15 min at room temperature. Cells were again centrifuged (1,000 × g for 5 min), resuspended gently in 10 ml of freshly prepared 3:1 methanol-glacial acetic acid, and stored at 4°C. Chromosome preparations were made by dropping the fragile cells onto chilled glass slides and staining them with 3% Giemsa stain for 4 min. Cells (n = 200) were examined for dicentric chromosomes by a blinded observer (C.A.M.).
Lymphocyte phenotyping.
The phenotype of freshly isolated PBMCs was determined by flow cytometry (FACScan; Becton Dickinson, Oxford, United Kingdom) in the lymphocyte gate with a panel of monoclonal antibodies (MAbs) specific to CD3, CD4, CD8, CD25, CD38, and HLA-DR (Becton Dickinson). A fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin F(ab)2 fragment was used as the second layer. In some assays, directly conjugated phycoerythrin- and fluorescein isothiocyanate-conjugated MAbs were used. To avoid nonspecific binding, the Fc receptor was blocked with human AB serum in all washes and incubations. Control preparations included omission of the first-layer MAb and its replacement by a mouse immunoglobulin of a matched isotype. Results were evaluated on a FACScan flow cytometer with a lymphocyte gate.
RT gene sequencing.
To search for mutations in the HTLV-1 pol gene that might confer resistance to lamivudine, we determined the sequence of a 2,843-bp region of HTLV-1 viral DNA encompassing the full coding region of RT and the initial 909 bp of INT. DNA was extracted from patient TAN’s PBMCs before treatment, during rebound of viral DNA while the patient was on lamivudine treatment, and when the viral DNA load had returned to baseline levels after 24 weeks of lamivudine therapy. The sequence of PCR-amplified DNA was determined, by using an ABI 377 automated DNA sequencer, in forward and reverse directions, by direct sequencing without cloning; Taq polymerase errors should therefore make a negligible contribution to the sequence obtained.
RESULTS
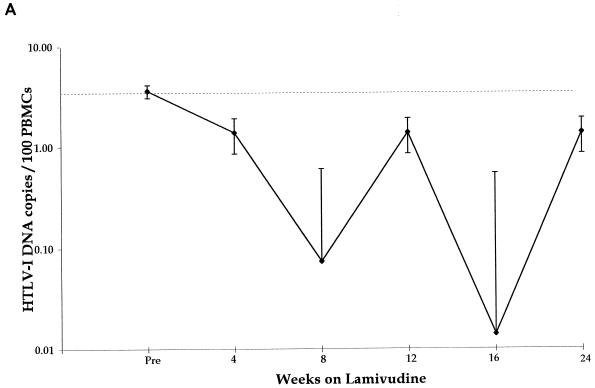
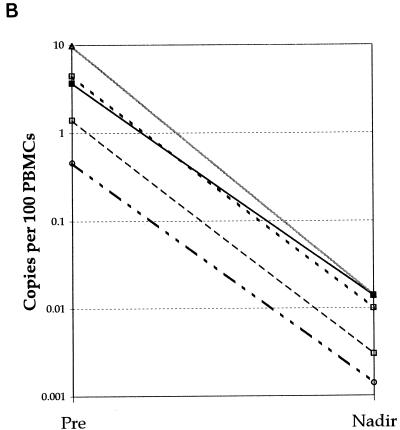
The change in HTLV-1 DNA copies per 100 PBMCs in all five patients, in relation to duration of therapy with lamivudine, is shown in Fig. 1A. The median pretreatment load was 2.2 HTLV-1 DNA copies/100 PBMCs. The median reduction in viral DNA copy number of all five patients was 1.1 log10. The median nadir in viral DNA load from pretreatment levels was 2.35 log10 but the time to reach the nadir varied between patients from 4 to 24 weeks (Fig. 1B).
FIG. 1.
All patients. (A) Median changes in HTLV-1 DNA copy number for five HTLV-1-infected subjects treated with lamivudine over 24 weeks. (B) Maximum reduction (nadir) in viral DNA copy number during lamivudine therapy in five patients with HAM/TSP: TAD (solid line with solid squares), TAL (short-dashed line with open squares), TAK (long-dashed line with open squares), TAF (dashed line with open circles), and TAN (shaded line with open triangle).
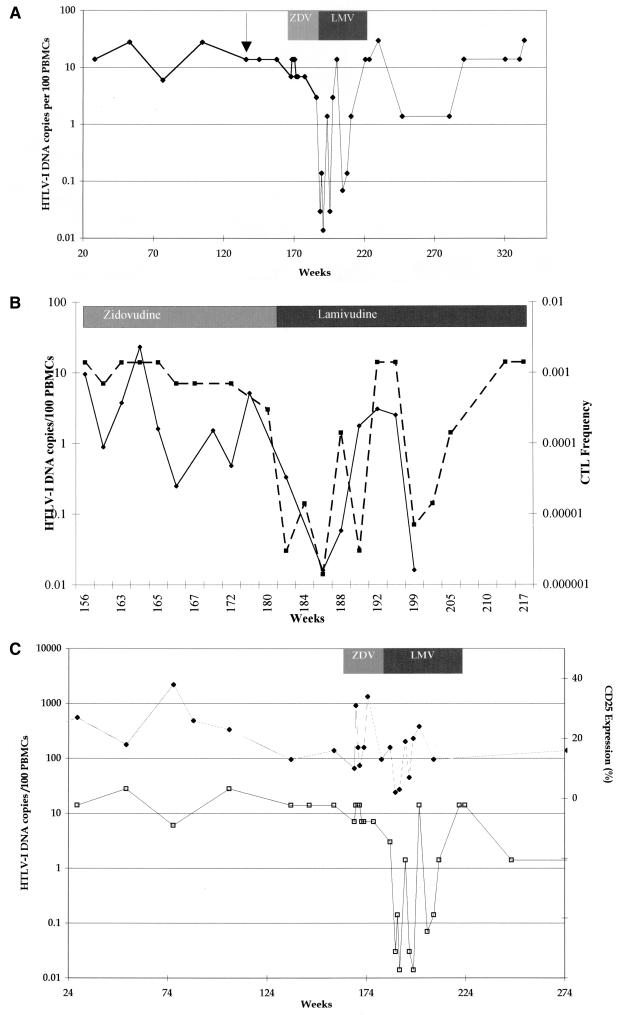
Patient TAN had been monitored prospectively over a period of 3 years prior to and during the development of HAM/TSP; the HTLV-1 DNA copy number in PBMCs was consistently high over this period, with a median of 14 (range, 7 to 28) copies of viral DNA per 100 PBMCs, compared with other asymptomatic carriers (median viral DNA, 0.28 copies per 100 PBMCs) (28), and this was not significantly reduced during 3 months’ treatment with the RT inhibitor zidovudine (median, 10.5 copies/100 PBMCs; range, 3 to 14 copies) (Fig. 2A). During the first 3 weeks of therapy with lamivudine (weeks 180 to 183), HTLV-1 viral DNA copy number fell from 3 to 0.03 per 100 PBMCs (Fig. 2A). While patient TAN was on continuous therapy with lamivudine, viral DNA load rose to baseline levels after 8 weeks and then fluctuated between baseline and nadir over 3 orders of magnitude for the full 24-week duration of therapy. After cessation of lamivudine, the fluctuations became smaller and the viral DNA load remained consistently above 1 copy per 100 PBMCs for a further 70 weeks (Fig. 2A). The rate of HTLV-1 viral DNA decline after the initiation of lamivudine indicated a viral DNA half-life of about 3.2 days. The estimated total number of HTLV-1-infected PBMCs prior to lamivudine treatment was 3 × 107. The observed half-life suggests that approximately 6 × 106 new cells become infected every day.
FIG. 2.
Patient TAN. (A) HTLV-1 viral DNA copy numbers from all available time points and their relationship to the onset of HAM/TSP (arrow) and therapy first with zidovudine (ZDV) and then with lamivudine (LMV) are shown for patient TAN. (B) Comparison of changes in the amount of HTLV-1 viral DNA (dashed line with squares) and HTLV-1 Tax-specific CTL effector frequency (solid line with diamonds) during therapy. (C) Comparison of the amount of HTLV-1 viral DNA (open squares) with changes in the percentage of lymphocytes expressing CD25 (solid diamonds) before, during, and after therapy with lamivudine. ZDV, zidovudine; LMV, lamivudine.
The consensus sequence of RT after 14 weeks of lamivudine therapy (week 201) did not differ from the pre-lamivudine (pre-zidovudine and during zidovudine) treatment sequences. However, the consensus RT sequence after 36 weeks of lamivudine therapy (week 224) differed at 11 nucleotide positions from the pretreatment sequences (Table 2). Of these 11 sequences, only one (A3982T) changed the predicted amino acid sequence of RT (Q488H); the new amino acid residue (H488) is present in seven of nine HTLV-1 sequences in the GenBank-EMBL database and is therefore unlikely to have been selected in patient TAN because it conferred resistance to lamivudine. In particular, no YMDD substitution was found. One further silent nucleotide substitution was found in the integrase coding region (T4588C); again, this substitution appeared for the first time after 24 weeks of lamivudine therapy.
TABLE 2.
Effect of treatment on HTLV-1 DNA sequencea
| Position and base (23) | Substitution | Amino acid change |
|---|---|---|
| 2699G | A | — |
| 2723A | G | — |
| 2972A | G | — |
| 3068T | A | — |
| 3098A | G | — |
| 3116G | A | — |
| 3227G | A | — |
| 3857C | T | — |
| 3911T | C | — |
| 3983A | T | Gln-His |
| 4148G | A | — |
| 4589T | C | — |
Sequencing revealed the emergence of a new dominant HTLV-1 DNA sequence with 11 nucleotide substitutions in the consensus sequence of the HTLV-1 pol gene from patient TAN. Each of these substitutions was first identified at the final time point, after 36 weeks of treatment with lamivudine, with the reexpansion of the amount of viral DNA. Before treatment, the nucleotide present at each of these positions was the same as that in the reference sequence (stub) (23). —, no amino acid change.
Prior to treatment, a high frequency of activated anti-Tax CD8+ lymphocytes recognizing several Tax epitopes simultaneously was present in the peripheral circulation of patient TAN. During treatment with lamivudine, the anti-Tax CTL frequency declined with the fall in viral DNA load and then rose again as the viral DNA copy number increased (Fig. 2B). The frequency of anti-Tax CTLs prior to lamivudine treatment was 7.9 × 10−4 CD8+ cells, declining to 1.6 × 10−6 CD8+ cells after 5 weeks of therapy. This is consistent with a CTL half-life of 3.9 days. Since the LDA is conducted over a period equal to 2.5 in vivo half-lives of the CTLs, the results may underestimate the frequency of Tax-specific CTLs; however, in vitro their half-life may be prolonged by the exogenous interleukin-2 support.
Prospective quantification of T-lymphocyte phenotypic markers was undertaken with patient TAN. There was no trend in the absolute counts or percentage of CD4+ or CD8+ T-lymphocyte counts or in the absolute number of circulating lymphocytes over time and no change in relation to lamivudine therapy. However, the proportion of CD25+ lymphocytes, which was elevated before and after lamivudine therapy, fell from 15 to 2%, coincident with the reduction in viral DNA copies on the initiation of lamivudine therapy and fluctuated in a manner parallel to that of the viral DNA copy number and anti-Tax CTL precursor frequency changes during therapy (Fig. 2C). The estimated half-life of CD25+ lymphocytes was 6.1 days.
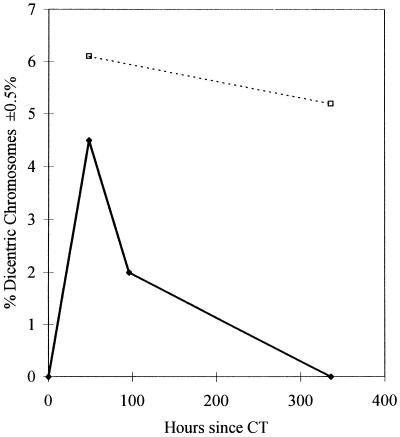
The presence of both abundant anti-Tax CTLs and the high viral DNA load suggested the possibility that there might be frequent CTL-mediated killing of infected CD4+ T lymphocytes in vivo. Lymphocyte half-lives can be estimated by measuring the rate of disappearance from the circulation of lymphocytes with stable, radiation-induced chromosome damage (15). We therefore counted the proportion of PBMCs that contained dicentric chromosomes, after patient TAN had undergone computerized tomography of the thorax, and their rate of disappearance from the blood (Fig. 3). The estimated half-life of 2.5 days reflects the rate at which the PBMCs were dividing or dying and is consistent with the observed rapid reduction in HTLV-1 viral DNA (estimated half-life, 3.2 days) and CTL frequency (estimated half-life, 3.9 days).
FIG. 3.
No dicentric chromosomes were found in cell preparations before computerized tomography (CT), 4.5% of cells contained dicentric chromosomes 48 h after computerized tomography, 2% dicentric chromosomes were detected 96 h post-computerized tomography, and none were detected at 336 h. No reduction in the total lymphocyte count occurred during this period. Solid diamonds, observed rate of disappearance of dicentric chromosomes for patient TAN. Open squares, rate of disappearance of dicentric chromosomes following radiotherapy for ankylosing spondylitis (15). The lymphocyte half-life is about 200 days.
The only clinical improvement was seen in the patient with recent-onset HAM/TSP, an improvement which persisted only during the period when lamivudine appeared to have reduced the viral burden. The patients with severe, long-standing, and stable disability showed no symptomatic improvement despite reduced HTLV-1 DNA load (27).
DISCUSSION
In this study, zidovudine treatment of patient TAN led to no change in HTLV-1 viral DNA copy number, CTL precursor frequency, or T-lymphocyte phenotype. Treatment was therefore changed to lamivudine, a cytosine analogue which has been found to be safe and effective with patients infected with HIV-1 (31) or hepatitis B virus (6). Other licensed nucleoside analogues, didanosine, stavudine, and zalcitabine, are associated with peripheral neuropathy during prolonged therapy (25). In cell proliferation assays (Glaxo Research and Development Ltd.) (12a), lamivudine does not affect the uptake of [3H]thymidine by uninfected and HTLV-1-infected PBMCs. The 50% infective dose of lamivudine in PBMCs was 2,533 μM (compared to 90 μM for zidovudine). Similar results were obtained for lymphocyte cell lines including HTLV-1 provirus-bearing C8166 cells. The administration of lamivudine orally, in the dose used in this study, results in a maximum concentration of lamivudine in serum which is 6 log10 below the antiproliferative concentration.
Initiation of lamivudine therapy was followed by an immediate reduction in viral DNA copy number. Subsequently, the viral DNA copy number oscillated, with the troughs gradually rising toward the baseline. CTL precursor frequency and CD25+ T lymphocytes also showed periodic oscillations during lamivudine therapy, which appeared to coincide with the changes in viral DNA load. Mathematical analysis shows that such oscillations in the quantity of viral DNA may result from the production of HTLV-1-infected cells from both cell division (not affected by lamivudine) and reverse transcription and from the interplay of viral production with CTL-mediated killing of infected cells (33). This model suggests that the high viral DNA load of HTLV-1 is indeed maintained by proviral expansion but that this is consistent with, and indeed may require, a high rate of HTLV-1 virion production (33). The subsequent return to pretreatment viral DNA levels in a compliant patient could be due to cellular resistance (a reduction in the uptake or phosphorylation of lamivudine) or the emergence of viral variants with decreased susceptibility to lamivudine. Lamivudine resistance in HIV-1 infection is often accompanied by a mutation in the active site of the HIV-1 RT (22). However, the corresponding region in the HTLV-1 RT gene was unchanged for patient TAN. The only coding change that appears in the RT gene following lamivudine treatment is commonly found in naturally occurring isolates of HTLV-1 and is therefore unlikely to have been selected because it confers resistance to lamivudine. However, the de novo appearance of 11 nucleotide substitutions in the consensus sequence of a 2,843-bp region of the HTLV-1 genome strongly suggests that the virus is replicating, causing the introduction of polymerase errors, and that the viral population passed through a bottleneck. The newly emergent consensus sequence represents a clone that (by chance) survived the genetic bottleneck and grew to replace the preexisting dominant sequence. Although we cannot exclude the possibility of drug resistance mutations in a minority of viruses, any such viruses cannot have made a major contribution to the recrudescence of HTLV-1 viral DNA in PBMCs.
Our previous finding of abundant activated anti-Tax CTLs in HTLV-1-infected patients (4) already suggested that there is persistent expression of the tax gene in the infected host. However, it was not clear whether expression was confined to the tax gene or whether it represented persistent replication of HTLV-1. The rapid fall in viral DNA for three patients during treatment with lamivudine suggests that the high pretreatment viral DNA copy number was maintained chiefly by active viral replication. Proliferation of infected lymphocytes, which has been suggested by others to be the main cause of high proviral load in HTLV-1 infection, is not sufficient to maintain the high viral DNA load in these patients. These results are in keeping with the recent observation that HTLV-1 mRNA was more frequently detected in patients with a high proviral load (19). However, for two patients a much more gradual decline in the amount of viral DNA was noted, suggesting that the relative importance of reverse transcription in the maintenance of viral DNA load differs between subjects.
Four independent techniques (limiting dilution analysis of CTL frequency, HTLV-1 viral DNA quantification, fluorescence-activated cell sorting analysis of lymphocyte phenotypes, and the clearance of cells containing dicentric chromosomes) during antiretroviral therapy each produced estimates of the average life expectancy of patient TAN’s PBMCs of 3 to 6 days. CTL-mediated killing of HTLV-1-infected cells may account for part of this rapid cell turnover. However, other factors must also have been involved, as even rapid killing of a minority population of PBMCs expressing Tax protein (at most, 14% of PBMCs) and high turnover of CD8+ HTLV-specific CTLs (which may constitute up to 10% of CD8+ PBMCs [16a]) would not reduce the average life expectancy of all PBMCs so far below the normal value of approximately 150 days (memory lymphocytes) or 3.5 years (naive lymphocytes) (15). Vigorous cell activation and proliferation, driven by HTLV-1 Tax protein, may have predisposed PBMCs to apoptosis (3).
In conclusion, lamivudine reduced the amount of HTLV-1 DNA in five of five patients with HAM/TSP. This reduction and the subsequent increase in the amount of viral DNA during treatment are consistent with active viral replication through reverse transcription. The viral kinetics indicate that the importance of viral replication in maintaining viral DNA load varies between patients. The results have important implications for our understanding of HTLV-1 pathogenesis and might open the way to specific therapy for HTLV-1-associated inflammatory disease.
ACKNOWLEDGMENTS
S.E.H. and C.R.M.B. are supported by The Wellcome Trust. G.P.T. was supported by the Jefferiss Trust. The HTLV European Research Network supported by the European Community Biomed Programme (BMH4 CT98-3781) provided a forum for critical appraisal of this work.
Jennifer Tosswill of the Central Public Health Laboratory, Colindale, London, United Kingdom, assisted with the HTLV-1 DNA measurements.
REFERENCES
- 1.Brun-Vezinet F, Boucher C, Loveday C, Descamps D, Fauveau V, Izopet J, Jeffries D, Kaye S, Krzyanowski C, Nunn A, Schuurman R, Seigneurin J M, Tamalet C, Tedder R, Weber J, Weverling G J. HIV-1 viral load, phenotype, and resistance in a subset of drug-naive participants from the Delta trial. The National Virology Groups. Delta Virology Working Group and Coordinating Committee. Lancet. 1997;350:983–990. doi: 10.1016/s0140-6736(97)03380-1. [DOI] [PubMed] [Google Scholar]
- 2.Cavrois M, Gessain A, Wain-Hobson S, Wattel E. Proliferation of HTLV-1 infected circulating cells in vivo in all asymptomatic carriers and patients with TSP/HAM. Oncogene. 1996;12:2419–2423. [PubMed] [Google Scholar]
- 3.Chlichlia K, Moldenhauer G, Daniel P T, Busslinger M, Gazzolo L, Schirrmacher V, Khazaie K. Immediate effects of reversible HTLV-I tax function: T-cell activation and apoptosis. Oncogene. 1995;10:269–277. [PubMed] [Google Scholar]
- 4.Daenke S, Kermonde A, Hall S E, Taylor G, Weber J, Nightingale S, Bangham C R M. High activated and memory cytotoxic T-cell responses to HTLV-1 in healthy carriers and patients with tropical spastic paraparesis. Virology. 1996;217:139–146. doi: 10.1006/viro.1996.0101. [DOI] [PubMed] [Google Scholar]
- 5.Dalgleish A, Richardson J, Matutes E, Cruickshank K, Newell A, Sinclair A, Thorpe R, Brasher M, Weber J, Catovsky D, et al. HTLV-I infection in tropical spastic paraparesis: lymphocyte culture and serological response. AIDS Res Hum Retroviruses. 1989;4:475–485. doi: 10.1089/aid.1988.4.475. [DOI] [PubMed] [Google Scholar]
- 6.Dienstag J L, Perrillo R P, Schiff E R, Bartholomew M, Vicary C, Rubin M. Preliminary trial of lamivudine for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 7.Gessain A, Vernant J C, Maurs L, Barin F, Gout O, Calender A, de Thé G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–409. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 8.Gout O, Gessain A, Iba-Zizen M, Kouzan S, Bolgert F, de Thé G, Lyon-Caen O. The effect of zidovudine on chronic myelopathy associated with HTLV-I. J Neurol. 1991;238:108–109. doi: 10.1007/BF00315691. [DOI] [PubMed] [Google Scholar]
- 9.International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. 67. Human immunodeficiency viruses and human T-cell lymphotrophic viruses. Lyon, France: International Agency for Research on Cancer; 1996. [PMC free article] [PubMed] [Google Scholar]
- 10.Isono T, Ogawa K, Seto A. Antiviral effect of zidovudine in the experimental model of adult T-cell leukaemia in rabbits. Leuk Res. 1990;14:841–847. doi: 10.1016/0145-2126(90)90172-6. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson S, Shida H, McFarlin D E, Fauci A S, Koenig S. Circulating CD8+ cytotoxic T lymphocytes specific for HTLV-I pX in patients with HTLV-I associated neurological disease. Nature. 1990;348:245–248. doi: 10.1038/348245a0. [DOI] [PubMed] [Google Scholar]
- 12.Koralnik I J, Boeri E, Saxinger W C, Lo Monico A, Fullen J, Gessain A, Guo H-G, Gallo R C, Markham P, Kalyanaraman V, Hirsch V, Allen J, Murthy K, Alford P, Slattery J P, O’Brien S J, Franchini G. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J Virol. 1994;68:2693–2707. doi: 10.1128/jvi.68.4.2693-2707.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Macchi, B. Personal communication.
- 13.Macchi B, Faraoni I, Zhang J, Grelli S, Favalli C, Mastino A, Bonmassar E. AZT inhibits the transmission of human T cell leukaemia/lymphoma virus type I to adult peripheral blood mononuclear cells in vitro. J Gen Virol. 1997;78:1007–1016. doi: 10.1099/0022-1317-78-5-1007. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita S, Mitsuya H, Reitz M S, Broder S. Pharmacological inhibition of in vitro infectivity of human T lymphotropic virus type I. J Clin Investig. 1987;80:394–400. doi: 10.1172/JCI113085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean A R, Michie C A. In vivo estimates of division and death rates of human T lymphocytes. Proc Natl Acad Sci USA. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai M, Usuku K, Matsumoto W, Kodama D, Takenouchi N, Moritoyo T, Hashiguchi S, Ichinose M, Bangham C R M, Izumo S, Osame M. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol. 1998;4:586–593. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- 16a.Navarrete, S., S. E. Hall, and C. R. M. Bangham. Unpublished data.
- 17.Niewiesk S, Daenke S, Parker C E, Taylor G P, Weber J N, Nightingale S, Bangham C R M. The transactivator gene of human T-cell leukemia virus type I is more variable within and between healthy carriers than patients with tropical spastic paraparesis. J Virol. 1994;68:6778–6781. doi: 10.1128/jvi.68.10.6778-6781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nusinoff-Lehrman S, St. Clair M, Miller R L, Broder S, Wilson H R, Bushby M, et al. Proceedings of the Second International Conference on AIDS, Paris, France, 23 to 25 June 1986. 1986. Azidothymidine: spectrum of in vitro antimicrobial activity, abstr. 556; p. 66. [Google Scholar]
- 19.Okayama A, Tachibana N, Ishihara S, Nagatomo Y, Murai K, Okamoto M, Shima T, Sagawa K, Tsubouchi H, Stuver S, Mueller N. Increased expression of interleukin-2 receptor alpha on peripheral blood mononuclear cells in HTLV-I tax/rex mRNA-positive asymptomatic carriers. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:70–75. doi: 10.1097/00042560-199705010-00011. [DOI] [PubMed] [Google Scholar]
- 20.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 21.Ruprecht R M, O’Brien L G, Rossoni L D, Nusinoff-Lehrman S. Suppression of mouse viraemia and retroviral disease by 3′-azido-3′-deoxythymidine. Nature. 1986;323:467–469. doi: 10.1038/323467a0. [DOI] [PubMed] [Google Scholar]
- 22.Schuurman R, Nijhuis M, van-Leeuwen L M, Schipper P, de-Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 23.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheremata W A, Benedict D, Squilacote D C, Sazant A, DeFreitas S. High-dose zidovudine induction in HTLV-I-associated myelopathy: safety and possible efficacy. Neurology. 1993;43:2125–2129. doi: 10.1212/wnl.43.10.2125. [DOI] [PubMed] [Google Scholar]
- 25.Simpson D M, Tagliati M. Nucleoside analogue-associated peripheral neuropathy in human immunodeficiency virus infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:153–161. [PubMed] [Google Scholar]
- 26.Tavares L, Roneker C, Johnston K, Lehrman S N, de Noronha F. 3′-Azido-3′-deoxythymidine in feline leukemia virus-infected cats: a model for therapy and prophylaxis of AIDS. Cancer Res. 1987;47:3190–3194. [PubMed] [Google Scholar]
- 27.Taylor G P, Hall S, Nowak M, Michie C, Rossor M, Davis R, Bangham C R M, Weber J N. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 1 to 5 February 1998. 1998. Reduction of HTLV-I proviral load in patients with HTLV-I associated myelopathy through lamivudine monotherapy, abstr. 508; p. 175. [Google Scholar]
- 28.Taylor G P, Tosswill J H C, Matutes E, Daenke S, Hall S, Bain B, Rossor M, Thomas D, Bangham C R M, Weber J N. Inflammatory consequences of HTLV-I infection in an initially asymptomatic UK cohort. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;22:92–100. doi: 10.1097/00042560-199909010-00012. [DOI] [PubMed] [Google Scholar]
- 29.Tosswill J H C, Taylor G P, Clewley J P, Weber J N. Quantification of proviral DNA load in human T-cell leukaemia virus type-I infections. J Virol Methods. 1998;75:21–26. doi: 10.1016/s0166-0934(98)00093-7. [DOI] [PubMed] [Google Scholar]
- 30.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukaemia: clinical and haematological features of 16 cases. Blood. 1977;50:481–492. [PubMed] [Google Scholar]
- 31.van-Leeuwen R, Katlam C, Kitchen V, Boucher C A, Tubiana R, McBride M, Ingrand D, Weber J, Hill A, McDade H, et al. Evaluation of safety and efficacy of 3TC (lamivudine) in patients with asymptomatic or mildly symptomatic human immunodeficiency virus infection: a phase I/II study. J Infect Dis. 1995;171:1166–1171. doi: 10.1093/infdis/171.5.1166. [DOI] [PubMed] [Google Scholar]
- 32.Wattel E, Cavrois M, Gessain A, Wain-Hobson S. Clonal expansion of infected cells. A way of life for HTLV-I. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):S92–S99. doi: 10.1097/00042560-199600001-00016. [DOI] [PubMed] [Google Scholar]
- 33.Wodarz D, Nowak M A, Bangham C R M. The dynamics of HTLV-I and the CTL response. Immunol Today. 1999;20:220–227. doi: 10.1016/s0167-5699(99)01446-2. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. WHO diagnostic guidelines of HAM. Virus diseases. Human T-lymphotropic virus type I, HTLV-I. Weekly Epidemiol Rec. 1989;49:382–383. [Google Scholar]