Conspectus
Creating a living system from nonliving matter is a great challenge in chemistry and biophysics. The early history of life can provide inspiration from the idea of the prebiotic “RNA World” established by ribozymes, in which all genetic and catalytic activities were executed by RNA. Such a system could be much simpler than the interdependent central dogma characterizing life today. At the same time, cooperative systems require a mechanism such as cellular compartmentalization in order to survive and evolve. Minimal cells might therefore consist of simple vesicles enclosing a prebiotic RNA metabolism.
The internal volume of a vesicle is a distinctive environment due to its closed boundary, which alters diffusion and available volume for macromolecules and changes effective molecular concentrations, among other considerations. These physical effects are mechanistically distinct from chemical interactions, such as electrostatic repulsion, that might also occur between the membrane boundary and encapsulated contents. Both indirect and direct interactions between the membrane and RNA can give rise to nonintuitive, “emergent” behaviors in the model protocell system. We have been examining how encapsulation inside membrane vesicles would affect the folding and activity of entrapped RNA.
Using biophysical techniques such as FRET, we characterized ribozyme folding and activity inside vesicles. Encapsulation inside model protocells generally promoted RNA folding, consistent with an excluded volume effect, independently of chemical interactions. This energetic stabilization translated into increased ribozyme activity in two different systems that were studied (hairpin ribozyme and self-aminoacylating RNAs). A particularly intriguing finding was that encapsulation could rescue the activity of mutant ribozymes, suggesting that encapsulation could affect not only folding and activity but also evolution. To study this further, we developed a high-throughput sequencing assay to measure the aminoacylation kinetics of many thousands of ribozyme variants in parallel. The results revealed an unexpected tendency for encapsulation to improve the better ribozyme variants more than worse variants. During evolution, this effect would create a tilted playing field, so to speak, that would give additional fitness gains to already-high-activity variants. According to Fisher’s Fundamental Theorem of Natural Selection, the increased variance in fitness should manifest as faster evolutionary adaptation. This prediction was borne out experimentally during in vitro evolution, where we observed that the initially diverse ribozyme population converged more quickly to the most active sequences when they were encapsulated inside vesicles.
The studies in this Account have expanded our understanding of emergent protocell behavior, by showing how simply entrapping an RNA inside a vesicle, which could occur spontaneously during vesicle formation, might profoundly affect the evolutionary landscape of the RNA. Because of the exponential dynamics of replication and selection, even small changes to activity and function could lead to major evolutionary consequences. By closely studying the details of minimal yet surprisingly complex protocells, we might one day trace a pathway from encapsulated RNA to a living system.
Key References
Saha R.; Verbanic S.; Chen I. A.. Lipid Vesicles Chaperone an Encapsulated RNA Aptamer. Nat. Commun. 2018, 9, 2313. .1 This study of the malachite green aptamer encapsulated inside model protocells showed that the confinement of the vesicle enhanced RNA aptamer activity, most likely through excluded volume effects.
Peng H.; Lelievre A.; Landenfeld A.; Müller S.; Chen I. A.. Vesicle Encapsulation Stabilizes Intermolecular Association and Structure Formation of Functional RNA and DNA. Curr. Biol. 2021, 32, 86–96.e6 .2 The effects of confinement were further probed by encapsulating several model nucleic acids as well as the hairpin ribozyme in model protocells. Encapsulation stabilized secondary and tertiary structures and rescued folding-deficient mutants of the hairpin ribozyme.
Lai Y.-C.; Liu Z.; Chen I. A.. Encapsulation of Ribozymes inside Model Protocells Leads to Faster Evolutionary Adaptation. Proc. Natl. Acad. Sci. USA 2021, 118, e2025054118. .3 An asymmetry in the effects of encapsulation on a wide variety of mutant ribozymes led to a “rich get richer” phenomenon in protocells, accelerating ribozyme evolution in vitro.
Introduction
Building minimal cells from nonliving materials is a fundamental challenge in synthetic biology, driven by a diverse set of motivations, from peering at our own origins to creating programmable biochemical factories. To approach this problem, many take inspiration from nature and biology. All cells come from cells (omnis cellula e cellula), except for the first cells, which must have somehow emerged from the chemistry of self-assembly and self-replication. The now-complex system of storing genetic information in DNA, transcribing into RNA, and translating into proteins via the genetic code, led early molecular biologists to propose that ancient life could have been much simpler: RNA might serve as the central, and possibly sole, biomolecular constituent of early life, simultaneously performing biocatalysis and carrying genetic information.4−9 This hypothesis is especially tempting since RNA now plays a myriad of fascinating and fundamental roles in biology, from catalyzing protein synthesis in the ribosome to carrying electrons for the cell in redox-active dinucleotides. The evidence for an early “RNA World” era is circumstantial, though, and there is little hope of finding direct evidence, such as fossils of primitive cells, owing to the age of these events (roughly 3–4 billion years ago). The difficulty of proving historical events at the origin of life has led to some criticism in the context of exobiology,10 which was skewered in a polemical essay by paleontologist G. Simpson as a field that “has yet to demonstrate that its subject matter exists”.11 However, such criticism misses a crucial point, namely that, like many other science and engineering disciplines, one of the main purposes of the field is to create its subject. The RNA World is an intellectual framework that guides bottom-up construction of simple cells, lacking the massive overhead of modern biology, regardless of whether an RNA World actually existed on the early earth.
Minimal life requires more than just evolving information. Specifically, RNA enzymes (ribozymes) must cooperate with one another because a functional, folded molecule cannot physically access its own entire sequence. At least two molecules are required to maintain a replication cycle, one to act as template and one to act as the catalyst. Such a system could be easily parasitized, though, by other sequences that act as perfectly good templates but have no catalytic activity. In this case, Darwinian selection would technically occur, possibly satisfying the widespread working definition of life put forth by a NASA working group (a “self-sustained chemical system capable of Darwinian evolution”),12 but would wind down to a dead end and loss of the replicator phenotype. Several mechanisms can be invoked to counter this tendency,13 generally involving a feedback mechanism whereby the ribozyme products affect the fitness of the ribozyme that made them.14 A simple mechanism is compartmentalization (including by protocell membranes), which keeps the replicating system together and prevents parasitic sequences from accessing their catalytic resources.15,16 Indeed, compartmentalization has been experimentally demonstrated to prevent parasitic takeover of an RNA replicator system,17−19 is theoretically advantageous compared to other mechanisms such as surfaced-based, cellular automata-like organization,20 and is validated by modern biology as a basic mechanism for defining genetic entities. In addition to these evolutionary effects, membrane encapsulation enables several key functions like nutrient transport, signal transduction, maintenance of chemical gradients, and growth and division of protocells.21,22 Therefore, synthetic protocells encapsulating RNA have emerged as a prominent goal for researchers interested in creating a minimal living system.14,23
A conceptually simple model of protocells may be constructed with prebiotically plausible single-chain amphiphiles, namely fatty acids and their derivatives, encapsulating catalytic RNA. These vesicles can be readily prepared in the laboratory. Interestingly, individual protocells can grow when “fed” with fatty acid molecules and divide when subjected to physical manipulations.24−26 During this primitive cell cycle, the protocells mostly maintain encapsulated material and thus create compartmentalized units for potential selection and evolution. Compared to diacyl phospholipids, which commonly compose the membranes of modern cells, fatty acid membranes provide greater permeability for relatively small molecules and display more rapid molecular dynamics. However, fatty acid vesicles, which are negatively charged overall, do show colloidal instability in the presence of divalent cations such as magnesium and calcium, which is problematic because ribozyme folding and catalysis often require divalent cations to interact with the negatively charged sugar–phosphate backbone of RNA.27−29 Strategies to stabilize fatty acid vesicles include adding significant percentages of phospholipids or other lipids, including prebiotically plausible single-chain amphiphiles, or adding partial chelation agents.1,30−33 Vesicles composed of simple lipids can already exhibit intriguing behaviors reminiscent of biological cells. For example, in addition to their ability to grow when fed, which derives from the thermodynamically downhill incorporation of lipids into the outer leaflet followed by rapid flip-flop of lipids to the inner leaflet, multilamellar vesicles can exhibit growth and division driven solely by compositional heterogeneity among individual vesicles.34 Fatty acid vesicles are also resistant to fusion under a variety of conditions that would cause fusion of phospholipid membranes, because monoacyl lipids cannot “splay” (in the way that diacyl lipids can) to bridge two hemifused vesicles. This behavior has the surprising consequence of helping to preserve the individual identities of their aqueous compartments.35 These observations indicate that an interplay of chemical and biophysical forces in even quite simple systems can lead to nontrivial behaviors that might be called “emergent”.36
When the vesicles encapsulate significant contents, such as RNA, to make protocells, the added complexity, though seemingly minor, also creates a new set of behaviors. For example, encapsulated RNA (or other solutes) causes vesicles to grow larger by exerting osmotic pressure on the membrane that can be relieved through incorporation of more lipid,37 thus linking together the amount of RNA to the growth for individual vesicles. An analogous phenomenon was observed in polypeptide-based giant vesicles, where gene expression increased the internal osmotic pressure and resulted in vesicle growth.38 These effects tie together internal metabolism to membrane growth, showing that these components of the protocell effectively work together even without specific mechanisms for interaction.14 In addition, protocells encapsulating RNA can exchange material during freeze–thaw cycles,39 leading to a kind of horizontal gene transfer, and encapsulation of random oligonucleotides create a mechanism for homeostasis in ribozyme activity.40 These life-like behaviors are not necessarily unique to RNA protocells. Growth, division, and phenotype-based selection can be observed when encapsulating DNA and enzymes,41,42 and even with an emulsified formose reaction solution.43 However, RNA is a system of choice for protocells. Its world-building potential comes not only from its genetic and catalytic possibilities, but also from its properties that tap into the nanoscale realm of colloid chemistry and macromolecular biophysics.
The modern cellular environment is densely crowded with macromolecules, including DNA, RNA, proteins, and lipids, as well as a large number of small molecules, creating a distinctive environment that differs significantly from dilute aqueous solutions.44,45 The geometry created by these macromolecules confined to the aqueous interior has multiple consequences, such as increased effective concentrations of small molecules due to the volume that is excluded by the macromolecules; hindered diffusion near the walls of a container, an effect that persists several particle lengths from the wall;46 and depletion forces, in which molecule centers are physically excluded from a significant region surrounding large particles, creating an osmotic gradient that, along with an entropic tendency to maximize the volume available to the molecules, creates an attractive force between large particles.47−49 These effects are distinct from chemical interactions such as van der Waals forces, electrostatic attraction or repulsion, hydrogen bonding, or the hydrophobic effect, but arise only from general physical considerations, particularly the inability of different molecules to occupy the same space. Thus, they must also occur inside protocells, which have a defined membrane boundary as well as macromolecular RNA, components that may interact either directly through chemical interactions, or indirectly through excluded volume and confinement effects. We have been exploring how simply physically encapsulating RNA might affect its properties and ultimately result in emergent protocell behaviors.
The Physical Importance of a Container for RNA
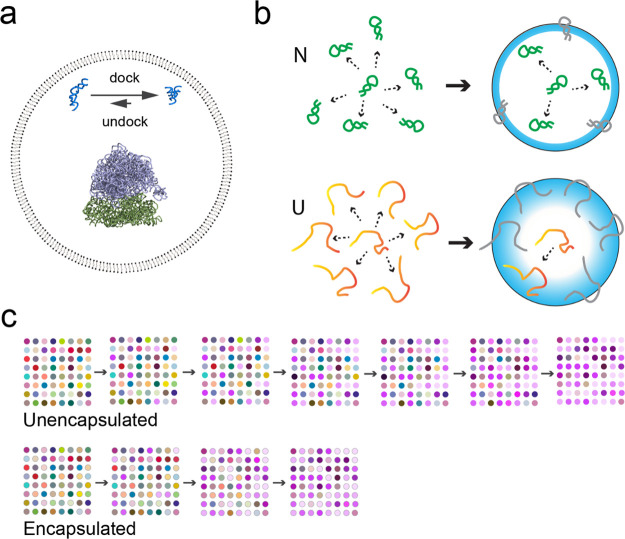
When fatty acid vesicles encapsulating RNA were first being developed as model protocells, a small self-cleaving ribozyme (the hammerhead ribozyme) was used to test whether the RNA would still be active once encapsulated. After accounting for a decrease in Mg2+ activity due to association with the negatively charged membrane, we found that the hammerhead self-cleavage rate constant dropped by about half when encapsulated.33 At the time, we were focused on ensuring chemical compatibility between the vesicles and ribozymes, which centered around various issues related to Mg2+, so we were satisfied by simply finding any conditions under which the ribozymes still worked inside vesicles. Over the next several years, a fascinating series of papers was published describing how crowding agents cause compaction and folding of RNA through excluded volume50−56 (these and later work are reviewed in51,57). Since the confining boundaries of vesicles would also exclude volume, we hypothesized that protocells might similarly enhance RNA folding (Figure 1a).58 (We had by then forgotten about our earlier result showing slightly lower hammerhead ribozyme activity inside vesicles, which turned out to be fortunate since it did not deter us from this line of inquiry, in which we dissected the separate importance of both chemical interactions and excluded volume effects.)
Figure 1.
Effects of protocell encapsulation on ribozymes. (a) Schematic drawing of a fatty acid vesicle (60 nm diameter1) encapsulating a hairpin ribozyme (PDB: 2OUE, docked conformation at top right) and a 70S ribosome (PDB: 4V42, purple/green structure in center). The undocked conformation of the hairpin ribozyme is an artistic illustration and is not based on a solved structure. Encapsulation shifts the equilibrium toward the docked state of the hairpin ribozyme due to physical confinement effects. Such effects are expected to be greater for larger multisubunit structures such as a ribosome. The molecules are drawn approximately to scale; for comparison, the diameter of an A-form helix is 2.3 nm and the thickness of a membrane bilayer is approximately 3 nm, depending on the lipid. (b) Illustration of the excluded volume effect from confinement inside a boundary membrane. The native (N), folded conformation of a ribozyme (green) has a specific compact shape that can be configured at multiple positions. When a boundary is present, some configurations (gray) are disallowed due to steric clashes with the membrane. The membrane boundary creates a volume from which the center of the molecule is excluded (blue zone). For the unfolded conformations (U), many configurations are possible, differing in both position and conformation (red/orange). When a boundary is present, as with N, many of these configurations are disallowed due to steric clashes (gray). However, a greater fraction of U configurations is affected compared to N, due to the extended nature of the U conformations. For U, the precise exclusion zone (blue) depends on the conformation, but is generally larger for U compared to N. The relative decrease in the number of accessible configurations for U is the basis for reduced entropy, and thus higher free energy of U relative to N, when encapsulated. (c) Encapsulation increases the rate of evolutionary adaptation of ribozymes, compared to unencapsulated ribozymes. In this drawing, each dot corresponds to a mutant ribozyme, with different colors representing different sequences (magenta colors being the highest fitness sequences). Encapsulation is represented by a gray circle around the dot. Beginning with a diverse set, without encapsulation (top row), the population adapts slowly, requiring several rounds of in vitro selection to converge on the fittest sequences (magenta shades). In contrast, the population of encapsulated RNAs converges quickly onto the fittest sequences (bottom row).
To study RNA activity, we chose an artificially selected RNA aptamer that binds the triphenylmethane dye malachite green (MG).59 While MG in solution is essentially nonfluorescent due to facile vibrational de-excitation, binding of the aptamer restricts vibrational modes and thus increases MG fluorescence almost 2360-fold.60 We found that MG was permeable to the membrane, so this “light-up” aptamer presented a straightforward assay for aptamer activity (although this aptamer choice would later turn out to be admittedly less straightforward in a different system involving minerals61). We first checked whether RNA exposed to vesicles, but not encapsulated (i.e., outside vesicles), would exhibit any change in binding constant (KD). To our initial surprise, mere exposure to lipids could decrease binding affinity significantly, especially when the headgroup was zwitterionic.1 Negatively charged lipids, including fatty acids, had a more modest effect, while positively charged lipids led to extensive aggregation in the presence of RNA,62 implicating electrostatic associations disrupting activity. The importance of chemical interactions between lipids and RNA has also been observed for tRNA,63 and is supported by the variety of effects we observed when adding different crowding agents to the MG aptamer, as also seen for the adenine riboswitch.64 Observing these chemical interactions clarified that the proper control to study a physical confinement effect for encapsulated RNA would be RNA “outside vesicles” (that is, exposed to the chemical environment of vesicles but not actually encapsulated), not RNA in dilute solution. This realization also solved the puzzle of why the hammerhead ribozyme had appeared to lose activity when encapsulated in our earlier study; the comparison to dilute solution had been confounded by the effect of exposure to lipids. Moving on to RNA encapsulated inside large unilamellar vesicles (generally 60–100 nm in diameter after extrusion), we indeed observed a ∼ 3-fold increase in binding affinity of the aptamer compared to the “outside vesicle” control, regardless of the vesicle type. Assuming random encapsulation, a simple calculation suggested that most vesicles would contain 0 to a few RNA molecules on average.1 These effects were approximately quantitatively consistent with a theoretical prediction of individual biopolymers folding inside spherical containers.49,65 The general idea is that the RNA exists in an ensemble of different conformations, some being compact and others being more extended. The presence of the boundary wall and its exclusion zone reduces the number of states available to more extended conformations, reducing their entropy. So, as the free energies of extended conformations are increased, compact conformations would be relatively stabilized within the ensemble (Figure 1b). For RNAs for which properly folded, functional conformations are compact relative to unfolded conformations, the overall effect of geometrical confinement can stabilize RNA folding. Thus, these results suggested that, in a prebiotic soup containing both RNA and lipids,66 RNAs that happened to be fortuitously encapsulated (or that promoted their own encapsulation) would benefit from improved folding and activity compared to their less fortunate counterparts left outside vesicles.
To directly probe the effect of encapsulation on RNA folding, we encapsulated model oligonucleotides, an RNA stem-loop and a DNA triplex, and used fluorescence assays to verify that encapsulation shifted the equilibrium toward the folded form, favoring formation of base pairs or noncanonical triple-base interactions, respectively.2 To determine whether effects on folding would translate directly into ribozyme activity, we then studied the hairpin ribozyme, which cleaves an RNA substrate following Michaelis–Menten enzyme kinetics. Confirming the generally denaturing effect of lipids, we noticed decreasing activity (kcat and kcat/Km values) with increasing lipid when RNA was outside vesicles, with complete loss of activity at higher lipid concentrations. However, encapsulation inside vesicles fully protected the ribozymes from this loss of activity.2 The catalytic mechanism of the hairpin ribozyme requires docking between two RNA stems to form an on-pathway tertiary contact, a step that is rate-limiting for the ribozyme.67 Following docking through FRET, encapsulated ribozymes showed both faster docking and a greater steady-state population of the docked intermediate, indicating stabilization of the docked conformation as well as the transition state. To push the limits of this effect, we tested known folding-deficient mutants of the hairpin ribozyme, that do not normally exhibit detectable docking or catalytic activity for an RNA ligation reaction. Encapsulation was able to rescue both docking and ligation activity in these mutants. Consistent with boundary confinement, encapsulation in larger vesicles (approximately 200 nm in diameter) gave a smaller effect.2 To sum up, encapsulation promoted a variety of secondary and tertiary interactions for RNA, which appears to be consistent with an excluded volume effect caused by confinement by the membrane boundary, and which results in better activity inside vesicles.
Protocells as Evolutionary Accelerant
Of particular interest to us was the observation that encapsulation profoundly affected the activity of the folding-deficient ribozyme mutants, bringing their activity close to wild-type levels. This signaled a potential connection between physical effects of encapsulation and ribozyme evolution. For over a decade, our laboratory has also been engaged in studying the sequence-activity relationship of ribozymes (known as the “fitness landscape”) to understand the emergence and evolution of RNA function.68−75 The fitness landscape is the function of fitness over multidimensional sequence space. For molecules, fitness is often equated with chemical activity, such as rate, meaning that the activity being studied is assumed to be important for the organism or system. Fitness landscapes for ribozymes are generally composed of “peaks” of high-activity (high fitness) sequences with the vast majority of sequences being very low in activity (i.e., nonfunctional). Around a wild-type ribozyme sequence, there exist mutants with varying degrees of activity, corresponding to a partially epistatic local landscape. While Darwinian evolution is often associated with survival of the fittest, mutational robustness, i.e., tolerance of mutations without loss of activity, can also be an important factor. That is, at relatively high mutation rates, a “survival of the flattest” would have also applied to evolution in error-prone primitive systems.76−78 We were intrigued by the possibility that excluded volume effects, brought about by vesicle confinement or crowding agents,53 might essentially flatten the fitness landscape by stabilizing the folding and thus increasing the fitness of ribozyme mutants. Perhaps more importantly for the emergence of function, raising the activity of mutant ribozymes would also simply increase the frequency of active ribozymes in sequence space, making ribozymes easier to discover during random sequence exploration.
While we had developed techniques to map complete fitness landscapes for RNA, they would be technically difficult to apply to encapsulated RNA due to relatively low encapsulation yields. Therefore, we focused on several families of self-aminoacylating RNAs that we had previously discovered.73 We generated a library of tens of thousands of ribozyme mutants and encapsulated them to test their activity inside vesicles compared to an outside-vesicle condition.3 This experiment was enabled by a massively parallel assay for ribozyme rates, k-Seq, which we had developed based on high-throughput sequencing,79 that allowed us to determine rate constants for many mutants in a small number of experiments. Consistent with our prior studies, there was nearly universal improvement in activity for the ribozymes when they were encapsulated, validating the generality of the confinement effect and the idea that encapsulation would increase the frequency of functional sequences. At the same time, a subtle feature caught our eye as well. When plotting ribozyme activity inside vesicles vs outside vesicles, the high-activity sequences improved noticeably more when encapsulated compared to the low-activity sequences. That is, while all sequences were more active when encapsulated, the greater the original activity of the sequence, the more benefit was conferred by encapsulation. This observation, based on data from thousands of self-aminoacylating ribozyme mutants, actually ran counter to our intuition that the fitness landscape would be “flattened” by encapsulation (which was based on the observed rescue of two folding-deficient mutants of the hairpin ribozyme), a point to which we will return later in this section. This phenomenon, in which high-activity sequences made greater fitness gains, is a type of Matthew effect, seen often in biology and sociology wherein “the rich get richer”.80 Fitter ribozymes gained an even greater advantage when encapsulated.
This observed asymmetry excited us because of its implication for natural selection. Fisher’s Fundamental Theorem of Natural Selection states that the rate of adaptation of a population is proportional to the variance of fitness, i.e., a population with large variance quickly converges to the fittest genotypes, while a population of many similarly fit variants converges slowly. By “helping” better ribozymes more than less-active ribozymes, encapsulation had the effect of increasing the fitness variance among the sequences. Fisher’s theorem is not widely applied in biology because it only accounts for genetic variation and natural selection, ignoring gene-environment interactions, feedback between the population and the fitness landscape, environmental changes, incomplete penetrance, genetic drift, mutation-selection balance,81 and other essential factors. However, in vitro evolution of ribozymes, which have a very tight genotype-phenotype link, has few confounding factors, and thus Fisher’s theorem could hold some weight in these experiments. To test the possible effect of encapsulation on the rate of adaptation, following upon work in our lab mapping RNA fitness landscapes,68,69,72−74 we applied in vitro selection to a pool of ribozyme mutants. The RNA was encapsulated, and the model protocells were incubated with a permeable biotinylated tyrosine analog (biotinyl-Tyr(Me)-oxazolone), producing aminoacyl-RNA inside the vesicles. Organic extraction lysed the vesicles and removed the lipids, and reacted RNAs were isolated by pull-down using streptavidin beads, amplified by RT-PCR, and transcribed for the next round of encapsulation and selection. Upon this selection for self-aminoacylating activity, we found that indeed, ribozymes adapted more quickly when they were encapsulated, compared to either free solution or when outside the vesicles (Figure 1c).3 These results imply that, in addition to improving ribozyme activity, protocell encapsulation would accelerate ribozyme evolution by natural selection.
Why should encapsulation lead to a Matthew effect for ribozyme activity? While the answer is a matter of speculation at this time, some perspective might be gained by considering the shape of the Boltzmann sigmoidal curve, which describes the fraction of molecules occupying one state in a two-state equilibrium (e.g., the fraction of folded RNA molecules) (Figure 2a).82 Different ribozymes sit at different positions on this curve. A well-folded ribozyme is positioned high on the curve, such that a given amount of energetic stabilization (from confinement) gives little increase in the fraction folded, a quantity which is already high. On this part of the curve, higher starting positions yield diminishing returns for a given energetic stabilization. A highly evolved ribozyme may occupy this portion of the curve, in which low-activity mutants benefit more, consistent with the rescue of folding-deficient mutants for the hairpin ribozyme, for which encapsulation brought up the fitness of mutants closer to the wild-type level. On the other hand, poorly folded ribozymes are positioned low on the curve, and the same amount of energetic stabilization could increase the fraction folded quite a bit. In this regime, starting a bit higher on the curve gives a greater boost in the fraction folded for the same energetic stabilization. It is reasonable to suppose that the self-aminoacylating ribozymes evolved in our lab, whose informational region had been deliberately designed to be quite small (21 nucleotides) in order to map a complete sequence space, are generally poorly folded and therefore occupy this lower portion of the curve. For example, the self-aminoacylating ribozymes are predicted to have a minimum free energy of −13 to −16 kcal/mol, compared to −25 kcal/mol for the hairpin ribozyme (based on the ViennaRNA83 nearest-neighbor model84 with 1 M salt, 37 °C). Thus, the positive curvature (positive second derivative) of this region of the sigmoid curve could result in a Matthew effect as observed.
Figure 2.
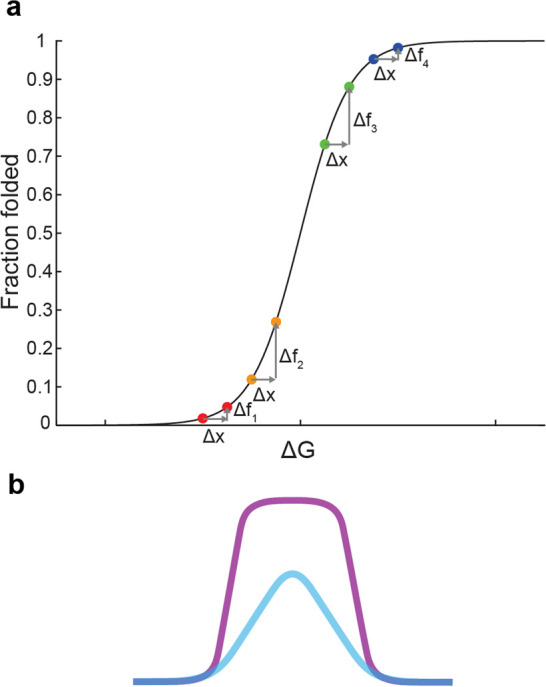
Confinement and evolutionary adaptation. (a) Boltzmann sigmoidal curve for two-state equilibrium. The fraction in one state (e.g., folded conformation) is shown as a function of free energy difference ΔG (Gunfolded – Gfolded). Confinement increases the energy of the unfolded state (Gunfolded) by + Δx, leading to an increase in the fraction of folded molecules (+Δf). If ribozymes are generally well-folded (high on the curve), confinement improves a better-folded variant (blue) less compared to a moderately folded variant (green) for the same increase Δx, i.e., Δf4< Δf3. On the other hand, if the ribozymes are generally poorly folded (low on the curve), confinement improves the better-folded variant (orange) more than it improves the poorly folded variant (red), i.e., Δf2> Δf1, consistent with our observations. (b) Illustrated comparison of a “peak” on a ribozyme fitness landscape with (purple) or without (blue) encapsulation. We suggest that confinement would sharpen the fitness landscape at low fitness, as observed for self-aminocylating RNAs, leading to greater slope in this region of sequence space. At the same time, confinement would rescue mutants close to high fitness, as observed for the hairpin ribozyme, leading to flattening of the peak apex.
While the curve is drawn with respect to free energy difference, a similar curve might apply to molecular fitness, such as if ribozyme fitness is proportional to the fraction folded or to the population of an intermediate state. In the case of ribozymes, our prior work has found that the frequency of RNA sequences at increasing fitness falls along a decreasing curve that fits the right side of a log-normal distribution, as inferred from experimental dynamics and high-throughput measurements of in vitro selection.71,73 In other words, a random population of RNAs contains many sequences of low fitness, and higher fitness sequences are increasingly scarce. Compounding this, models of prebiotic synthesis of polymer sequences suggest that abundance generally decreases with length, but a minimal length is usually required for functional activity (see85 for discussion). Therefore, one may speculate that an emerging biochemical system would begin with short, poorly functional RNAs, and therefore encapsulation would initially accelerate adaptation, as we observed for the self-aminoacylating RNAs. Later, once more advanced, well-folded RNAs evolved, encapsulation would slow down adaptation by effectively buffering the sequences against deleterious mutations, as we observed for the hairpin ribozyme. In other words, encapsulation would “sharpen” the fitness landscape at low fitness, and “flatten” the fitness landscape at high fitness, resulting in both a faster climb and greater diversity once the peak is attained (Figure 2b).
Outlook
Looking back at various emergent properties exhibited by protocells encapsulating RNA, one might be delighted by the range of life-like behaviors. The work described here also brings up additional possibilities. For example, a predicted consequence of the experimental Matthew effect observed for self-aminoacylating RNAs is that encapsulation should also increase the information capacity of the ribozyme genome. This expectation is based on the principle that the amount of information (Lmax) that can be stably maintained in a replicating system is limited by both the mutation rate (μ) and the relative fitness difference (f) (in the classic error threshold model, Lmax = (ln f)/μ).86,87 Thus, higher fitness advantages brought about by encapsulation should allow greater complexity to evolve. At the same time, one might also fairly ask whether these behaviors, such as accelerated evolution and mutational tolerance, could have been predicted in advance, and if so, whether such systems are truly life-like. While behaviors should always be physically rationalizable post hoc, protocells are approaching a fascinating point where the chemical system becomes complex enough to be fundamentally unpredictable due to a combinatorial explosion in the number of possible sequence and chemical states. In addition to the RNA sequences (whose potential space grows exponentially with molecular length), the membranes and their effects on RNA also represent a rich source of diversity and dynamism. This might be appreciated in the already richly altered evolutionary behavior anticipated on the basis of the physical confinement of encapsulation. While we have focused on membrane-bound protocells, protocells have also been modeled using membraneless aqueous two-phase systems (e.g., complex coacervates; reviewed elsewhere88−90) or, more recently, membrane-encapsulated coacervates.91 The biophysical environment of these compartments is also significantly altered and may have surprising effects on the RNA system.92−95 The history of life is a story of exceptions rather than averages, and the synthetic biology of protocells may soon join this categorization. Fortunately, technical innovations are continuously extending our analytical reach, stimulating the community of researchers dedicated to the humble, yet exceptional, protocell.
Acknowledgments
Recent funding for the work described in this Account has been granted by NASA (80NSSC21K0595), NSF (2318736, 1935087), the Simons Foundation (290356FY18), and the Sloan (AWD103574) and Moore (AWD103653) Foundations. We thank Jack Szostak, Ken Dill, Sarah Woodson, Anna Wang, Kevin Plaxco, and the members of the Simons Collaboration on the Origins of Life for insights and encouragement for these projects. We thank past and present members of the Chen lab, including Yei-Chen Lai, Huan Peng, Samuel Verbanic, Yuning Shen, Celia Blanco, Alberto Vázquez-Salazar, Evan Janzen, Abe Pressman, Jose Jimenez, Ramon Xulvi-Brunet, Greg Campbell, and Sudha Rajamani, and our collaborators Ziwei Liu, Sabine Müller, Gerald Joyce, Sheref Mansy, Ulrich Müller, Burckhard Seelig, Robert Pascal, Nate Charest, Joan-Emma Shea, and Xiao Heng for their expertise and enthusiasm in our joint endeavors.
Biographies
Ranajay Saha is currently an Assistant Project Scientist in Irene A. Chen’s laboratory at the University of California, Los Angeles. He studied Biotechnology at Jadavpur University, India, and received his Ph.D. in Biochemistry from the University of Calcutta, India. He carried out postdoctoral research (with Irene A. Chen) at the University of California, Santa Barbara. His current research interests are focused on the emergence of catalytic RNAs during the origin of life.
Jongseok A. Choi is currently a Ph.D. candidate in Irene Chen’s laboratory at the University of California, Los Angeles. He earned his B.S. degree in Chemical and Biomolecular Engineering from the same institute. His primary research interests lie in the areas of the origin of life, RNA World theory, and protocells.
Irene A. Chen is a Professor of Chemical and Biomolecular Engineering and Chemistry and Biochemistry at the University of California, Los Angeles. She attended Harvard University, earning degrees in Chemistry (A.B.), Biophysics (Ph.D., advised by Jack Szostak), and Medicine (M.D.), and completing a postdoctoral Bauer Fellowship in systems biology. Her laboratory studies synthetic protocells and phage engineering for both fundamental understanding and biomedical applications.
Author Contributions
CRediT: Ranajay Saha conceptualization, investigation, visualization, writing-original draft, writing-review & editing; Jongseok A Choi conceptualization, visualization, writing-original draft, writing-review & editing; Irene A. Chen conceptualization, funding acquisition, supervision, writing-original draft, writing-review & editing.
The authors declare no competing financial interest.
Special Issue
Published as part of Accounts of Chemical Researchvirtual special issue “Prebiotic Catalysis”.
References
- Saha R.; Verbanic S.; Chen I. A. Lipid Vesicles Chaperone an Encapsulated RNA Aptamer. Nat. Commun. 2018, 9 (1), 2313. 10.1038/s41467-018-04783-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H.; Lelievre A.; Landenfeld K.; Müller S.; Chen I. A. Vesicle Encapsulation Stabilizes Intermolecular Association and Structure Formation of Functional RNA and DNA. Curr. Biol. 2022, 32 (1), 86–96.e6. 10.1016/j.cub.2021.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.-C.; Liu Z.; Chen I. A. Encapsulation of Ribozymes inside Model Protocells Leads to Faster Evolutionary Adaptation. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (21), e2025054118 10.1073/pnas.2025054118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. C. The Origin of the Genetic Code. J. Mol. Biol. 1968, 38 (3), 367–379. 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Woese C. R.The Genetic Code: The Molecular Basis for Genetic Expression; Harper & Row, 1967. [Google Scholar]
- Orgel L. E. Evolution of the Genetic Apparatus. J. Mol. Biol. 1968, 38 (3), 381–393. 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Robertson M. P.; Joyce G. F. The Origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4 (5), a003608–a003608. 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs P. G.; Lehman N. The RNA World: Molecular Cooperation at the Origins of Life. Nat. Rev. Genet. 2015, 16 (1), 7–17. 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- Pressman A.; Blanco C.; Chen I. A. The RNA World as a Model System to Study the Origin of Life. Curr. Biol. 2015, 25 (19), R953–R963. 10.1016/j.cub.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Jeancolas C.; Gillen C.; McMahon S.; Vickers P. Is Astrobiology Serious Science?. Nat. Astron. 2024, 8 (1), 5–7. 10.1038/s41550-023-02165-9. [DOI] [Google Scholar]
- Simpson G. G. The Nonprevalence of Humanoids: We Can Learn More about Life from Terrestrial Forms than We Can from Hypothetical Extraterrestrial Forms. Science 1964, 143 (3608), 769–775. 10.1126/science.143.3608.769. [DOI] [PubMed] [Google Scholar]
- Origins of Life: The Central Concepts; Deamer D. W., Fleischaker G. R., Eds.; Jones and Bartlett Publishers: Boston, 1994. [Google Scholar]
- Nowak M. A.; Highfield R.. SuperCooperators: Altruism, Evolution, and Why We Need Each Other to Succeed, 1. Free Press trade paperback ed.; Free Press: New York, NY, 2012. [Google Scholar]
- Joyce G. F.; Szostak J. W. Protocells and RNA Self-Replication. Cold Spring Harb. Perspect. Biol. 2018, 10 (9), a034801. 10.1101/cshperspect.a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmáry E.; Demeter L. Group Selection of Early Replicators and the Origin of Life. J. Theor. Biol. 1987, 128 (4), 463–486. 10.1016/S0022-5193(87)80191-1. [DOI] [PubMed] [Google Scholar]
- Traulsen A.; Nowak M. A. Evolution of Cooperation by Multilevel Selection. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (29), 10952–10955. 10.1073/pnas.0602530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansho Y.; Furubayashi T.; Ichihashi N.; Yomo T. Host-Parasite Oscillation Dynamics and Evolution in a Compartmentalized RNA Replication System. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (15), 4045–4050. 10.1073/pnas.1524404113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S.; Kun Á.; Ryckelynck M.; Coldren F.; Szilágyi A.; Jossinet F.; Rick C.; Nghe P.; Szathmáry E.; Griffiths A. D. Transient Compartmentalization of RNA Replicators Prevents Extinction Due to Parasites. Science 2016, 354 (6317), 1293–1296. 10.1126/science.aag1582. [DOI] [PubMed] [Google Scholar]
- Ichihashi N.; Usui K.; Kazuta Y.; Sunami T.; Matsuura T.; Yomo T. Darwinian Evolution in a Translation-Coupled RNA Replication System within a Cell-like Compartment. Nat. Commun. 2013, 4 (1), 2494. 10.1038/ncomms3494. [DOI] [PubMed] [Google Scholar]
- Shah; De Bouter; Pauli; Tupper; Higgs Survival of RNA Replicators Is Much Easier in Protocells than in Surface-Based, Spatial Systems. Life 2019, 9 (3), 65. 10.3390/life9030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.; Szostak J. W. Lipid Constituents of Model Protocell Membranes. Emerg. Top. Life Sci. 2019, 3 (5), 537–542. 10.1042/ETLS20190021. [DOI] [PubMed] [Google Scholar]
- Li H.; Yan Y.; Chen J.; Shi K.; Song C.; Ji Y.; Jia L.; Li J.; Qiao Y.; Lin Y. Artificial Receptor-Mediated Phototransduction toward Protocellular Subcompartmentalization and Signaling-Encoded Logic Gates. Sci. Adv. 2023, 9 (9), eade5853 10.1126/sciadv.ade5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W.; Bartel D. P.; Luisi P. L. Synthesizing Life. Nature 2001, 409 (6818), 387–390. 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Zhu T. F.; Szostak J. W. Coupled Growth and Division of Model Protocell Membranes. J. Am. Chem. Soc. 2009, 131 (15), 5705–5713. 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczyc M. M.; Fujikawa S. M.; Szostak J. W. Experimental Models of Primitive Cellular Compartments: Encapsulation, Growth, and Division. Science 2003, 302 (5645), 618–622. 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A.; Walde P. From Self-Assembled Vesicles to Protocells. Cold Spring Harb. Perspect. Biol. 2010, 2 (7), a002170. 10.1101/cshperspect.a002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard P.-A.; Apel C. L.; Kanavarioti A.; Deamer D. W. Influence of Ionic Inorganic Solutes on Self-Assembly and Polymerization Processes Related to Early Forms of Life: Implications for a Prebiotic Aqueous Medium. Astrobiology 2002, 2 (2), 139–152. 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]
- Monnard P.-A.; Walde P. Current Ideas about Prebiological Compartmentalization. Life 2015, 5 (2), 1239–1263. 10.3390/life5021239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Mirazo K.; Briones C.; De La Escosura A. Prebiotic Systems Chemistry: New Perspectives for the Origins of Life. Chem. Rev. 2014, 114 (1), 285–366. 10.1021/cr2004844. [DOI] [PubMed] [Google Scholar]
- O’Flaherty D. K.; Kamat N. P.; Mirza F. N.; Li L.; Prywes N.; Szostak J. W. Copying of Mixed-Sequence RNA Templates inside Model Protocells. J. Am. Chem. Soc. 2018, 140 (15), 5171–5178. 10.1021/jacs.8b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K.; Szostak J. W. Nonenzymatic Template-Directed RNA Synthesis Inside Model Protocells. Science 2013, 342 (6162), 1098–1100. 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toparlak Ö. D.; Sebastianelli L.; Egas Ortuno V.; Karki M.; Xing Y.; Szostak J. W.; Krishnamurthy R.; Mansy S. S. Cyclophospholipids Enable a Protocellular Life Cycle. ACS Nano 2023, 17 (23), 23772–23783. 10.1021/acsnano.3c07706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A.; Salehi-Ashtiani K.; Szostak J. W. RNA Catalysis in Model Protocell Vesicles. J. Am. Chem. Soc. 2005, 127 (38), 13213–13219. 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toparlak Ö. D.; Wang A.; Mansy S. S. Population-Level Membrane Diversity Triggers Growth and Division of Protocells. JACS Au 2021, 1 (5), 560–568. 10.1021/jacsau.0c00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.; Deckel Y.; Lowe L. A.; Loo D. W. K.; Yomo T.; Szostak J. W.; Nisler C.; Wang A. Lipid Exchange Promotes Fusion of Model Protocells. Small Methods 2023, 7 (12), 2300126. 10.1002/smtd.202300126. [DOI] [PubMed] [Google Scholar]
- Walde P.; Umakoshi H.; Stano P.; Mavelli F. Emergent Properties Arising from the Assembly of Amphiphiles. Artificial Vesicle Membranes as Reaction Promoters and Regulators. Chem. Commun. 2014, 50 (71), 10177–10197. 10.1039/C4CC02812K. [DOI] [PubMed] [Google Scholar]
- Chen I. A.; Roberts R. W.; Szostak J. W. The Emergence of Competition Between Model Protocells. Science 2004, 305 (5689), 1474–1476. 10.1126/science.1100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank T.; Vogele K.; Dupin A.; Simmel F. C.; Pirzer T. Growth of Giant Peptide Vesicles Driven by Compartmentalized Transcription-Translation Activity. Chem. - Eur. J. 2020, 26 (72), 17356–17360. 10.1002/chem.202003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salibi E.; Peter B.; Schwille P.; Mutschler H. Periodic Temperature Changes Drive the Proliferation of Self-Replicating RNAs in Vesicle Populations. Nat. Commun. 2023, 14 (1), 1222. 10.1038/s41467-023-36940-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart A. E.; Adamala K. P.; Szostak J. W. A Simple Physical Mechanism Enables Homeostasis in Primitive Cells. Nat. Chem. 2016, 8 (5), 448–453. 10.1038/nchem.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara K.; Okura Y.; Matsuo M.; Toyota T.; Suzuki K.; Sugawara T. A Recursive Vesicle-Based Model Protocell with a Primitive Model Cell Cycle. Nat. Commun. 2015, 6 (1), 8352. 10.1038/ncomms9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M.; Kan Y.; Kurihara K.; Jimbo T.; Imai M.; Toyota T.; Hirata Y.; Suzuki K.; Sugawara T. DNA Length-Dependent Division of a Giant Vesicle-Based Model Protocell. Sci. Rep. 2019, 9 (1), 6916. 10.1038/s41598-019-43367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.; Blokhuis A.; Turk-MacLeod R.; Karuppusamy J.; Franconi A.; Woronoff G.; Jeancolas C.; Abrishamkar A.; Loire E.; Ferrage F.; Pelupessy P.; Jullien L.; Szathmary E.; Nghe P.; Griffiths A. D. Small-Molecule Autocatalysis Drives Compartment Growth, Competition and Reproduction. Nat. Chem. 2024, 16 (1), 70–78. 10.1038/s41557-023-01276-0. [DOI] [PubMed] [Google Scholar]
- Minton A. P. The Influence of Macromolecular Crowding and Macromolecular Confinement on Biochemical Reactions in Physiological Media. J. Biol. Chem. 2001, 276 (14), 10577–10580. 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B.; Trach S. O. Estimation of Macromolecule Concentrations and Excluded Volume Effects for the Cytoplasm of Escherichia Coli. J. Mol. Biol. 1991, 222 (3), 599–620. 10.1016/0022-2836(91)90499-V. [DOI] [PubMed] [Google Scholar]
- Eral H. B.; Oh J. M.; Van Den Ende D.; Mugele F.; Duits M. H. G. Anisotropic and Hindered Diffusion of Colloidal Particles in a Closed Cylinder. Langmuir 2010, 26 (22), 16722–16729. 10.1021/la102273n. [DOI] [PubMed] [Google Scholar]
- Minton A. P. Excluded Volume as a Determinant of Macromolecular Structure and Reactivity. Biopolymers 1981, 20 (10), 2093–2120. 10.1002/bip.1981.360201006. [DOI] [Google Scholar]
- Marenduzzo D.; Finan K.; Cook P. R. The Depletion Attraction: An Underappreciated Force Driving Cellular Organization. J. Cell Biol. 2006, 175 (5), 681–686. 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X.; Rivas G.; Minton A. P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37, 375–397. 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel B. P.; Rueda D. Molecular Crowding Accelerates Ribozyme Docking and Catalysis. J. Am. Chem. Soc. 2014, 136 (48), 16700–16703. 10.1021/ja5073146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy K. A.; Assmann S. M.; Mathews D. H.; Bevilacqua P. C. Bridging the Gap between in Vitro and in Vivo RNA Folding. Q. Rev. Biophys. 2016, 49, e10 10.1017/S003358351600007X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta S.; Zhang S.; Szostak J. W. Molecular Crowding Facilitates Ribozyme-Catalyzed RNA Assembly. ACS Cent. Sci. 2023, 9 (8), 1670–1678. 10.1021/acscentsci.3c00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-T.; Kilburn D.; Behrouzi R.; Briber R. M.; Woodson S. A. Molecular Crowding Overcomes the Destabilizing Effects of Mutations in a Bacterial Ribozyme. Nucleic Acids Res. 2015, 43 (2), 1170–1176. 10.1093/nar/gku1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D.; Roh J. H.; Behrouzi R.; Briber R. M.; Woodson S. A. Crowders Perturb the Entropy of RNA Energy Landscapes to Favor Folding. J. Am. Chem. Soc. 2013, 135 (27), 10055–10063. 10.1021/ja4030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D.; Roh J. H.; Guo L.; Briber R. M.; Woodson S. A. Molecular Crowding Stabilizes Folded RNA Structure by the Excluded Volume Effect. J. Am. Chem. Soc. 2010, 132 (25), 8690–8696. 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R.; Kilburn D.; Lee H.-T.; Woodson S. A. Increased Ribozyme Activity in Crowded Solutions. J. Biol. Chem. 2014, 289 (5), 2972–2977. 10.1074/jbc.M113.527861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta S. Molecular Crowding and RNA Catalysis. Org. Biomol. Chem. 2020, 18 (39), 7724–7739. 10.1039/D0OB01695K. [DOI] [PubMed] [Google Scholar]
- Saha R.; Pohorille A.; Chen I. A. Molecular Crowding and Early Evolution. Orig. Life Evol. Biospheres 2014, 44 (4), 319–324. 10.1007/s11084-014-9392-3. [DOI] [PubMed] [Google Scholar]
- Wilson D. S.; Szostak J. W. In Vitro Selection of Functional Nucleic Acids. Annu. Rev. Biochem. 1999, 68 (1), 611–647. 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Babendure J. R.; Adams S. R.; Tsien R. Y. Aptamers Switch on Fluorescence of Triphenylmethane Dyes. J. Am. Chem. Soc. 2003, 125 (48), 14716–14717. 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- Saha R.; Kao W.-L.; Malady B.; Heng X.; Chen I. A. Effect of Montmorillonite K10 Clay on RNA Structure and Function. Biophys. J. 2023, 123 (4), 451–463. 10.1016/j.bpj.2023.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza T. P.; Bossa G. V.; Stano P.; Steiniger F.; May S.; Luisi P. L.; Fahr A. Vesicle Aggregates as a Model for Primitive Cellular Assemblies. Phys. Chem. Chem. Phys. 2017, 19 (30), 20082–20092. 10.1039/C7CP03751A. [DOI] [PubMed] [Google Scholar]
- Suga K.; Tanabe T.; Tomita H.; Shimanouchi T.; Umakoshi H. Conformational Change of Single-Stranded RNAs Induced by Liposome Binding. Nucleic Acids Res. 2011, 39 (20), 8891–8900. 10.1093/nar/gkr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell J.; Weeks K. M.; Pielak G. J. Challenge of Mimicking the Influences of the Cellular Environment on RNA Structure by PEG-Induced Macromolecular Crowding. Biochemistry 2015, 54 (42), 6447–6453. 10.1021/acs.biochem.5b00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X.; Dill K. A. Stabilization of Proteins in Confined Spaces. Biochemistry 2001, 40 (38), 11289–11293. 10.1021/bi0155504. [DOI] [PubMed] [Google Scholar]
- Patel B. H.; Percivalle C.; Ritson D. J.; Duffy C. D.; Sutherland J. D. Common Origins of RNA, Protein and Lipid Precursors in a Cyanosulfidic Protometabolism. Nat. Chem. 2015, 7 (4), 301–307. 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X.; Kim H.; Pereira M. J. B.; Babcock H. P.; Walter N. G.; Chu S. Correlating Structural Dynamics and Function in Single Ribozyme Molecules. Science 2002, 296 (5572), 1473–1476. 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- Athavale S. S.; Spicer B.; Chen I. A. Experimental Fitness Landscapes to Understand the Molecular Evolution of RNA-Based Life. Curr. Opin. Chem. Biol. 2014, 22, 35–39. 10.1016/j.cbpa.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Jiménez J. I.; Xulvi-Brunet R.; Campbell G. W.; Turk-MacLeod R.; Chen I. A. Comprehensive Experimental Fitness Landscape and Evolutionary Network for Small RNA. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (37), 14984–14989. 10.1073/pnas.1307604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xulvi-Brunet R.; Campbell G. W.; Rajamani S.; Jiménez J. I.; Chen I. A. Computational Analysis of Fitness Landscapes and Evolutionary Networks from in Vitro Evolution Experiments. Methods 2016, 106, 86–96. 10.1016/j.ymeth.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Pressman A.; Moretti J. E.; Campbell G. W.; Müller U. F.; Chen I. A. Analysis of in Vitro Evolution Reveals the Underlying Distribution of Catalytic Activity among Random Sequences. Nucleic Acids Res. 2017, 45 (14), 8167–8179. 10.1093/nar/gkx540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco C.; Janzen E.; Pressman A.; Saha R.; Chen I. A. Molecular Fitness Landscapes from High-Coverage Sequence Profiling. Annu. Rev. Biophys. 2019, 48, 1–18. 10.1146/annurev-biophys-052118-115333. [DOI] [PubMed] [Google Scholar]
- Pressman A. D.; Liu Z.; Janzen E.; Blanco C.; Müller U. F.; Joyce G. F.; Pascal R.; Chen I. A. Mapping a Systematic Ribozyme Fitness Landscape Reveals a Frustrated Evolutionary Network for Self-Aminoacylating RNA. J. Am. Chem. Soc. 2019, 141 (15), 6213–6223. 10.1021/jacs.8b13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen E.; Shen Y.; Vázquez-Salazar A.; Liu Z.; Blanco C.; Kenchel J.; Chen I. A. Emergent Properties as By-Products of Prebiotic Evolution of Aminoacylation Ribozymes. Nat. Commun. 2022, 13 (1), 3631. 10.1038/s41467-022-31387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest N.; Shen Y.; Lai Y.-C.; Chen I. A.; Shea J.-E. Discovering Pathways through Ribozyme Fitness Landscapes Using Information Theoretic Quantification of Epistasis. RNA 2023, 29 (11), 1644–1657. 10.1261/rna.079541.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani S.; Ichida J. K.; Antal T.; Treco D. A.; Leu K.; Nowak M. A.; Szostak J. W.; Chen I. A. Effect of Stalling after Mismatches on the Error Catastrophe in Nonenzymatic Nucleic Acid Replication. J. Am. Chem. Soc. 2010, 132 (16), 5880–5885. 10.1021/ja100780p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke C. O.; Wang J. L.; Ofria C.; Lenski R. E.; Adami C. Evolution of Digital Organisms at High Mutation Rates Leads to Survival of the Flattest. Nature 2001, 412 (6844), 331–333. 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- Codoñer F. M.; Darós J.-A.; Solé R. V.; Elena S. F. The Fittest versus the Flattest: Experimental Confirmation of the Quasispecies Effect with Subviral Pathogens. PLoS Pathog. 2006, 2 (12), e136 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Pressman A.; Janzen E.; Chen I. A. Kinetic Sequencing (k-Seq) as a Massively Parallel Assay for Ribozyme Kinetics: Utility and Critical Parameters. Nucleic Acids Res. 2021, 49 (12), e67 10.1093/nar/gkab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merton R. K. The Matthew Effect in Science: The Reward and Communication Systems of Science Are Considered. Science 1968, 159 (3810), 56–63. 10.1126/science.159.3810.56. [DOI] [PubMed] [Google Scholar]
- Basener W. F.; Sanford J. C. The Fundamental Theorem of Natural Selection with Mutations. J. Math. Biol. 2018, 76 (7), 1589–1622. 10.1007/s00285-017-1190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J.-M.; Ouanounou G.; Rouzaire-Dubois B. The Boltzmann Equation in Molecular Biology. Prog. Biophys. Mol. Biol. 2009, 99 (2–3), 87–93. 10.1016/j.pbiomolbio.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Gruber A. R.; Lorenz R.; Bernhart S. H.; Neubock R.; Hofacker I. L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H.; Disney M. D.; Childs J. L.; Schroeder S. J.; Zuker M.; Turner D. H. Incorporating Chemical Modification Constraints into a Dynamic Programming Algorithm for Prediction of RNA Secondary Structure. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (19), 7287–7292. 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr J.; Manapat M. L.; Rajamani S.; Leu K.; Xulvi-Brunet R.; Joseph I.; Nowak M. A.; Chen I. A. Prebiotically Plausible Mechanisms Increase Compositional Diversity of Nucleic Acid Sequences. Nucleic Acids Res. 2012, 40 (10), 4711–4722. 10.1093/nar/gks065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften 1971, 58 (10), 465–523. 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Tupper A. S.; Higgs P. G. Error Thresholds for RNA Replication in the Presence of Both Point Mutations and Premature Termination Errors. J. Theor. Biol. 2017, 428, 34–42. 10.1016/j.jtbi.2017.05.037. [DOI] [PubMed] [Google Scholar]
- Lin Z.; Beneyton T.; Baret J.-C.; Martin N. Coacervate Droplets for Synthetic Cells. Small Methods 2023, 7 (12), e2300496 10.1002/smtd.202300496. [DOI] [PubMed] [Google Scholar]
- Abbas M.; Lipiński W. P.; Wang J.; Spruijt E. Peptide-Based Coacervates as Biomimetic Protocells. Chem. Soc. Rev. 2021, 50 (6), 3690–3705. 10.1039/D0CS00307G. [DOI] [PubMed] [Google Scholar]
- Crowe C. D.; Keating C. D. Liquid-Liquid Phase Separation in Artificial Cells. Interface Focus 2018, 8 (5), 20180032. 10.1098/rsfs.2018.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N.; Mann S. Membranized Coacervate Microdroplets: From Versatile Protocell Models to Cytomimetic Materials. Acc. Chem. Res. 2023, 56 (3), 297–307. 10.1021/acs.accounts.2c00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobot B.; Iglesias-Artola J. M.; Le Vay K.; Mayr V.; Kar M.; Kreysing M.; Mutschler H.; Tang T.-Y. D. Compartmentalised RNA Catalysis in Membrane-Free Coacervate Protocells. Nat. Commun. 2018, 9 (1), 3643. 10.1038/s41467-018-06072-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudyal R. R.; Guth-Metzler R. M.; Veenis A. J.; Frankel E. A.; Keating C. D.; Bevilacqua P. C. Template-Directed RNA Polymerization and Enhanced Ribozyme Catalysis inside Membraneless Compartments Formed by Coacervates. Nat. Commun. 2019, 10 (1), 490. 10.1038/s41467-019-08353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vay K. K.; Salibi E.; Ghosh B.; Tang T. Y. D.; Mutschler H. Ribozyme Activity Modulates the Physical Properties of RNA-Peptide Coacervates. eLife 2023, 12, e83543 10.7554/eLife.83543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson C. A.; Molden R. C.; Keating C. D.; Bevilacqua P. C. RNA Catalysis through Compartmentalization. Nat. Chem. 2012, 4 (11), 941–946. 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]