Abstract
Background
Previous research has revealed the potential impact of circadian rhythms on pulmonary diseases; however, the connection between circadian rhythm-associated Thyrotroph Embryonic Factor (TEF) and Pulmonary Arterial Hypertension (PAH) remains unclear. We aim to assess the genetic causal relationship between TEF and PAH by utilizing two sets of genetic instrumental variables (IV) and publicly available Pulmonary Arterial Hypertension Genome-Wide Association Studies (GWAS).
Methods
Total of 23 independent TEF genetic IVs from recent MR reports and PAH GWAS including 162,962 European individuals were used to perform this two-sample MR study. Gain- and loss-of-function experiments were used to demonstrate the role of TEF in PAH.
Results
Our analysis revealed that as TEF levels increased genetically, there was a corresponding increase in the risk of PAH, as evidenced by IVW (OR = 1.233, 95% CI: 1.054–1.441; P = 0.00871) and weighted median (OR = 1.292, 95% CI for OR: 1.064–1.568; P = 0.00964) methods. Additionally, the up-regulation of TEF expression was associated with a significantly higher likelihood of abnormal circadian rhythm (IVW: P = 0.0024733, β = 0.05239). However, we did not observe a significant positive correlation between circadian rhythm and PAH (IVW: P = 0.3454942, β = 1.4980398). In addition, our in vitro experiments demonstrated that TEF is significantly overexpressed in pulmonary artery smooth muscle cells (PASMCs). And overexpression of TEF promotes PASMC viability and migratory capacity, as well as upregulates the levels of inflammatory cytokines.
Conclusion
Our analysis suggests a causal relationship between genetically increased TEF levels and an elevated risk of both PAH and abnormal circadian rhythm. Consequently, higher TEF levels may represent a risk factor for individuals with PAH.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12931-024-02934-8.
Keywords: Mendelian randomization (MR), Thyrotroph embryonic factor (TEF), Pulmonary arterial hypertension (PAH), Circadian rhythm, GWAS
Introduction
Pulmonary arterial hypertension (PAH) is a pathobiological condition, defined by hemodynamics as an elevation in the average pulmonary artery pressure exceeding 25 or 30 mmHg at rest [1–3]. It represents a rare and severe chronic pulmonary vascular disease. The most common types of PAH include idiopathic PAH and PAH associated with connective tissue diseases [4]. The symptoms of PAH lack specificity, leading to a delay of months or even years from symptom onset to diagnosis [5]. Notably, even minor exertion can lead to frequent syncope in patients with pulmonary hypertension, clearly indicating a life-threatening high mortality rate of PAH [6]. Currently, the therapeutic approach for PAH aims to rectify the imbalance between vascular activity and vasodilatory mediators while restoring endothelial cell function [7]. And the traditional Pharmacological treatments include prostacyclin analogs, Phosphodiesterase (PDE)-5 inhibitors, endothelin receptor antagonists, and cGMP activators [8–10]. Despite some progress in PAH treatment, it remains an exceptionally serious disease, posing challenges in diagnosis, treatment, and prevention. Clearly, further investigation is warranted to identify the risk factors leading to pulmonary hypertension and evaluate high-risk patients, enabling timely prevention and early intervention.
Thyrotroph embryonic factor (TEF) is a crucial regulatory transcription factor within the circadian rhythm pathway. TEF combines with various other important transcription activators associated with circadian rhythms at the D-box promoter site to enhance the transcription of numerous clock-controlled genes and pivotal genes within the core circadian rhythm feedback loop, such as NR1D1, NR1D2, PER, and CRY [11, 12]. There is increasing evidence to suggest that lung function is significantly influenced by circadian rhythms [13]. It is widely believed that the disruption of the biological clock, whether caused by genetics or environmental factors, is perceived as a stressor by organisms and can have both short-term and long-term effects on health, increasing the risk of pulmonary diseases and metabolic syndrome [13]. To date, research on circadian rhythms has revealed how they play a role in the pathogenesis of various diseases, including cancer, Alzheimer’s disease, and cardiovascular diseases [14–16]. However, there is currently no evidence to suggest a connection between TEF and PAH.
Observational research often comes with inherent limitations stemming from factors like measurement errors, potential bias, and the possibility of reverse causality. Mendelian randomization (MR) analysis is a genetic approach used for assessing variables and typically leverages single nucleotide polymorphisms (SNPs) in genome-wide association studies (GWAS) to unveil causal relationships between a given phenotype and its resulting outcome [17]. In this study, we validated the association between TEF and PAH through Mendelian randomization analysis, providing novel insights and therapeutic targets for PAH treatment.
Methods
Data sources
The PAH rat datasets presented in our study can be found in Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA732522/). And the CTEPH samples were obtained from TCGA (GSE221511). The All genome-wide association study (GWAS) summary data used in this study were obtained from the IEU GWAS Database (https://gwas.mrcieu.ac.uk/). This research was carried out within a European population, involving the extraction of data related to “TEF” and “PAH” from GWAS datasets. And the specific information is provided in Table 1.
Table 1.
Genome-wide association study (GWAS) of pulmonary arterial hypertension (PAH) and TEF
| GWAS ID | Year | Trait | sample | nsnp | Population | Sex |
|---|---|---|---|---|---|---|
| eqtl-a-ENSG00000167074 | 2018 | TEF | 31,684 | 17,655 | European | Males and Females |
| finn-b-I9_HYPTENSPUL | 2021 | Hypertension, Pulmonary Arterial | 162,962 | 16,380,163 | European | Males and Females |
| GCST003837 | 2016 | Circadian rhythm | 89,283 | 16,760,980 | European | NA |
Mendelian randomization analysis
In this study, a bidirectional Mendelian randomization approach was employed using a two-sample instrumental variable analysis to evaluate the causal relationship between TEF and PAH. Instrumental variables were chosen from exposure factors. Subsequently, we applied the PLINK clustering method with a clustering threshold (r2 < 0.001, kb = 100) to eliminate SNPs with bias due to linkage disequilibrium (LD), and low-quality SNPs were removed based on allele frequencies and incompatible alleles. We retained only those SNPs that showed significant associations with the phenotypes of “TEF” and “PAH”.
Pleiotropy and heterogeneity test
To assess heterogeneity and test for pleiotropy effects, we employed three different MR methods, including inverse variance weighted (IVW), MR Egger, and weighted median, with IVW being a widely accepted analytical approach. Additionally, we conducted Cochran’s Q statistic test to evaluate heterogeneity, where lower heterogeneity indicates more reliable MR results. Horizontal pleiotropy assessment was performed using the MR Egger algorithm, and we also conducted leave-one-out sensitivity analysis to assess the influence of individual SNPs on causal effect estimates.
Cell culture
Primary VSMCs were obtained from the pulmonary aorta of SPF Sprague-Dawley (SD) rats (weighing 150–180 g) using the tissue explant method, as previously described [18]. The rat vascular smooth muscle cells (rVSMCs) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, C11995500BT, Gibco) containing 20% fetal bovine serum (FBS, 10270-106, Gibco) and 100 U/ml penicillin/streptomycin (15140-122, Gibco). Primary rat VSMCs from passages 4 to 7 were used for the experiments. VSMCs were treated with PDGF-BB (25 ng/ml) to construct an in vitro PAH model for subsequent experiments.
Western blot
Collect the pulmonary artery tissues from rats in each group and add RIPA lysis buffer (Biosharp, China) containing 1 mM protein phosphatase inhibitor for protein extraction. Then, centrifuge the samples at 12,000 rpm for 15 min at 4 °C to obtain the supernatant, and determine the protein concentration using a BCA protein assay kit (ThermoFisher Scientific). Separate the proteins on a 10% sodium dodecyl sulfate-polyacrylamide gel and transfer them onto a PVDF membrane (Bio-Rad, Hercules, CA, USA). Block the membrane with 5% non-fat milk at room temperature for 1 h, followed by incubation with the TEF antibody (1:1000, PA5-106495, ThermoFisher Scientific). Next, wash the membrane three times with TBST and incubate it with the secondary antibody (Goat Anti-Rabbit IgG (H + L)-HRP, 1:5000, ab205718, Abcam) at room temperature for 1 h. Perform chemiluminescence detection using a ChemiDoc XRS + imaging system (Tanon, Shanghai, China). Quantify the protein using ImageJ software.
Immunofluorescence
Fix rVSMCs with 4% PFA for 20 min, then wash with PBS and permeabilize the cells using 0.4% Triton X-100. Next, block the cells with 1% BSA for 30 min, followed by incubation with α-SMA (1:1000, #48938, Cell Signaling Technology) and TEF (1:1000, PA5-106495, ThermoFisher Scientific) antibody overnight at 4 °C. Subsequently, incubate the cells with secondary antibodies (Goat Anti-Mouse IgG (H + L), 1:1000, ab150113, Abcam, and Goat Anti-Rabbit IgG (H + L), 1:1000, 4413 S, Cell Signaling Technology). Finally, stain the nuclei using Hoechst 33,258 (Sigma, 94403). Image the samples using a laser confocal scanning microscope (Olympus FV3000, Tokyo, Japan).
CFDA-SE assay
After culturing rVSMCs under the specified conditions, wash the rVSMCs once with PBS, then add the CFDA-SE working solution to achieve a final concentration of 10 µM, and incubate at 37 °C for 15–30 min. After staining, wash the cells 3 times with PBS, and then incubate with fresh culture medium at 37 °C for 30 min. Finally, observe and analyze the stained cells using a fluorescence microscope.
Enzyme-linked immunosorbent assays (ELISAs)
The levels of IL-6 and IL-1β in the rVSMCs were detected by ELISA kits (EK306 and EK301B, Multi Sciences, China) according to the manufacturer’s instructions.
Transwell assay
A total of 1 × 10^4 cells/100 µL were seeded onto polycarbonate filters (pore size, 8 μm; Corning, NY, USA) and cultured in 200 µL of serum-free DMEM medium. Medium containing 10% FBS was added to the lower chamber, and the two chambers were incubated at 37 °C for 12 h. Subsequently, the upper side of the membrane was fixed with 4% paraformaldehyde at room temperature for 30 min, the top layer of the filter was scrubbed with a sterile cotton swab, and stained with crystal violet (cat. no. G1014, Servicebio, Wuhan, China) at room temperature for 30 min. The cells were examined and imaged using a digital microscope.
Statistical analysis
Statistical analyses were conducted using R software version 4.0. Data were presented as mean ± standard deviation (SD). The IVW, MR Egger, and weighted median analyses were executed through the “TwoSampleMR” package (Version: 0.5.6) and the “MRPRESSO” package (Version: 1.0) within the R software. Statistical significance was defined as P < 0.05.
Results
Effect of TEF on the risk of PAH
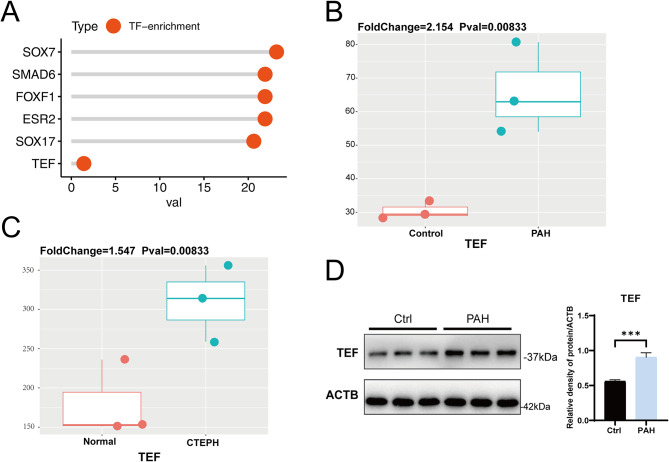
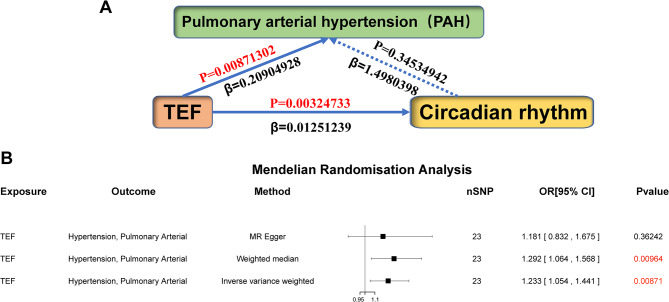
Initially, we identified 124 PAH-related genes and transcription factor analysis revealed TEF as a significantly enriched transcription factor associated with PAH-related genes (Fig. 1A). Through the assessment of TEF expression in human and rat samples, we observed a significant upregulation of TEF in both human and rat PAH samples when compared to the control group (Fig. 1B-D). Subsequently, we collected genetic variation information related to “TEF” and “PAH” associated genes from GWAS and conducted analyses. We selected SNPs strongly correlated with the exposure factors as instrumental variables, and detailed SNP information is provided in Table 1 and Supplementary Table 1. The Mendelian randomization analysis was performed after removing linkage disequilibrium and various confounding factors. As we all know, the positive/negative sign of the β value typically indicates a positive/negative correlation between the exposure factor and the outcome in the MR analysis. And in our study, we observed that the βvalue between TEF and PAH was 0.20904928, suggesting a positive correlation between TEF and PAH (Fig. 2A). Furthermore, our results also unveiled that the high expression of TEF (OR = 1.233, 95% CI, 1.054–1.441, P = 0.00871) could increase the risk of PAH (Fig. 2B).
Fig. 1.
TEF differential expression between PAH and normal samples in rat and human tissues. (A) Enrichment analysis of TEF in the genes related to PAH. (B) The expression of TEF between samples from the Pulmonary Arterial Hypertension (PAH) and control rats. (C) The expression of TEF between the samples from the Chronic Thromboembolic Pulmonary Hypertension (CTEPH) and normal patients. (D) The protein expression of TEF between the samples from the control rat models and PAH rat models
Fig. 2.
Effect of TEF on the risk of pulmonary arterial hypertension (PAH). (A) A schematic diagram illustrating the causal relationship between PAH, TEF, and circadian rhythm. (B) A Mendelian randomization analysis was conducted using TEF as the exposure factor and PAH as the outcome. The results indicated a positive correlation between TEF and PAH, suggesting that the presence of high exposure to TEF increases the risk of PAH
A significant positive correlation between TEF and PAH
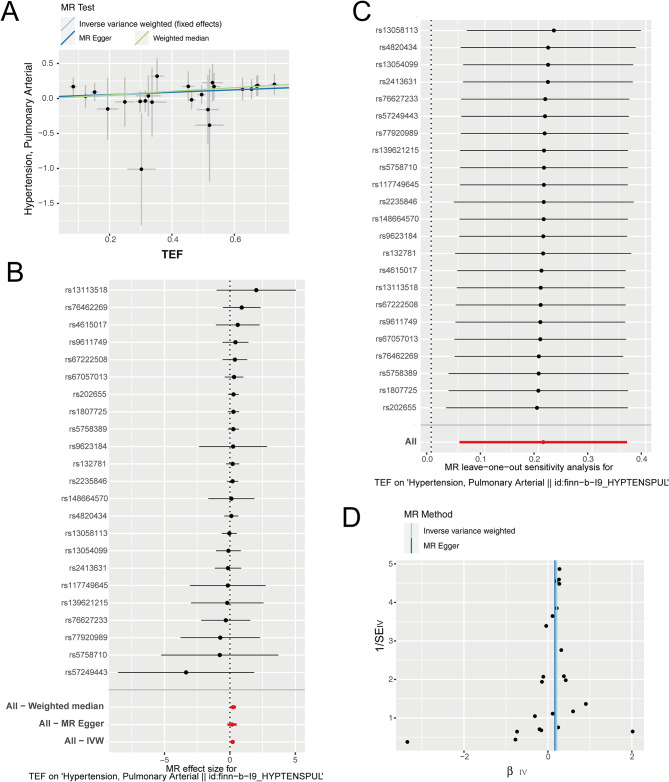
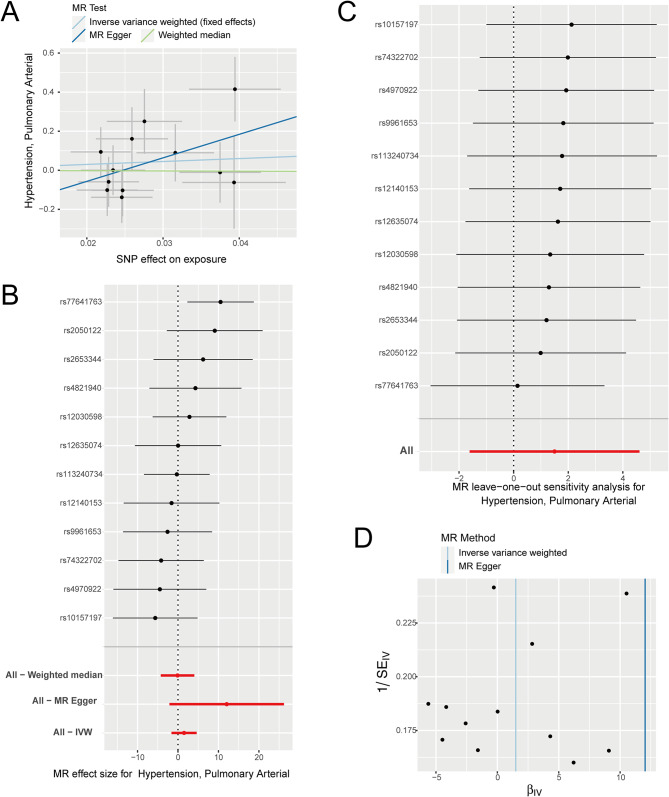
Next, we designate “TEF” as the exposure factor and “PAH” as the outcome factor and proceeded to investigate the correlation between exposure factors and the outcomes. Our results revealed a significant positive correlation between an increased risk of PAH and the exposure factor of TEF (Fig. 3A-B; Table 2). Subsequently, we conducted leave-one-out sensitivity tests to examine whether causal estimates were driven by any single SNP. We observed that the removal of any SNP did not fundamentally impact the results, thereby demonstrating the reliability of our findings (Fig. 3C). To assess heterogeneity among the selected SNPs, we further conducted Cochran’s Q-test. The results indicated no significant differences in either MR Egger or IVW analysis (Fig. 3D; Table 2). Taken together, our findings demonstrate a causal relationship between TEF and PAH. The increased exposure to the factor TEF significantly elevates the risk of PAH.
Fig. 3.
The MR analysis results of TEF as the exposure. (A) Scatter plot of MR analysis, showing the distribution of individual rate estimates for PAH as a result of TEF. Each scatter plot includes trend lines derived from three different MR methods, indicating a positive causal relationship. (B) A forest plot depicts the association between PAH and TEF in the MR analysis. Each circle next to a SNP represents the causal estimate for each IV, with the bottom three circles indicating the results from multi-instrument MR analysis using Weighted Median, Egger Regression, and IVW. The horizontal lines represent the 95% confidence intervals for OR. (C) The leave-one-out sensitivity analysis of the SNPs, with each black dot representing an IVW estimate and red dots representing estimates using all IVs. The horizontal lines represent the 95% confidence intervals. (D) Funnel plot for PAH on MR analysis
Table 2.
The causal association of TEF with pulmonary arterial hypertension (PAH)
| Exposure | Outcome | Method | nsnp | b | pval | egger_intercept | pval | Q | Q_pval |
|---|---|---|---|---|---|---|---|---|---|
| TEF | “Hypertension, Pulmonary Arterial” (no controls excluded) | MR Egger | 23 | 0.14600826 | 0.42065036 | 0.024743302 | 0.77483804 | 7.69271482 | 0.99631514 |
| TEF | “Hypertension, Pulmonary Arterial” (no controls excluded) | Weighted median | 23 | 0.22223992 | 0.02827423 | ||||
| TEF | “Hypertension, Pulmonary Arterial” (no controls excluded) | Inverse variance weighted | 23 | 0.19210706 | 0.01540639 | 7.77667835 | 0.99770367 |
MR analysis between TEF and circadian rhythm
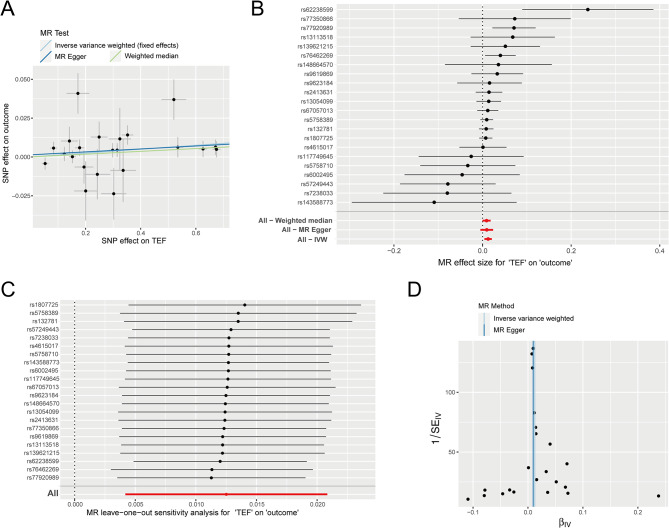
As TEF is one of several key regulatory transcription factors in circadian rhythm pathway, we further explored the correlation between TEF and circadian rhythm. Setting “TEF” as the exposure factor and “circadian rhythm” as the result, the MR analysis showed that there was a significant positive correlation between TEF and circadian rhythm (IVW: P = 0.00324733, β = 0.01251239). The results from the Weighted Median Regression (WMR) (P = 0.04726759, β = 0.00901459) align with those of the IVW method. In addition, the Egger’s intercept shows that the tool variable we selected does not have horizontal pleiotropy. In a word, our results substantiate the relationship between TEF and circadian rhythm (Fig. 4; Table 3).
Fig. 4.
The MR analysis between TEF and circadian rhythm. (A) Scatter plot of MR analysis, showing the distribution of individual rate estimates for circadian rhythm as a result of TEF. Each scatter plot includes trend lines derived from three different MR methods, indicating a positive causal relationship. (B) Forest plot shows the result of MR analysis pleiotropy test. Each circle next to a SNP represents the causal estimate for each IV, with the bottom three circles indicating the results from multi-instrument MR analysis using Weighted Median, Egger Regression, and IVW. The horizontal lines represent the 95% confidence intervals for OR. (C) The leave-one-out sensitivity analysis of the SNPs, with each black dot representing an IVW estimate and red dots representing estimates using all IVs. The horizontal lines represent the 95% confidence intervals. (D) Funnel plot for circadian rhythm on MR analysis
Table 3.
The causal association of TEF with circadian rhythm
| exposure | outcome | method | nsnp | b | pval | egger_intercept | pval | Q | Q_pval |
|---|---|---|---|---|---|---|---|---|---|
| TEF | Circadian rhythm | MR Egger | 22 | 0.00948703 | 0.20895783 | 0.001364577 | 0.613006092 | 29.2128681 | 0.08365176 |
| TEF | Circadian rhythm | Weighted median | 22 | 0.00901459 | 0.04726759 | ||||
| TEF | Circadian rhythm | Inverse variance weighted | 22 | 0.01251239 | 0.00324733 | 29.5984985 | 0.10035802 |
MR analysis between circadian rhythm and PAH
Next, we further analyze the relationship between circadian rhythm and PAH. Utilizing 12 genetic variations as instrumental variables in the MR analysis, with rhythm as the exposure factor, our findings indicated no discernible correlation between PAH and circadian rhythm (IVW: P = 0.34534942, β = 1.4980398). This conclusion was further corroborated by WMR (P = 0.95989182, β=-0.1095441). Additionally, as per the Egger intercept, the selected SNPs exhibited no potential pleiotropic effects. Taken together, our comprehensive analysis suggests the absence of a causal relationship between circadian rhythm and PAH (Fig. 5; Table 4).
Fig. 5.
The MR analysis between circadian rhythm and PAH. (A) Scatter plot of MR analysis, showing the distribution of individual rate estimates for PAH as a result of circadian rhythm. Each scatter plot includes trend lines derived from three different MR methods, indicating a causal relationship. (B) Forest plot shows the result of MR analysis pleiotropy test. Each circle next to a SNP represents the causal estimate for each IV, with the bottom three circles indicating the results from multi-instrument MR analysis using Weighted Median, Egger Regression, and IVW. The horizontal lines represent the 95% confidence intervals for OR. (C) The leave-one-out sensitivity analysis of the SNPs, with each black dot representing an IVW estimate and red dots representing estimates using all IVs. The horizontal lines represent the 95% confidence intervals (D) Funnel plot for PAH on MR analysis
Table 4.
The causal association of circadian rhythm with PAH
| exposure | outcome | method | nsnp | b | pval | egger_intercept | pval | Q | Q_pval |
|---|---|---|---|---|---|---|---|---|---|
| Circadian rhythm | "Hypertension, Pulmonary Arterial" (no controls excluded) | MR Egger | 12 | 12.0384487 | 0.12652557 | 0.29751504 | 0.166422967 | 9.86687668 | 0.45224937 |
| Circadian rhythm | "Hypertension, Pulmonary Arterial" (no controls excluded) | Weighted median | 12 | -0.1095441 | 0.95989182 | ||||
| Circadian rhythm | "Hypertension, Pulmonary Arterial" (no controls excluded) | Inverse variance weighted | 12 | 1.4980398 | 0.34534942 | 12.0944991 | 0.35658212 |
TEF drives the progression of PAH by affecting PASMCs
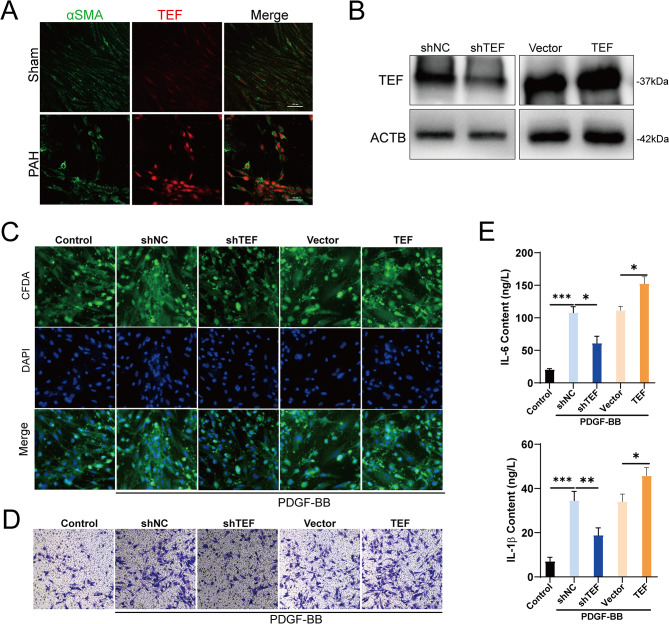
To elucidate the role of TEF in PAH, we isolated PASMCs from rat pulmonary artery tissues and conducted IF double staining to determine the expression and distribution of TEF. Our results demonstrated that TEF colocalizes with α-SMA and is significantly upregulated in PASMCs derived from PAH, suggesting that TEF may mediate PAH progression by influencing PASMCs function (Fig. 6A). Further, we constructed PASMCs with silenced and overexpressed TEF (Fig. 6B). Under PDGF-BB treatment, TEF overexpression markedly promoted PASMCs viability and migratory capacity, whereas TEF silencing had the opposite effect (Fig. 6C-D). Notably, we also found that TEF overexpression significantly enhanced the expression of IL-6 and IL-1β in PASMCs, whereas TEF silencing significantly inhibited the expression of these inflammatory cytokines in PASMC (Fig. 6E). Taken together, our results indicate that TEF is significantly upregulated in the PASMCs of PAH and mediates the release of inflammatory factors, which may be a critical factor in mediating PAH progression.
Fig. 6.
Functional validation of TEF in PAH. (A) Representative immunofluorescence staining of α-SMA (green) and TEF (red) in PASMCs from the control and PAH groups (scale bar: 100 μm). (B) Efficiency of TEF silencing and overexpression in PASMCs. (C) Representative immunofluorescence staining of CFDA-SE (green) and DAPI (blue) in PASMCs, with or without treatment with PDGF-BB. CFDA-SE, Cell proliferation and tracking dye. (D) Representative images of PASMCs migration in different groupsassessed by Transwell. (E) Expression levels of IL6 and IL-1β in different groups assessed by Specific ELISA kits
Discussion
The present study provides further evidence supporting the causal relationship between genetically increased TEF levels and an increased risk of Pulmonary arterial hypertension (PAH). Therefore, individuals with a genetic predisposition to higher TEF levels may also have a higher risk of developing PAH.
Thyrotrophic Embryonic Factor (TEF) is a transcription factor associated with the regulation of circadian rhythms, exhibiting widespread expression in cells and tissues [19]. Research has revealed its ability to inhibit cell growth by downregulating the expression of the common β-chain of cytokine receptors and its association with the oncogenic E2A-HLF fusion protein [20]. Dysregulation of TEF has been reported to be correlated with various diseases. For instance, Hua et al.‘s study [21] suggests that the polymorphism rs738499 of TEF may serve as a predictive marker for depression and sleep disorders in Parkinson’s patients. Furthermore, employing a multivariate generalized linear model, the research team found that Tef is not only a risk factor for depressive symptoms in Parkinson’s disease but also a risk factor for Major Depressive Disorder (MDD) in the Chinese population [22]. The disruption of circadian rhythms in chronic lung diseases has been demonstrated to play a crucial role in oxidative stress enhancement, inflammatory responses, metabolic imbalance, hypoxia/hyperoxia, mucus secretion, dysregulated autophagy, and altered lung function [13]. However, the connection between TEF and PAH remains unclear. Over the past decade, the importance of diurnal rhythmic changes in lung health has exponentially grown. Our study unveils a causal relationship between an inherited increase in TEF levels and an elevated risk of PAH. Therefore, TEF may be considered a risk factor contributing to the development of pulmonary arterial hypertension. Our results address the current gap in research regarding the relationship between TEF and PAH.
Oxygen is considered a deterministic resetting signal for the circadian rhythm system, and chronic hypoxia is identified as one of the most prevalent causes of PAH [23]. Consequently, in research, hypoxia is commonly employed to induce PAH in mouse models. Recent evidence suggests that both hypoxia and hypoxia-inducible factor 1 (HIF-1) signaling can significantly modulate the circadian rhythm [24, 25]. Adding to the complexity, hypoxia-induced circadian dysregulation exhibits both tissue-specific and time-specific characteristics. For instance, an immediate response to hypoxia results in a phase delay in the liver, while causing a phase advance in the lungs and kidneys, leading to a misalignment of organ tissues within the body. However, unlike the liver, which rapidly returns to its initial phase following hypoxic events and feeding behaviors, the lungs maintain their hypoxia-induced phase [26]. Additionally, compared to the liver and kidneys, the lungs demonstrate the highest gene expression level changes at all time points, with the lowest time-dependent variation, underscoring the crucial role of oxygen in maintaining the lung’s biological clock. Our finding showed corroborating evidence that TEF was increased in both lung tissues from PAH patients and PAH rats. This phenomenon is most likely caused by the hypoxic environment in PAH inducing the circadian rhythm disorder of the lung, which further induces the abnormal upregulation of TEF to correspond to the circadian rhythm, leading to the deterioration of PAH. This warrants further investigation of the circadian variability medicated by TEF in lung vasoconstriction and the sustained vascular remodeling response during these hypoxic episodes.
The direct role of circadian clock molecules in the pathogenesis of PAH remains poorly understood. In a recent study, another circadian clock-regulated gene, REV-ERBα, exhibited a significant downregulation in protein abundance and transcription levels in pulmonary artery smooth muscle cells (PASMC) of PAH patients. The pathological features of PAH include endothelial dysfunction, leading to abnormal smooth muscle proliferation associated with reduced apoptosis. Treatment with a REV-ERBα agonist (SR9011) substantially reduced PASMC proliferation and increased sensitivity to apoptosis, manifesting an improvement in the PAH phenotype in a monocrotaline-induced rat model [27]. Our experiments further elucidate the role of TEF in PAH. We found that TEF is highly expressed in PASMCs, and this overexpression correlates with increased PASMC viability and proliferation. Additionally, Rev-erbα demonstrated the ability to inhibit IL-6 expression, and the excessive expression of IL-6 is associated with the development and progression of vascular remodeling and anti-apoptotic mechanisms [28, 29]. Previous studies have indicated an association between circadian clock genes, including clock-controlled genes DBP, TEF, HIF, and period genes Per1, Per2, and Per3, and IL-6 [30]. Notably, in our study, inflammatory cytokines such as IL-6 and IL-1β were also significantly upregulated in TEF-overexpressing PASMCs. Therefore, it can be speculated that during the progression of pulmonary arterial hypertension, TEF may participate in the advancement of PAH by regulating IL-6 expression through a novel PASMC-specific signaling pathway. This suggests that TEF could be a potential therapeutic target for modulating inflammation and abnormal smooth muscle proliferation in PAH.
Low statistical power is a major limitation in MR study. To improve the statistical power, we used two sets of genetic instrumental variants (IVs) including 23 independent TEF genetic IVs (31684 European individuals) and, to date, the largest PAH genome-wide association studies (GWAS) with total sample size (162,962 European individuals). Because of a higher statistical power, we found this causal association of genetically increased TEF levels with increased risk of PAH. As TEF levels genetically increased, the risk of PAH increased using IVW (β = 0.209, OR = 1.233, 95% CI for OR: 1.054–1.441; P = 0.00871), weighted median (β = 0.256, OR = 1.292, 95% CI for OR: 1.064–1.568; P = 0.00964), MR-Egger (β = 0.166, OR = 1.818, 95% CI for OR: 0.832–1.675; P = 0.362). Our results suggest the importance of TEF as a risk factor for PAH disease. Similarly, TEF gene expressions might be used as potential biomarkers for bladder cancer [31]. Thus, future studies need to consider the complex role of TEF and its trans-signaling in the PAH disease process might be worthwhile. Furthermore, our findings unveil a substantial positive genetic causal association between TEF and circadian rhythm. As TEF expression up-regulates, the likelihood of abnormal rhythm also notably increases (IVW: P = 0.0024733, β = 0.05239). Regrettably, we did not observe a statistically significant positive correlation between circadian rhythm and PAH. While our results suggest a positive link between circadian rhythm and PAH, it does not reach statistical significance (IVW: P = 0.3454942, β = 1.4980398).
There were several strengths in this study. First, 23 independent TEF genetic IVs were chosen from a previously reported large-scale TEF GWAS of 3,1684 European individuals and have first used in our MR reports. Second, 23 of 24 independent TEF genetic IVs were successively extracted from PAH GWAS. Third, we used four different MR analysis methods demonstrated no significant pleiotropy or heterogeneity of TEF genetic IVs as the effective IVs. Fourth, two MR analysis methods including IVW, and weighted median proved the causal link between genetically increased TEF and increased risk of PAH, and also proved that the up-regulation of TEF expression may be the key factor leading to circadian rhythm abnormality. Fifth, all three methods demonstrated that each effect of TEF SNPs on PAH and circadian rhythm was no obvious bias.
This study also has several limitations. First, our TEF IVs and PAH GWAS datasets are from European ancestry. Therefore, our results need be proven in other ancestries. Second, randomized controlled trials are required to clarify whether blockade of TEF could is effective in some subsets of PAH. Third, the underlying mechanism by which TEF genetically affects the risk of PAH is still unclear and worth to explore in the future.
In summary, our results indicate that elevated hereditary TEF levels are causally associated not only with an increased risk of PAH but also with disruptions in circadian rhythm. Higher TEF levels may pose a risk factor for individuals with PAH, while lower TEF levels may act as a protective factor for PAH patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank ieu open gwas project (https://gwas.mrcieu.ac.uk/datasets/) for providing summary results data for these analyses.
Abbreviations
- PAH
Pulmonary arterial hypertension
- TEF
Thyrotroph embryonic factor
- GWAS
Genome-wide association study
- MR
Mendelian randomization
- SNP
Single nucleotide polymorphism
- IVW
Inverse variance weighted
Author contributions
Guan Lihua and Wenzhi Pan conceived, designed and supervised the whole study; Lifan Yang and Mingfei Li operation experiment; Qi Jin, Xiaochun Zhang and Lei Zhang performed the analyses; Dandan Chen and and Qi Jin wrote the manuscript; Lihua Guan, Daxin Zhou, Junbo Ge audit data and revised the manuscript. All authors provided critical comments and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation for youth of China (82200062), the Yangfan Project of Science and Technology Commission of Shanghai Municipality (22YF1439500) and the Youth Fund of Zhongshan Hospital, Fudan University (2021-016).
Data availability
The summary statistics of pulmonary arterial hypertension (PAH) GWAS (GWAS ID: finn-b-I9_HYPTENSPUL) is available on ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.
Declarations
Ethics approval and consent to participate
The study was reviewed and approved by the Ethics Committee of Shanghai Fudan University and Institutional Animal Care and Use Committee of Shanghai Fudan University.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dandan Chen and Qi Jin contributed equally to this work.
Contributor Information
Daxin Zhou, Email: zhou.daxin@zs-hospital.sh.cn.
Junbo Ge, Email: jbge@zs-hospital.sh.cn.
Lihua Guan, Email: guanlihua2023@163.com.
References
- 1.Badesch DB, Champion HC, Gomez Sanchez MA, Hoeper MM, Loyd JE, Manes A, et al. Diagnosis and assessment of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54(1 Suppl):S55–66. 10.1016/j.jacc.2009.04.011 [DOI] [PubMed] [Google Scholar]
- 2.Galie N, Hoeper MM, Humbert M, Torbicki A, Vachiery JL, Barbera JA, et al. Guidelines for the diagnosis and treatment of pulmonary hypertension: the Task Force for the diagnosis and treatment of pulmonary hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS), endorsed by the International Society of Heart and Lung Transplantation (ISHLT). Eur Heart J. 2009;30(20):2493–537. 10.1093/eurheartj/ehp297 [DOI] [PubMed] [Google Scholar]
- 3.Virsinskaite R, Karia N, Kotecha T, Schreiber BE, Coghlan JG, Knight DS. Pulmonary hypertension - the latest updates for physicians. Clin Med (Lond). 2023;23(5):449–54. 10.7861/clinmed.2023-23.5.Cardio4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vonk Noordegraaf A, Groeneveldt JA, Bogaard HJ. Pulmonary hypertension. Eur Respir Rev. 2016;25(139):4–11. 10.1183/16000617.0096-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poch D, Mandel J. Pulmonary hypertension. Ann Intern Med. 2021;174(4):ITC49–64. 10.7326/AITC202104200 [DOI] [PubMed] [Google Scholar]
- 6.Hoeper MM, Humbert M, Souza R, Idrees M, Kawut SM, Sliwa-Hahnle K, et al. A global view of pulmonary hypertension. Lancet Respir Med. 2016;4(4):306–22. 10.1016/S2213-2600(15)00543-3 [DOI] [PubMed] [Google Scholar]
- 7.Gredic M, Hadzic S. Pulmonary hypertension: unveiling Molecular mechanisms, diagnosis, and therapeutic targets. J Pers Med. 2023;13(10). [DOI] [PMC free article] [PubMed]
- 8.McLaughlin VV, Davis M, Cornwell W. Pulmonary arterial hypertension. Curr Probl Cardiol. 2011;36(12):461–517. 10.1016/j.cpcardiol.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 9.Mandras SA, Mehta HS, Vaidya A. Pulmonary hypertension: a brief guide for clinicians. Mayo Clin Proc. 2020;95(9):1978–88. [DOI] [PubMed]
- 10.Lajoie AC, Lauziere G, Lega JC, Lacasse Y, Martin S, Simard S, et al. Combination therapy versus monotherapy for pulmonary arterial hypertension: a meta-analysis. Lancet Respir Med. 2016;4(4):291–305. 10.1016/S2213-2600(16)00027-8 [DOI] [PubMed] [Google Scholar]
- 11.Grennan KS, Chen C, Gershon ES, Liu C. Molecular network analysis enhances understanding of the biology of mental disorders. BioEssays. 2014;36(6):606–16. 10.1002/bies.201300147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozek K, Relogio A, Kielbasa SM, Heine M, Dame C, Kramer A, et al. Regulation of clock-controlled genes in mammals. PLoS ONE. 2009;4(3):e4882. 10.1371/journal.pone.0004882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giri A, Wang Q, Rahman I, Sundar IK. Circadian molecular clock disruption in chronic pulmonary diseases. Trends Mol Med. 2022;28(6):513–27. 10.1016/j.molmed.2022.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat Med. 2018;24(12):1795–803. 10.1038/s41591-018-0271-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musiek ES, Holtzman DM. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science. 2016;354(6315):1004–8. 10.1126/science.aah4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crnko S, Du Pre BC, Sluijter JPG, Van Laake LW. Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol. 2019;16(7):437–47. 10.1038/s41569-019-0167-4 [DOI] [PubMed] [Google Scholar]
- 17.Emdin CA, Khera AV, Kathiresan S, Mendelian Randomization. JAMA. 2017;318(19):1925–6. 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- 18.Chi J, Meng L, Pan S, Lin H, Zhai X, Liu L, et al. Primary culture of rat aortic vascular smooth muscle cells: a New Method. Med Sci Monit. 2017;23:4014–20. 10.12659/MSM.902816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gachon F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann Med. 2007;39(8):562–71. 10.1080/07853890701491034 [DOI] [PubMed] [Google Scholar]
- 20.Inukai T, Inaba T, Dang J, Kuribara R, Ozawa K, Miyajima A, et al. TEF, an antiapoptotic bZIP transcription factor related to the oncogenic E2A-HLF chimera, inhibits cell growth by down-regulating expression of the common beta chain of cytokine receptors. Blood. 2005;105(11):4437–44. 10.1182/blood-2004-08-2976 [DOI] [PubMed] [Google Scholar]
- 21.Hua P, Cui C, Chen Y, Yao Y, Yu CY, Xu LG, et al. Thyrotroph embryonic factor polymorphism predicts faster progression of Parkinson’s disease in a longitudinal study. J Integr Neurosci. 2021;20(1):95–101. 10.31083/j.jin.2021.01.366 [DOI] [PubMed] [Google Scholar]
- 22.Hua P, Liu W, Chen D, Zhao Y, Chen L, Zhang N, et al. Cry1 and tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J Affect Disord. 2014;157:100–3. 10.1016/j.jad.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pak O, Aldashev A, Welsh D, Peacock A. The effects of hypoxia on the cells of the pulmonary vasculature. Eur Respir J. 2007;30(2):364–72. 10.1183/09031936.00128706 [DOI] [PubMed] [Google Scholar]
- 24.Wu G, Lee YY, Gulla EM, Potter A, Kitzmiller J, Ruben MD et al. Short-term exposure to intermittent hypoxia leads to changes in gene expression seen in chronic pulmonary disease. eLife. 2021;10. [DOI] [PMC free article] [PubMed]
- 25.O’Connell EJ, Martinez CA, Liang YG, Cistulli PA, Cook KM. Out of breath, out of time: interactions between HIF and circadian rhythms. Am J Physiol Cell Physiol. 2020;319(3):C533–40. 10.1152/ajpcell.00305.2020 [DOI] [PubMed] [Google Scholar]
- 26.Manella G, Aviram R, Bolshette N, Muvkadi S, Golik M, Smith DF, et al. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc Natl Acad Sci USA. 2020;117(1):779–86. 10.1073/pnas.1914112117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potus F, Frump AL, Umar S, Al Ghouleh RRV, Lai I. Recent advancements in pulmonary arterial hypertension and right heart failure research: overview of selected abstracts from ATS2020 and emerging COVID-19 research. Pulmonary Circulation. 2021;11(3):20458940211037274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steiner MK, Syrkina OL, Kolliputi N, Mark EJ, Hales CA, Waxman AB. Interleukin-6 overexpression induces pulmonary hypertension. Circul Res. 2009;104(2):236–44. 28p following 44. 10.1161/CIRCRESAHA.108.182014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato S, Sakurai T, Ogasawara J, Shirato K, Ishibashi Y, Oh-ishi S, et al. Direct and indirect suppression of interleukin-6 gene expression in murine macrophages by nuclear orphan receptor REV-ERBα. TheScientificWorldJournal. 2014;2014:685854. 10.1155/2014/685854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, et al. TNF-alpha suppresses the expression of clock genes by interfering with e-box-mediated transcription. Proc Natl Acad Sci USA. 2007;104(31):12843–8. 10.1073/pnas.0701466104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saleh AA, Gohar SF, Hemida AS, Elgharbawy M, Soliman SE. Evaluation of ASPM and TEF Gene expressions as potential biomarkers for bladder Cancer. Biochem Genet. 2020;58(3):490–507. 10.1007/s10528-020-09962-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary statistics of pulmonary arterial hypertension (PAH) GWAS (GWAS ID: finn-b-I9_HYPTENSPUL) is available on ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.