Abstract
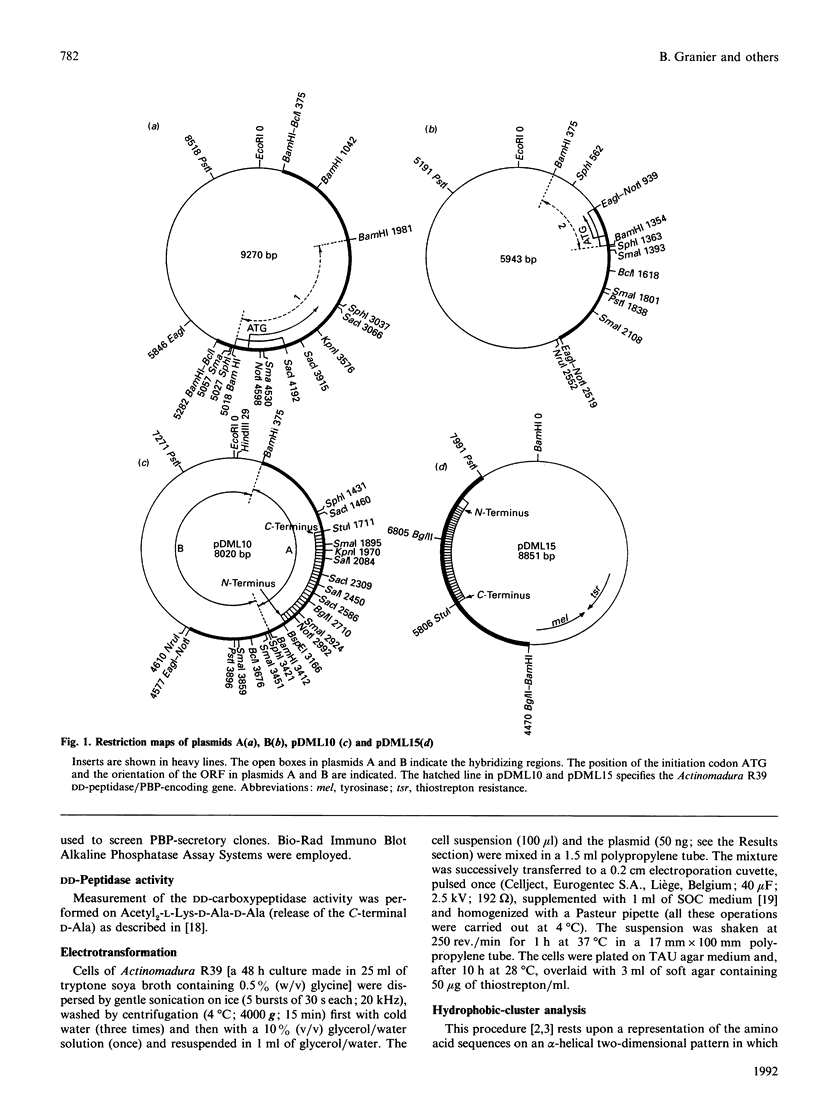
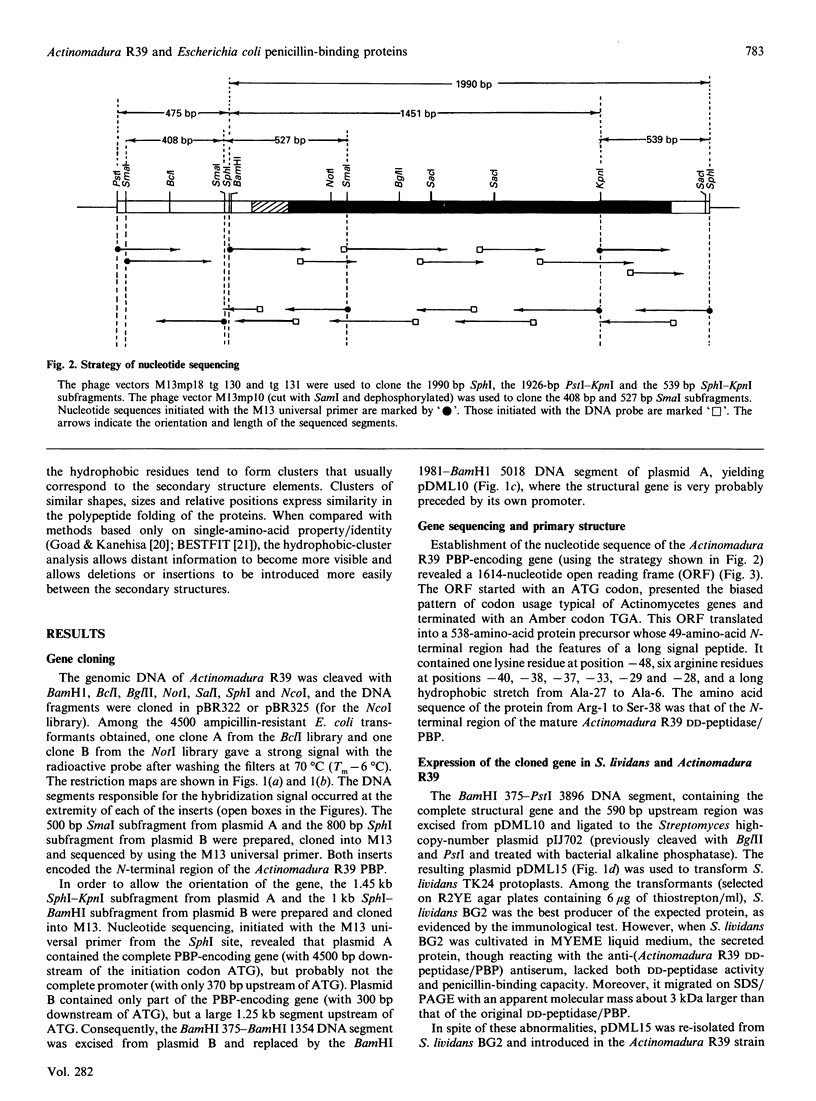
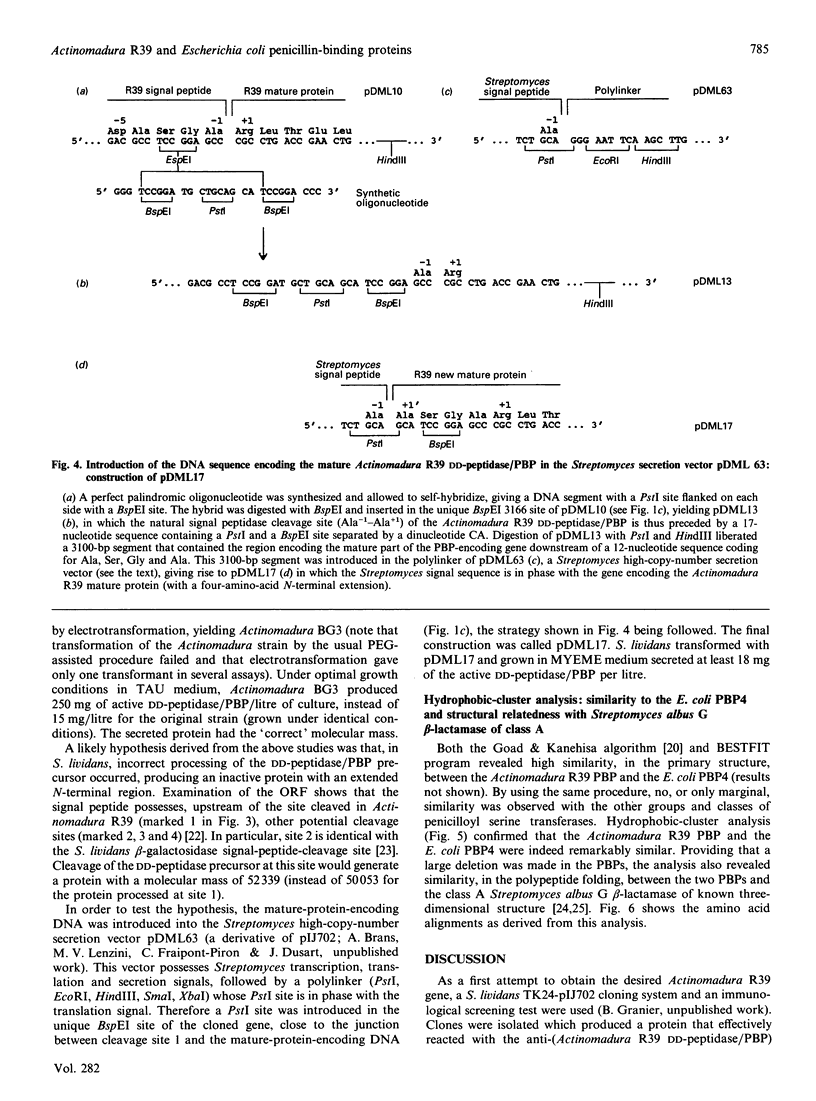
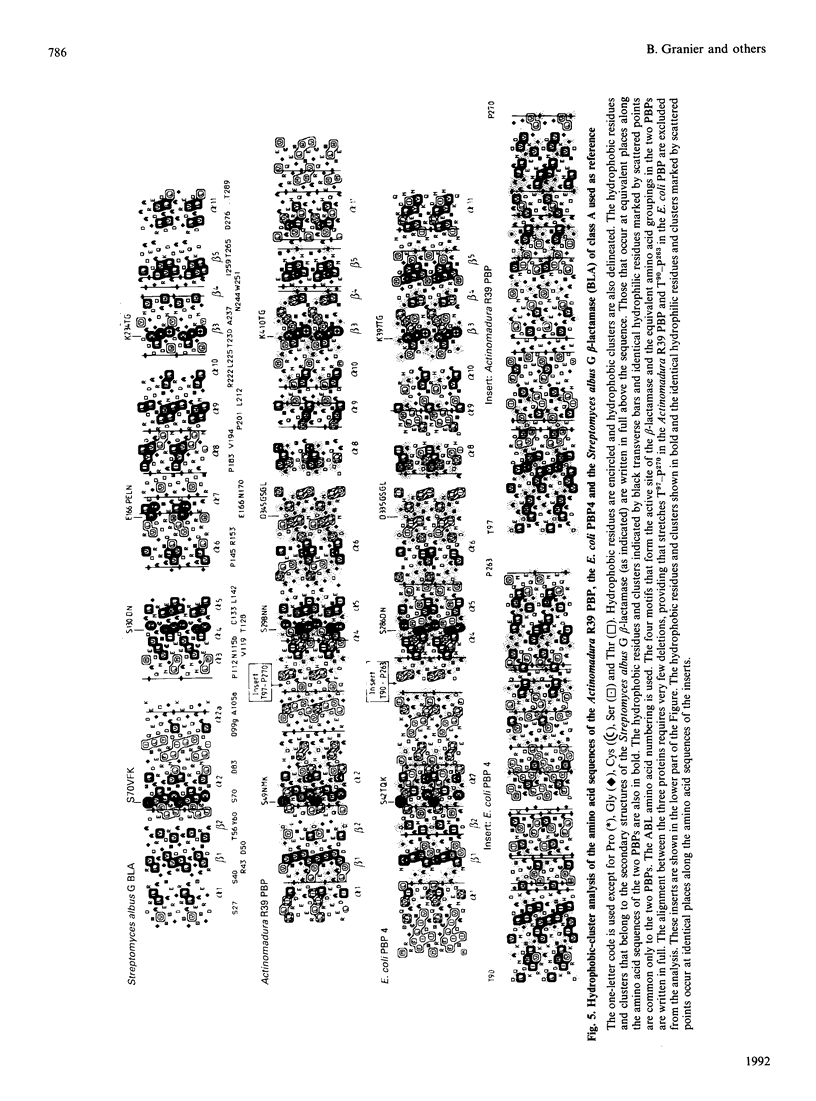
As derived from gene cloning and sequencing, the 489-amino-acid DD-peptidase/penicillin-binding protein (PBP) produced by Actinomadura R39 has a primary structure very similar to that of the Escherichia coli PBP4 [Mottl, Terpstra & Keck (1991) FEMS Microbiol. Lett. 78, 213-220]. Hydrophobic-cluster analysis of the two proteins shows that, providing that a large 174-amino-acid stretch is excluded from the analysis, the bulk of the two polypeptide chains possesses homologues of the active-site motifs and secondary structures found in the class A beta-lactamase of Streptomyces albus G of known three-dimensional structure. The 174-amino-acid insert occurs at equivalent places in the two PBPs, between helices alpha 2 and alpha 3, away from the active site. Such an insert is unique among the penicilloyl serine transferases. It is proposed that the Actinomadura R39 PBP and E. coli PBP4 form a special class, class C, of low-Mr PBPs/DD-peptidases. A vector has been constructed and introduced by electrotransformation in the original Actinomadura R39 strain, allowing high-level expression and secretion of the DD-peptidase/PBP (250 mg.l-1). The gene encoding the desired protein is processed differently in Actinomadura R39 and Streptomyces lividans. Incorrect processing in Streptomyces lividans leads to a secreted protein which is inert in terms of DD-peptidase activity and penicillin-binding capacity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez C., Joris B., Frère J. M., Ghuysen J. M., Van Beeumen J. The penicillin-binding site in the exocellular DD-carboxypeptidase-transpeptidase of Actinomadura R39. Biochem J. 1981 Jan 1;193(1):83–86. doi: 10.1042/bj1930083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T., Strickler J., Gorniak L., Burnett W. V., Fare L. R. Characterization of the promoter, signal sequence, and amino terminus of a secreted beta-galactosidase from "Streptomyces lividans". J Bacteriol. 1987 Sep;169(9):4249–4256. doi: 10.1128/jb.169.9.4249-4256.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère J. M., Ghuysen J. M., Reynolds P. E., Moreno R. Binding of beta-lactam antibiotics to the exocellular DD-carboxypeptidase-transpeptidase of Streptomyces R39. Biochem J. 1974 Oct;143(1):241–249. doi: 10.1042/bj1430241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère J. M., Joris B. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit Rev Microbiol. 1985;11(4):299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- Frére J. M., Leyh-Bouille M., Ghuysen J. M., Nieto M., Perkins H. R. Exocellular DD-carboxypeptidases-transpeptidases from Streptomyces. Methods Enzymol. 1976;45:610–636. doi: 10.1016/s0076-6879(76)45054-1. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Ghuysen J. M., Reynolds P. E., Perkins H. R., Frère J. M., Moreno R. Effects of donor and acceptor peptides on concomitant hydrolysis and transfer reactions catalyzed by the exocellular DD-carboxypeptidase-transpeptidase from Streptomyces R39. Biochemistry. 1974 Jun 4;13(12):2539–2547. doi: 10.1021/bi00709a010. [DOI] [PubMed] [Google Scholar]
- Ghuysen J. M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Saloheimo M., Lavaitte S., Knowles J. K. Structural homology among the peroxidase enzyme family revealed by hydrophobic cluster analysis. Proteins. 1990;8(3):251–257. doi: 10.1002/prot.340080307. [DOI] [PubMed] [Google Scholar]
- Houba S., Willem S., Duez C., Molitor C., Dusart J., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the active-site serine beta-lactamase from Actinomadura R39. FEMS Microbiol Lett. 1989 Dec;53(3):241–246. doi: 10.1016/0378-1097(89)90224-3. [DOI] [PubMed] [Google Scholar]
- Korat B., Mottl H., Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991 Mar;5(3):675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottl H., Terpstra P., Keck W. Penicillin-binding protein 4 of Escherichia coli shows a novel type of primary structure among penicillin-interacting proteins. FEMS Microbiol Lett. 1991 Mar 1;62(2-3):213–220. doi: 10.1016/0378-1097(91)90160-c. [DOI] [PubMed] [Google Scholar]
- Palomeque-Messia P., Englebert S., Leyh-Bouille M., Nguyen-Distèche M., Duez C., Houba S., Dideberg O., Van Beeumen J., Ghuysen J. M. Amino acid sequence of the penicillin-binding protein/DD-peptidase of Streptomyces K15. Predicted secondary structures of the low Mr penicillin-binding proteins of class A. Biochem J. 1991 Oct 1;279(Pt 1):223–230. doi: 10.1042/bj2790223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J. M., Jackson M. E., Holland I. B. The C terminus of penicillin-binding protein 5 is essential for localisation to the E. coli inner membrane. EMBO J. 1986 Sep;5(9):2399–2405. doi: 10.1002/j.1460-2075.1986.tb04510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]



