Abstract
Although infection by human immunodeficiency virus (HIV) typically requires an interaction between the viral envelope glycoprotein (Env), CD4, and a chemokine receptor, CD4-independent isolates of HIV and simian immunodeficiency virus have been described. The structural basis and underlying mechanisms for this phenotype are unknown. We have derived a variant of HIV-1/IIIB, termed IIIBx, that acquired the ability to utilize CXCR4 without CD4. This virus infected CD4-negative T and B cells and fused with murine 3T3 cells that expressed human CXCR4 alone. A functional IIIBx env clone exhibited several mutations compared to the CD4-dependent HXBc2 env, including the striking loss of five glycosylation sites. By constructing env chimeras with HXBc2, the determinants for CD4 independence were shown to map outside the V1/V2 and V3 hypervariable loops, which determine chemokine receptor specificity, and at least partly within an area on the gp120 core that has been implicated in forming a conserved chemokine receptor binding site. We also identified a point mutation in the C4 domain that could render the IIIBx env clone completely CD4 dependent. Mutations in the transmembrane protein (TM) were also required for CD4 independence. Remarkably, when the V3 loop of a CCR5-tropic Env was substituted for the IIIBx Env, the resulting chimera was found to utilize CCR5 but remained CD4 independent. These findings show that Env determinants for chemokine receptor specificity are distinct from those that mediate CD4-independent use of that receptor for cell fusion and provide functional evidence for multiple steps in the interaction of Env with chemokine receptors. Combined with our observation that the conserved chemokine receptor binding site on gp120 is more exposed on the IIIBx gp120 (T. L. Hoffman, C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms, Proc. Natl. Acad. Sci. USA 96:6359–6364, 1999), the findings from this study suggest novel approaches to derive and design Envs with exposed chemokine receptor binding sites for vaccine purposes.
Human immunodeficiency virus (HIV) entry is known to require an interaction of the viral envelope glycoprotein (Env) with CD4 and cellular chemokine receptors. Differential use of chemokine receptors by HIV and simian immunodeficiency virus (SIV) has largely explained differences in tropism among various isolates (4, 27). While a number of chemokine receptors and orphan members of this family of proteins can serve as coreceptors for HIV or SIV (10, 14, 20, 22, 37, 56), CCR5 and CXCR4 appear to be the principal coreceptors for HIV-1 (69, 70). Isolates of HIV that first establish infection target peripheral blood lymphocytes and macrophages by using CCR5 (2, 13, 15, 16), while viruses that are generally associated with progression to AIDS and that can infect T-cell lines in vitro acquire the ability to use CXCR4 (10, 12, 23). An understanding of the mechanism by which HIV uses CD4 and chemokine receptors to enter cells is of central importance in developing immunologic and pharmacologic strategies to prevent infection.
Following binding to CD4, Env undergoes poorly understood conformational changes that enable gp120 to bind to a chemokine receptor and lead to fusion of the viral and cellular membranes (32, 42, 66, 68). Immunologic and mutagenesis approaches have indicated that these changes involve movement of V1/V2 and V3 hypervariable loops on gp120 (42, 66, 68), which play a critical role in the specificity of chemokine receptor utilization (9–11, 29, 53, 59). The recent crystallographic resolution of a gp120 core structure bound to CD4 has revealed an intervening β sheet comprised of conserved residues between the inner and outer domains of gp120 that may serve as a major contact site for the chemokine receptor (52, 67).
Although CD4 is generally required for gp120 to associate with a chemokine receptor, the identification of CD4-independent isolates of HIV type 1 (HIV-1), HIV-2, and SIV has clearly indicated that functional interactions with chemokine receptors can occur in the absence of CD4 (18, 20, 21, 49). The determinants for this phenotype have been mapped to Env, but the underlying mechanisms are unknown. It has been proposed that mutations may increase the exposure and/or the affinity of the chemokine receptor binding site on gp120, thus circumventing the need for CD4 (21). Biochemical assays have also shown that mutated or deglycosylated gp120 can bind directly to chemokine receptors, suggesting that domains normally activated by CD4 can be exposed artificially (3, 26, 38, 40). A greater understanding of the determinants responsible for CD4 independence could provide insight into the Env domains that mediate and modulate interactions with chemokine receptors and ultimately govern viral entry.
In this report we describe the derivation and molecular characterization of a variant of HIV-1/IIIB, termed IIIBx, which acquired the ability to utilize CXCR4 in the absence of CD4. A functional IIIBx env clone (8x) was used to construct chimeras with a closely related but CD4-dependent env, and the determinants in gp120 required for CD4 independence were shown to map outside the variable loops and, at least in part, to residues flanking the putative chemokine receptor binding site on the gp120 core. Remarkably, when 8x contained the V3 loop of a CCR5-tropic Env, it was shown to utilize CCR5 but remained CD4 independent. These findings provide further evidence that CD4 binding likely exposes a domain on the gp120 core that can interact with genetically divergent chemokine receptors. This work may have important implications for designing HIV-1 Env proteins with exposed chemokine receptor binding sites that could exhibit novel biochemical and immunogenic properties.
MATERIALS AND METHODS
Cells, viruses, and infectivity assays.
Hut-78 and SupT1 are immortalized CD4+ T-cell lines. BC7 is a CD4-negative line derived from SupT1 (21). Uncloned HIV-1/IIIB was obtained from R. C. Gallo (48) in chronically infected Hut-78 cells. Virus from this culture was serially passaged onto SupT1 from which IIIBx was isolated by subsequent passage onto BC7. The IIIB/Sup virus was derived from an early passage of HIV-1/IIIB in SupT1. NIH 3T3 cells, untransfected or stably transfected with human CXCR4, were obtained from D. Littman. For neutralization assays, BC7 cells were preincubated with various concentrations of anti-CXCR4 monoclonal antibody (MAb) 12G5 (21) for 30 min at 37°C, inoculated with IIIBx (10 50% tissue culture infective doses), and monitored for reverse transcriptase (RT) activity.
RT assays.
Productive infection of cells was documented by detection of RT activity in the culture supernatant as previously described (30). Briefly, virus from 1 ml of clarified culture supernatant was pelleted at 100,000 × g for 30 min at 4°C and solubilized in 100 μl of solubilizing buffer (0.15 M Tris [pH 8], 0.4 M NaCl, 0.25% Triton X-100, 10% glycerol, 0.5 mM dithiothreitol). Duplicate 20-μl aliquots were mixed with 85 μl of RT cocktail {67.5 mM Tris [pH 7.5]–1.3 mM dithiothreitol–1 mM ATP–13.5 mM MgCl2 containing 0.05 U of polyr(A) and 12.5 μCi of [3H]dTTP} and incubated for 1 h at 37°C. Tubes were placed on ice, 225 μg of tRNA was added, and RNA was precipitated with cold 10% trichloroacetic acid. Precipitated RNA was captured on a glass fiber filter, washed with trichloroacetic acid, and ethanol, and dried, and radioactivity (counts per minute) was determined in a scintillation counter (LKB/Wallac).
PCR, cloning, virus production, and chimera construction.
Full-length env coding regions were amplified by PCR from genomic DNA of chronically infected cells (sense primer, 5′-CGCAACCTATACCAATAGTAGCAA-3′; antisense primer, 5′-CAGTAAGCCATCCAATCACACTAC-3′) in a BioCycler (Ericomp). The PCR product was TA cloned into pCDNA3.1 (Invitrogen) and tested in a reporter gene fusion assay described below. Functional env clones of IIIBx (8x) and IIIB/Sup (S10) were sequenced in an automated sequencer (Applied Biotechnologies Inc.). Clones were also subcloned into pSP73 (Promega) that contained the HXBc2 env, using Asp718 and BamHI (29).
env clones were subcloned into the 3′ hemigenome of pNL4-3 (from the EcoRI site), using unique NdeI and BamHI restriction sites in env which encompass the mutations in the 8x env clone of IIIBx. Virus was generated by digesting 20 μg of each of the 5′ pNL4-3 Δvpr hemigenome (to the EcoRI site) (24) and the various 3′ hemigenome constructs with EcoRI. Constructs were extracted with phenol and coprecipitated before transfection into BC7 and SupT1 by electroporation. Cells were monitored for syncytium formation, and virus was harvested from supernatants to generate virus stocks. HIV-1/IIIB stocks were frozen at −70°C in 1-ml aliquots. Stocks of IIIBx and NL4-3 containing an 8x Env were frozen at −140°C in 5% sucrose to preserve infectivity. Chimeras between 8x and HXBc2 were constructed by using a BsaBI site (nucleotide [nt] 7673) to isolate changes in the surface protein from those in the transmembrane protein (TM) and DraIII (nt 6714), StuI (nt 6948), and Bsu36I (nt 7430) to isolate V1-V2, V3, and V4/C4 regions, respectively (29). Clones containing the V3 loop of an R5 virus were constructed by subcloning the Asp718-BamHI fragment from a proviral clone of HXB with the V3 loop of BaL (31) into pSP73-HXBc2 (29). A version of 8x containing the V3 loop of BaL was made in a similar fashion, by inserting the StuI-Bsu36I fragment of this provirus into pSP73-8x.
Mutagenesis.
Point mutations were engineered into Env constructs in pSP73 by using a Quickchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s specifications. The following primer pair produced the D368R mutation to ablate CD4 binding: D368R-forward (5′-CCTCAGGAGGGGACCCAGAAATTGTAACGC-3′) plus D368R-reverse (5′-GCGTTACAATTTCTGGGTCCCCTCCTGAGG-3′). Reciprocal exchange of residues at position 431 in 8x and S10 was accomplished with two sets of oligonucleotides: 8X-G431E-forward (5′-GGCAGGAAGTAGAAAAAGCAATGTATGCCCC-3′) plus 8X-G431E-reverse (5′-GGGGCATACATTGCTTTTTCTACTTCCTGCC-3′); and S10-E431G-forward (5′-GGCAGGAAGTAGGAAAAGCAATGTATGCCCC-3′) plus S10-E431G-reverse (5′-GGGGCATACATTGCTTTTCCTACTTCCTGCC-3′).
Cell-cell fusion assay and flow cytometric analysis of Env surface expression.
The ability of env genes to mediate cell-cell fusion was evaluated by using a luciferase-based gene reporter assay (55). Briefly, quail QT6 cells were cotransfected with plasmids containing HIV env genes by CaPO4 and infected with a vaccinia virus expressing T7 RNA polymerase (1). These cells were mixed with quail QT6 cells transiently expressing human CXCR4 or CCR5 with or without human CD4 and the luciferase gene under the control of the T7 promoter. Fusion was quantified by lysing the cells 7 to 8 h after combining the cells and measuring luciferase expression (represented in figures as mean relative light units [RLU] ± standard error of the mean [SEM]) with a luminometer. For experiments in which Env expression was assessed, aliquots of cells (105 per sample) were incubated (4°C, 30 min) in staining buffer (phosphate-buffered saline, 0.1% bovine serum albumin) with a 1/103 dilution of normal human serum (NHS) or serum from an HIV-1-infected individual and stained with a 1/40 dilution of fluorescein isothiocyanate-conjugated F(ab)′2 goat anti-human immunoglobulin G (BioSource International, Burlingame, Calif.). Cells were fixed in 2% paraformaldehyde and analyzed with a FACScan analyzer (Becton Dickinson).
Western blotting.
Virus from supernatant of infected cell culture was pelleted at 100,000 × g for 90 min at 4°C, and resuspended in lysis buffer (20 mM Tris [pH 8.0], 120 mM NaCl, 0.2% sodium deoxycholate, 0.5% NP-40, 0.2 mM EGTA, 0.2 mM NaF, 1 μM pepstatin, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml) on ice. Equal volumes of lysate and 2× sample buffer (50 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 30% glycerol, 10% β-mercaptoethanol, 0.2% pyronine Y), were mixed, boiled for 7 min, chilled on ice for 7 min, and run on an SDS–12% polyacrylamide gel. Proteins were transferred to nitrocellulose (Bio-Rad) by using a Multiphor II semidry electrotransfer apparatus (Pharmacia-LKB). HIV-1 transmembrane proteins were detected by using MAb D12 (19) followed by biotinylated sheep anti-mouse immunoglobulin (heavy and light chains; Jackson Immunoresearch), streptavidin-horseradish peroxidase (Amersham), and chemiluminescence substrate (Pierce).
Nucleotide sequence accession numbers.
The GenBank accession number for the 8x env clone is AF189158, and that for the S10 env clone is AF189159.
RESULTS
Derivation of a CD4-independent variant of HIV-1/IIIB.
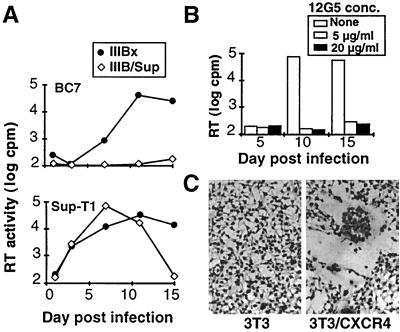
A CD4-independent variant of HIV-1/IIIB was derived by serial passage of an uncloned stock of HIV-1/IIIB in SupT1 and then inoculating BC7, a CD4-negative cell line derived from SupT1 (21). In one experiment, approximately 5% of BC7 cells were noted to be positive for viral p24gag by immunofluorescence assay. Virus from this culture was passaged twice onto uninfected BC7 cells, and a chronically infected line was established. Virus from this line, termed IIIBx, was compared to an earlier passage of HIV-1/IIIB in SupT1 cells, designated IIIB/SupT1. As shown in Fig. 1A, only IIIBx could infect BC7, while both viruses were able to infect SupT1. IIIBx was also able to infect Raji cells, a CD4-negative B-lymphoblastoid cell line (not shown). Infection of BC7 could be completely inhibited by the anti-CXCR4 MAb 12G5 (Fig. 1B), indicating that this infection was likely mediated by CXCR4. Moreover, IIIBx-infected BC7 cells induced syncytia when cocultured with murine 3T3 fibroblasts that stably expressed human CXCR4, while no fusion was induced on untransfected 3T3 cells (Fig. 1C). In addition, no fusion was observed when HIV-1/IIIB-infected HUT-78 cells were cocultured with the CXCR4-expressing 3T3 cells (not shown). Taken together, these data indicate that the IIIBx variant can utilize CXCR4 as a primary receptor in the absence of CD4 on T- and B-lymphoid cell lines and murine fibroblasts.
FIG. 1.
Derivation and characterization of CD4-independent HIV-1. (A) Viral replication in CD4-negative T cells. SupT1 and CD4-negative BC7 cells were inoculated with equal amounts of HIV-1/IIIB or IIIBx, and viral replication was determined by RT activity in culture supernatants. (B) Inhibition of IIIBx by anti-CXCR4 MAb. BC7 cells were inoculated with IIIBx in the presence or absence of 12G5, and RT activity determined at the indicated time points. (C) IIIBx-induced fusion on murine cells expressing CXCR4. IIIBx-infected BC7 cells were cocultured for 24 h with murine 3T3 cells or 3T3 cells that expressed human CXCR4 and stained for syncytium formation as described previously (21).
Cloning and characterization of a functional IIIBx env.
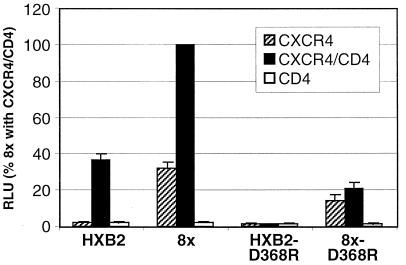
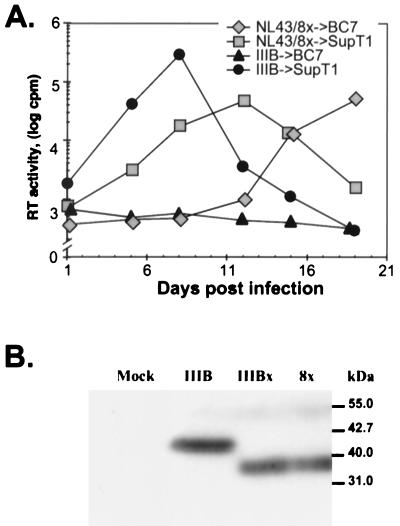
A full-length env clone of IIIBx (designated 8x) was amplified by PCR from infected BC7 cells, cloned into pSP73, and compared to a prototypic CD4-dependent IIIB clone (HXBc2) in a cell fusion assay. Both 8x and HXBc2 were able to mediate fusion on quail QT6 cells expressing both CD4 and CXCR4, but only 8x could fuse with cells that expressed CXCR4 alone (Fig. 2). Of note, 8x fusion was enhanced when CD4 and CXCR4 were coexpressed, indicating that the 8x Env was likely still able to interact with CD4. Additionally, the 8x env cloned into the pNL4-3 provirus was able to generate a replication-competent virus that could infect BC7 cells as well as SupT1 (Fig. 3A), providing further proof that the 8x Env was able to utilize CXCR4 in the absence of CD4 and that this clone was representative of the uncloned parental IIIBx.
FIG. 2.
Evaluation of a IIIBx env clone in fusion assays. env genes indicated were cloned into pSP73, transfected into QT6 cells, and evaluated in fusion assays on QT6 cells expressing CD4 plus CXCR4, CXCR4 alone, or CD4 alone (55). Results are expressed as RLU (mean + SEM) normalized to the activity of 8x on CXCR4+ CD4+ cells. Also shown are fusion activities for 8x and HXBc2 Envs that contain a D368R mutation in gp120, which ablates the CD4 binding site (44).
FIG. 3.
Generation of a replication-competent virus with a IIIBx Env and evaluation of its TM size. (A) The 8x env was inserted into pNL4-3, and a virus stock was generated after transfection into BC7 cells. Equal amounts of the resulting virus (designated NL43/8x) and HIV-1/IIIB were inoculated onto SupT1 and BC7 cells, and RT levels were monitored over time. (B) Viral lysates from HIV-1/IIIB-infected SupT1 cells (IIIB), IIIBx-infected BC7 cells (IIIBx), and NL43/8x-infected BC7 cells (8x) were evaluated by Western blotting using an anti-TM MAb, D12. “Mock” viral lysate was prepared from the supernatant of uninfected BC7 cells. Consistent with sequence analysis shown in Fig. 4, both IIIBx and NL43/8x have a truncated TM.
The CD4 independence of the 8x Env was evaluated further by introducing an Asp to Arg change at amino acid 368 in gp120. This Asp is highly conserved among all HIV-1 isolates and has been shown to be a critical determinant for CD4 binding by forming a salt bridge with Arg-59 in the CDR2 loop of CD4 (34). Mutations at this position have been shown to completely ablate CD4 binding (44). As shown in Fig. 2, although a D368R mutation abrogated fusion by HXBc2, this mutation did not prevent 8x from fusing with target cells that expressed CXCR4 either with or without CD4. Interestingly, unlike the case for the 8x clone, fusion of 8x-D368R was not enhanced when CD4 and CXCR4 were coexpressed, confirming that this Env was unable to interact with CD4. Thus, the 8x Env could not only utilize CXCR4 in the absence of CD4 but could also tolerate a mutation that destroyed the CD4 binding site on gp120.
Sequence analysis of 8x env.
The sequence of 8x was compared to the published sequence of HXBc2 (Fig. 4). While the number of mutations in 8x is large (17 in gp120 and 7 in TM), 4 of the mutations in gp120 and 2 in TM have been observed in other env clones derived from HIV-1/IIIB and are thus not likely to be involved in the CD4-independent phenotype. In gp120, 9 of the 13 unique mutations were in the hypervariable loops V1/V2 (S143G, I165K, G167S, Q170K, and T188P), V3 (R298K, Q310H, and I320V), and V4 (N386K). Three mutations were in the gp120 core (D62E, N339S, and I423V), and one (S29N) was in the leader sequence. Interestingly, five mutations in the gp120 resulted in the loss of potential N-linked glycosylation sites, and four of these (S143G, T188P, N339S, and N386K) were unique to 8x. The four 8x-specific mutations in the external domain of TM were located within the two regions that form coiled coils (T536A, L544S, N651I, and K655M). Remarkably, 8x also contained a single nucleotide deletion in the TM membrane-spanning domain that introduced a frameshift at position 706 and is predicted to generate a divergent cytoplasmic tail of only 30 amino acids. This feature is surprising since HIV-1 isolates with truncated cytoplasmic tails typically have been attenuated or noninfectious (8, 17, 58). Nonetheless, as noted above, the 8x env was able to generate a replication-competent virus that could infect SupT1 and BC7 cells (Fig. 3A). Moreover, Western blots of viral lysates from uncloned IIIBx as well as NL4-3 containing the 8x env demonstrated a TM of approximately 35 kDa, compared to 41 kDa for parental HIV-1/IIIB (Fig. 3B).
FIG. 4.
Sequence analysis for IIIBx env clones 8x and S10 in comparison to HXBc2. Shaded regions indicate mutations that are also found in other clones from HIV-1/IIIB. Predicted N-linked glycosylation sites are indicated ( ), as are the positions of variable loops, the gp120/gp41 cleavage site, and the TM membrane-spanning domain (msd). 8x contains a frameshift mutation at position 706 resulting in a prematurely truncated TM cytoplasmic tail. S10 contains a deletion of 50 nt, which also leads to frameshift and a prematurely truncated TM cytoplasmic tail.
), as are the positions of variable loops, the gp120/gp41 cleavage site, and the TM membrane-spanning domain (msd). 8x contains a frameshift mutation at position 706 resulting in a prematurely truncated TM cytoplasmic tail. S10 contains a deletion of 50 nt, which also leads to frameshift and a prematurely truncated TM cytoplasmic tail.
Mapping CD4 independence by using chimeric Env proteins.
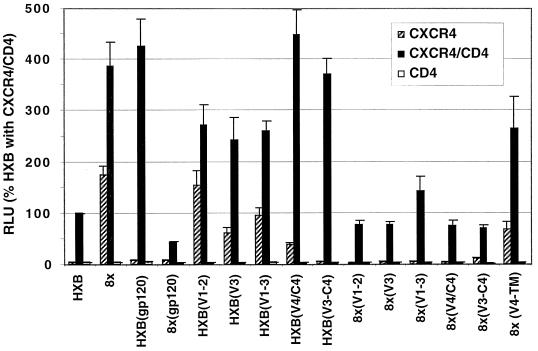
To identify the determinants of CD4 independence, a set of reciprocal chimeras was generated between 8x and HXBc2. Unique restriction sites were chosen to isolate mutations in gp120 from those in TM and to define the effects of mutations in V1/V2, V3, and the V4/C4 subdomains (Fig. 5). Chimeras were cloned into pSP73 and analyzed in fusion assays as described above on target cells expressing CXCR4 alone or with CD4. Results for each chimera are expressed as the percentage of HXBc2 Env fusion activity on target cells that expressed both CD4 and CXCR4. All chimeras were functional on CXCR4+ CD4+ target cells, although considerable quantitative differences were seen with activities ranging from approximately 50 to 500% of that of HXBc2 (Fig. 6). Reciprocal chimeras that exchanged the entire gp120 and TM were CD4 dependent, although an assessment of 8x(gp120), which contained the 8x gp120 and the HXBc2 TM, was somewhat limited due to the poor overall fusion activity of this Env. Of note, chimeras that contained the 8x TM were in general more fusogenic than HXBc2 or chimeras that contained an HXBc2 TM, suggesting that determinants in the 8x TM ectodomain and/or the prematurely truncated cytoplasmic tail were contributing to the increased fusogenicity of these clones. Nonetheless, these findings indicated that determinants for CD4 independence were not entirely restricted to the 8x gp120 or TM.
FIG. 5.
Construction of chimeric Env proteins for fusion assays. Diagrams representing HXBc2, 8x, and S10 env genes are shown along with chimeras constructed by using the indicated restriction sites. Mutations present in 8x are indicated above the top schematic. Chimeras were cloned into pSP73 and evaluated in cell fusion assays described in the legend to Fig. 6.
FIG. 6.
Evaluation of chimeric Env proteins in fusion assays. Chimeric Env proteins between HXBc2 and 8x shown in Fig. 5 were evaluated in fusion assays on QT6 target cells expressing CXCR4 alone, CXCR4 and CD4, or CD4 alone. Results are expressed as luciferase activity (RLU) relative to that of HXBc2 on CXCR4+ CD4+ cells. Bars indicate the mean RLU for at least five experiments + SEM.
Among chimeras that introduced subdomains of HXBc2 gp120 into an 8x background, replacement of V1/V2 and V3 loops either individually or in combination failed to eliminate CD4 independence [Fig. 6, chimeras HX(V1/V2), HX(V3), HX(V1-V3)]. This finding was of interest given the importance of these loops as determinants of chemokine receptor specificity (9–11, 29, 53, 59). For these chimeras, fusion activity relative to 8x was reduced on both CD4-negative and CD4-positive cells, although CD4-dependent fusion was still greater than that seen with HXBc2. In contrast, fusion activity of HX(V4/C4), which contained the HXBc2 V4/C4, was considerably reduced relative to 8x on CD4-negative cells but was unchanged on CD4+ cells. Interestingly, when both the V3 and V4/C4 domains of 8x were replaced with those of HXBc2 [HX(V3-C4)], CD4-independent fusion was completely abrogated, while CD4-dependent fusion was unaffected.
For chimeras in which domains of 8x were introduced into an HXBc2 background, no single region of gp120 was able to confer CD4 independence to HXBc2, consistent with evidence noted above that determinants in both gp120 and TM were required (Fig. 6). However, an HXBc2 chimera that contained both the V4/C4 and TM domains from 8x [8x(V4-TM)] was highly competent for both CD4-dependent and CD4-independent fusion. Collectively, these findings with 8x- and HXBc2-based chimeras indicate that determinants in the 8x gp120 V3, and in particular the V4/C4 domain, contribute to the CD4-independent phenotype of 8x, but only in the presence of an 8x TM.
Surface expression of Env proteins.
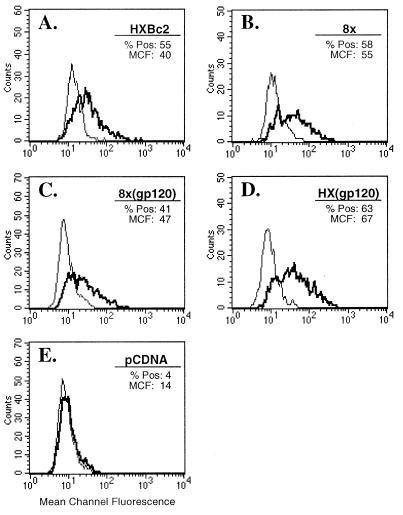
One striking alteration in the TM of the CD4-independent 8x is the frameshift mutation in the membrane-spanning domain that results in a divergent, truncated TM cytoplasmic tail. In light of previous studies showing that the cytoplasmic tail of TM can affect levels of Env on the cell surface (35, 54, 57), it seemed possible that the 8x TM might increase surface levels of Env and contribute to its CD4-independent fusion and/or to the increased fusogenicity of chimeras that contained an 8x TM. To examine this possibility, QT6 cells were transfected with Env-expressing plasmids shown in Fig. 5 and were then divided and used for fusion assays as described above and for FACS analysis of Env surface expression, using NHS or serum from an HIV-1-infected individual. Analysis gates were selected to optimize signal-to-noise ratios and adjusted to give <5% positivity for cells stained with NHS. When QT6 cells were transfected with either parental HXBc2 or 8x or with the gp120-TM chimera 8x(gp120) or HX(gp120) (Fig. 5), only minimal differences in levels of Env were seen, with no consistent differences observed in several experiments. Histograms from a representative experiment are shown in Fig. 7. As described above, striking differences in fusogenicity had been noted between HX(gp120) and 8x(gp120) (Fig. 6). Similar results were seen when these Envs were expressed in 293T cells (not shown). In four experiments in which the entire panel of chimeras shown in Fig. 5 were evaluated, no consistent differences in Env surface expression were seen between CD4-dependent and -independent Env proteins or among chimeras showing greater fusogenicity (not shown). These findings indicate that neither CD4 independence, the increased fusogenicity of chimeras containing an 8x TM, nor the poor fusogenicity of the 8x(gp120) chimera could be explained by different levels of Env on the cell surface.
FIG. 7.
Surface expression of CD4-dependent and -independent Env proteins on transfected cells. QT6 cells were transiently transfected with the Env proteins indicated or a vector-only control (pCDNA), and surface expression was quantitated by FACS. Shown for each Env protein is a histogram for cells labeled with NHS (thin line) or serum from an HIV-1-infected patient (bold line). The percent cells positive (% Pos) and the mean channel of fluorescence intensity (MCF) are indicated for cells stained with anti-HIV serum. Thresholds for positivity were defined so that <5% of cells stained with NHS were considered reactive.
Evaluation of a CD4-dependent clone from IIIBx-infected cells.
We also derived a CD4-dependent Env from IIIBx-infected SupT1 cells. This clone, termed S10, was able to mediate fusion on QT6 cells that coexpressed CXCR4 and CD4 but was unable to fuse in the absence of CD4 (Fig. 8). Sequence analysis showed that S10 shared several mutations with 8x relative to HXBc2 (eight in gp120 and three in gp41 [Fig. 4]). In addition, S10 contained several unique mutations: in gp120, G431E in C4 and S461N in V5; in TM, six additional amino acid changes in the ectodomain and membrane-spanning domain, including the loss of predicted N-linked glycosylation sites at positions 611 and 674 (Fig. 4). S10 also contained a 55-nt deletion in the TM cytoplasmic tail that, similar to 8x, produced a frameshift mutation and a prematurely truncated cytoplasmic tail. This deletion also disrupts the rev open reading frame by eliminating the nuclear localization signal at the N terminus and introducing a frameshift mutation that truncates the protein before the Rev response element binding site (47). Finally, S10 lacked several changes that were present in the 8x gp120 (S143G, G167S, Q310H, and I423V) and TM (N651I). Even though S10 was functional in fusion assays on CD4+ CXCR4+ target cells, this env was unable to generate infectious virus when cloned into NL4-3 (not shown), likely as a result of its nonfunctional Rev protein.
FIG. 8.
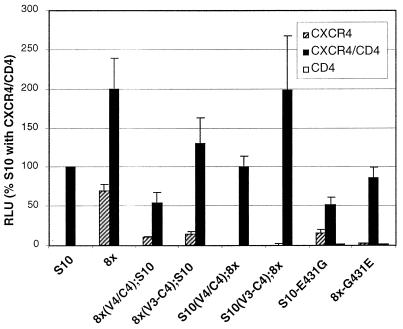
Mapping determinants for a CD4-dependent clone of IIIBx. Fusion activity is shown for the CD4-dependent S10 clone of IIIBx and S10-8x chimeras indicated in Fig. 5. In addition, activity is shown for an S10 Env in which the G431E mutation in the C4 domain was corrected (S10-431G) and for an 8x Env that contained this mutation (8x-431E). Results are expressed as the percentage of S10 luciferase activity on target cells that coexpressed CXCR4 and CD4.
Because the S10 Env shared several mutations with 8x but was completely CD4 dependent, we made a set of chimeras between 8x and S10 to identify the determinants for this change. As shown in Fig. 8, chimeras that contained the 8x V3 and V4/C4 [8x(V3-C4);S10] or V4/C4 alone [8x(V4/C4);S10] on an S10 background exhibited some CD4 independence, while the reciprocal chimeras on an 8x background [S10(V3-C4);8x] and [S10(V4/C4);8x] were completely CD4 dependent. Because the S10 V4/C4 domain contained a unique G431E mutation, we considered the possibility that this change could have a negative effect on CD4 independence. Indeed, when a Gly was restored at this position in S10 (S10-E431G), this clone exhibited a limited degree of CD4 independence on CXCR4-expressing target cells. Moreover, when the G431E mutation was introduced into 8x (8x-G431E), this clone remained fusion competent but became completely CD4 dependent (Fig. 8). Thus, a charge change within the C4 domain was sufficient to abrogate CD4 independence of 8x, supporting mapping data from the 8x-HXBc2 chimeras described above that implicate this region as being critical to the CD4-independent phenotype.
CCR5-tropic V3 loop alters chemokine receptor specificity but not CD4 independence.
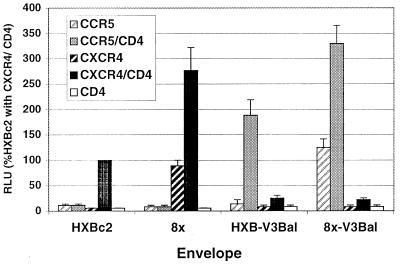
Given the importance of the V3 loop in determining chemokine receptor specificity and the evidence that determinants for CD4 independence were located outside this domain, we sought to determine the extent to which tropism and CD4 independence of 8x could be dissociated. An HXBc2 gp120 that contained the V3 loop from the macrophage/CCR5-tropic isolate HIV-1/BaL (HXB2-V3BaL) (31) was used to introduce the BaL V3 loop into 8x. The resulting chimera (8x-V3BaL) was compared to 8x, HXBc2, and HXB2-V3BaL in fusion assays on target cells that expressed CXCR4 or CCR5, in the presence or absence of CD4 (Fig. 9). As expected, HXBc2 and HXB2-V3BaL fused with CXCR4- and CCR5-expressing cells, respectively, and their activity was strictly CD4 dependent. In contrast, the 8x-V3BaL chimera was both CCR5 tropic and CD4 independent. Thus, determinants for the CD4 independence of 8x are functionally distinct from those that mediate tropism for chemokine receptors.
FIG. 9.
Construction of a CCR5-tropic, CD4-independent Env. 8x and HXB2 Env proteins containing V3 loop from the CCR5-tropic Env, HIV-1/BaL, were constructed, and their fusion activities were compared to those of parental 8x and HXBc2 Envs on target cells that expressed CCR5 or CXCR4 with or without CD4. Fusion activity is expressed as the percentage of luciferase activity of HXBc2 on target cells that expressed both CXCR4 and CD4. Bars indicate mean + SEM.
DISCUSSION
In this report, we describe the derivation and characterization of a CD4-independent variant of HIV-1/IIIB, termed IIIBx, that could utilize CXCR4 in the absence of CD4. The 8x env clone of IIIBx was able to generate a replication-competent, CD4-independent virus when cloned into an HIV-1 provirus and could mediate fusion on CXCR4-expressing quail cells in the absence of CD4. This clone remained fusion competent when Arg was substituted for Asp at gp120 position 368, a residue previously shown to be critical to the formation of the CD4 binding site (34, 44). Sequence analysis of 8x revealed 18 mutations that have not been described in other HIV-1/IIIB proviral clones and a remarkable net loss of five glycosylation sites on gp120. Reciprocal chimeras between 8x and a related CD4-dependent clone, HXBc2, indicated that the determinants for CD4 independence mapped at least in part to regions outside the hypervariable V1/V2 and V3 loops. An HXBc2 chimera that contained both the V4/C4 and TM domains of 8x was CD4 independent, while chimeras that contained either domain alone were CD4 dependent. In addition, a CD4-dependent clone from the IIIBx swarm, S10, that contained a unique G431E mutation in the gp120 C4 domain became CD4 independent when this mutation was corrected. Introduction of the G431E mutation into 8x rendered this Env completely CD4 dependent, indicating that a charge change at this position was sufficient to disrupt CD4-independent but not CD4-dependent utilization of CXCR4. In a recent publication, we also demonstrated that the 8x gp120 bound directly to cells expressing CXCR4 in the absence of CD4, while the HXBc2 gp120 required pretreatment with soluble CD4 for binding to occur (28). Collectively, these findings indicate that a chemokine receptor binding site exists on the gp120 core and that mutations in this region can, in association with alterations in TM, render an HIV-1 Env CD4 independent.
The HIV-1 V3 loop has been shown to be a principal determinant for chemokine receptor specificity for CCR5 and CXCR4 (9–11, 59, 64, 66). More recently, the V1/V2 region has also been shown, in the context of an appropriate V3, to mediate use of additional chemokine receptors including CCR3, CCR2b, STRL33, and APJ (29, 53), suggesting that cooperative interactions between V1/V2 and V3 are involved in chemokine receptor recognition. These loops are known to undergo conformational changes following CD4 binding (32, 42, 66, 68) that may facilitate an interaction with a particular chemokine receptor (32, 66, 68). While these findings have suggested that V3 itself may contain a chemokine receptor binding site, the marked genetic diversity of V3 loops among CCR5- or CXCR4-tropic viruses indicates either that these loops contain a common structural element or that other regions on Env also contribute to chemokine receptor utilization. Recently, mutagenesis of a CCR5-tropic HIV-1 gp120 has identified a probable CCR5 binding site on Env that is formed by a bridging sheet connecting the inner and outer domains of the gp120 core. This region is located between the bases of the V1/V2 and V3 loops and is predicted to be oriented toward the cell membrane following CD4 binding (52). The remarkable conservation of amino acids in this region among CCR5- and CXCR4-tropic Envs suggested that this site could represent a generic chemokine receptor binding domain capable of interacting with multiple chemokine receptors. These findings are consistent with a model in which CD4 induces movement of the V1/V2 and V3 loops, facilitating an initial interaction with a specific chemokine receptor and exposing this conserved binding site, which is required for fusion to occur (52, 67).
Because determinants for CD4 independence of the 8x clone mapped outside regions required for chemokine receptor specificity, we hypothesized that a different V3 might change the chemokine receptor tropism of 8x without affecting its CD4 independence. Remarkably, when the V3 loop from a CCR5-tropic Env (HIV-1/BaL) was inserted into 8x, the resulting chimera was able to mediate CD4-independent fusion on CCR5-expressing cells. No fusion on CXCR4-expressing cells with or without CD4 was observed for this chimera. In contrast, a chimera containing the HIV-1/BaL V3 loop on an HXBc2 background utilized CCR5 but was completely CD4 dependent. Similar results have recently been shown for soluble gp120s from these Env proteins in binding assays on CXCR4- and CCR5-expressing cells (28). These findings have clearly indicated that chemokine receptor specificity and CD4-independent utilization of that chemokine receptor are mediated by distinct regions of gp120. Moreover, our data also provide direct evidence that although specificity determinants on V3 are still required, a region on the gp120 core that is rendered functional on CD4-independent viruses is able to mediate fusion by using genetically divergent chemokine receptors.
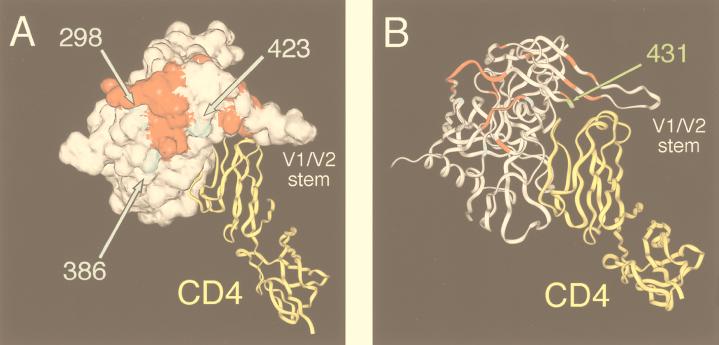
The bridging sheet on gp120 noted above is made up largely of amino acids from the C4 domain and the V1/V2 stem (52). This region has also been shown to contribute to the formation of gp120 epitopes that are induced by CD4 binding (34, 63). Interestingly, the two mutations in the 8x V4/C4 domain (N386K and I423V) and a third mutation near the base of the V3 loop (R298K) map to positions that immediately flank this area (Fig. 10A). As we have shown, an 8x chimera that included the corresponding V3 and V4/C4 domains from HXBc2 and that lacked these mutations was highly competent for fusion but was completely CD4 dependent (Fig. 6). The remarkable proximity of R298K, N386K, and I423V to the putative chemokine receptor binding domain suggests that they may expose this site and/or help to present it to the chemokine receptor during viral attachment. Recent findings from our lab have demonstrated that recombinant 8x gp120 is able to bind directly to CXCR4-expressing cells independently of CD4 and that CD4-induced epitopes that are partially contained within the gp120 chemokine receptor binding domain are stably exposed in the absence of CD4 binding (28). In addition, the G431E mutation in C4, which was sufficient to abrogate CD4 independence on S10 and 8x, is shown by the crystal structure of the gp120 core to be juxtaposed to residues at the base of the V1/V2 stem that contribute to the chemokine receptor binding site (Fig. 10B). The acquisition of a negative charge at this residue could alter the orientation of the V1/V2 loops and/or affect the conformation of the chemokine receptor binding site. Regardless of the mechanism, it is apparent that mutations in or around this chemokine receptor binding site can positively and negatively affect the ability of the 8x Env to function without CD4 and is consistent with the view that CD4 binding improves the overall efficiency and/or avidity of chemokine receptor utilization.
FIG. 10.
Locations of HIV-1/IIIBx mutations on the gp120 crystal structure. (A) A space-filling model of the HIV-1/HXB2 gp120 core crystal structure is shown (white) in conjunction with a ribbon diagram for CD4 (yellow) (34). Amino acid sites at which mutations produced a 50% decrease or increase gp120 binding to CCR5 are shown in red (52). Of the six mutations in 8x that could be mapped onto the gp120 core, three (shown in blue) are located immediately adjacent to this putative chemokine receptor binding site. (B) A ribbon diagram of the gp120-CD4 complex is depicted in a slightly different orientation to indicate the position of the G431E mutation, which was sufficient to abrogate CD4-independent but not -dependent fusion of the 8x clone.
While the V4/C4 domain is clearly involved with CD4 independence of IIIBx, it is apparent that other regions of the Env also contribute. A chimera that contained the 8x V4/C4 on an HXBc2 background was CD4 independent only when it also contained the 8x TM (Fig. 6). In a previous study of the CD4-independent HIV-2 isolate ROD-B, Reeves and Schulz observed that mutations in both gp120 and TM (i.e., a Leu-to-Phe mutation just proximal to the analogous V4 loop and two mutations in the first heptad repeat of the TM ectodomain) were required for this phenotype (49). Although the underlying mechanism for this effect is unclear, regions of the HIV-1 TM have been implicated in a number of cooperative interactions with gp120 that could affect its binding to CD4 and/or chemokine receptors (5, 7, 39, 45). Of note, the putative chemokine receptor binding site on gp120 noted above is located near the predicted trimer axis of the assembled Env oligomer where interactions with TM are likely to occur (34). In recent experiments, we observed that an HXBc2 chimera containing only the 8x V4/C4 and the 8x frameshift mutation in TM was able to mediate modest CD4-independent fusion (unpublished observations). Although the mechanism for this effect is unclear, there is precedent for changes in the TM cytoplasmic tail inducing structural alterations in the TM ectodomain (51, 61) and/or gp120 (5, 7, 39). As we demonstrated, although mutations in the TM cytoplasmic tail may modulate Env expression on the cell surface (35, 43, 60), our findings indicate that neither increased surface expression nor increased fusogenicity per se can account for the CD4 independence of these clones. Finally, 8x also contains mutations that are predicted to eliminate five glycosylation sites in gp120, including N386K noted above, which lies adjacent to the putative chemokine receptor binding site. Carbohydrates have recently been implicated in modifying the immunogenicity of SIV gp120 and in masking neutralization epitopes (50), and it is possible that the loss of one or more glycosylation sites is involved in helping to expose this chemokine receptor binding site.
Although our studies have implicated mutations in the IIIBx V4/C4 and TM as determinants for CD4 independence, it should be noted that mutations in different regions of gp120 have been associated with CD4 independence for other HIV-1 isolates. A CD4-independent variant of HIV-1/NDK that could infect CXCR4+ CD4− HeLa cells by virtue of a combination of mutations in the C2, C3, and V3 domains has been described (18). Recent findings by Kolchinsky et al. showed that determinants for a CD4-independent, CCR5-tropic variant of HIV-1/ADA mapped to point mutations in the distal region of the V1/V2 stem (33). Despite these genetic differences, CD4-independent viruses could have similar structural bases for this phenotype. In this regard, at least some of the changes in CD4-independent HIV-1/NDK and HIV-1/ADA are similar to those in IIIBx, being located near the gp120 bridging sheet where they could affect the presentation of this region to a chemokine receptor.
HIV has evolved strategies that enable viral replication to continue in spite of a vigorous host immune response (46, 65). Neutralizing antibodies typically arise late in the course of infection, if at all, and are frequently directed at type-specific rather than group-specific determinants on gp120 (25, 41, 67). The deduced crystal structure of the gp120 core has suggested that the CD4 binding domain and the chemokine receptor binding site are poorly accessible and/or are concealed within the Env oligomer (52, 67). In contrast, the exposed surfaces of gp120 contain hypervariable domains and carbohydrates that may serve as immunologic decoys for the humoral immune response (6, 50, 62). Approaches to expose these conserved and functionally critical domains may enable qualitatively different and perhaps more efficacious immune responses to be generated. Recent studies by LaCasse and coworkers have demonstrated that a fusion-activated form of Env in which conserved neutralization epitopes on gp120 and gp41 were apparently exposed was able to generate a potent and broadly cross-neutralizing antibody response in mice (36). As an alternate or complementary approach, CD4-independent Envs that are derived or designed may provide a means to identify and present these domains in a biologically relevant context. As noted above, we have recently shown that the soluble 8x gp120 protein exhibits a number of novel immunological and biochemical properties including the stable exposure of CD4-induced epitopes and the ability to bind to CXCR4 in the absence of CD4 (28). These studies have also shown that IIIBx is highly sensitive to neutralization by sera from some HIV-infected individuals and to MAbs reactive with CD4-induced epitopes, suggesting that functional domains that contribute to CD4 independence are also neutralizing sites (28). Future studies of IIIBx and additional CD4-independent isolates should provide powerful tools to probe the structure and function of the viral envelope glycoprotein and lead to the rational design of gp120 molecules with altered immunogenic properties.
ACKNOWLEDGMENTS
We thank D. Littman for 3T3 cells expressing human CXCR4, B. Cullen for the HXB2-V3BaL chimeric Env, and J. Sodroski for providing unpublished data. HIV-1NL4-3 hemigenome clones p210-19 and p83-10 were supplied by Ronald Desrosiers through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Receptor clones for expression were kindly provided by Dennis Kolson (CD4), Marc Parmentier (CCR5), and Steve Peiper (CXCR4). The T7 polymerase recombinant vaccinia virus was a generous gift from Bernard Moss.
This work was supported by grants R21 AI44308 (C.C.L.), RO1-AI40880 (R.W.D.), and RO1-AI45378 (J.A.H.). T.L.H. was supported by the University of Pennsylvania Franklin Scholars Program and the Pine Family Foundation.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Bandres J C, Wang Q F, O’Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 5.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen S S, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 9.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 12.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 14.Deng H K, Unutmaz D, KewelRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 15.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 16.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 17.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl P L, Broder C C, Doms R W, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edinger A L, Mankowski J L, Doranz B J, Marguilies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent SIV strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endres M J, Clapham P R, Marsh M, Ahuja M, Davis Turner J, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 22.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 25.Haigwood N L, Misher L, Chin S M, Blair M, Planelles V, Scandella C J, Steimer K S, Gardner M B, Yilma T, Hirsh V M. Characterization of group specific antibodies in primates: studies with SIV envelope in macaques. J Med Primatol. 1992;21:82–90. [PubMed] [Google Scholar]
- 26.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12(Suppl. A):S17–S26. [PubMed] [Google Scholar]
- 28.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoxie J A, Haggarty B S, Rackowski J L, Pilsbury N, Levy J A. Persistent noncytopathic infection of human lymphocytes with AIDS-associated retrovirus. Science. 1985;229:1400–1402. doi: 10.1126/science.2994222. [DOI] [PubMed] [Google Scholar]
- 31.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 32.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and coreceptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 33.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on cells. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 37.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 39.Matthews T J, Wild C, Chen C H, Bolognesi D P, Greenberg M L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 40.Misse D, Cerutti M, Schmidt I, Jansen A, Devauchelle G, Jansen F, Veas F. Dissociation of the CD4 and CXCR4 binding properties of human immunodeficiency virus type 1 gp120 by deletion of the first putative alpha-helical conserved structure. J Virol. 1998;72:7280–7288. doi: 10.1128/jvi.72.9.7280-7288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore J P, Ho D D. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS. 1995;9(Suppl. A):S117–S136. [PubMed] [Google Scholar]
- 42.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulligan M J, Yamshchikov G V, Ritter G D, Jr, Gao F, Jin M J, Nail C D, Spies C P, Hahn B H, Compans R W. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olshevsky U, Helseth E, Furman D, Li J, Haseltine W A, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–5705. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park E J, Quinnan G V., Jr Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J Virol. 1999;73:5707–5713. doi: 10.1128/jvi.73.7.5707-5713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 47.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 48.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 49.Reeves J D, Schulz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1996;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 51.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 52.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 53.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of the HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 55.Rucker J, Doranz B J, Edinger A L, Long D, Berson J F, Doms R W. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 56.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M F, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizu H, Hasebe F, Tsuchie H, Morikawa S, Ushijima H, Kitamura T. Analysis of a human immunodeficiency virus type 1 isolate carrying a truncated transmembrane glycoprotein. Virology. 1992;189:534–546. doi: 10.1016/0042-6822(92)90577-c. [DOI] [PubMed] [Google Scholar]
- 59.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spies C P, Compans R W. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology. 1994;203:8–19. doi: 10.1006/viro.1994.1449. [DOI] [PubMed] [Google Scholar]
- 61.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trkola A, Ketas T, KewalRamani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H. Viral dynamics in human immunodeficiency virus type 1 infection. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 66.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 67.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 68.Wyatt R, Thali M, Tilley S, Pinter A, Posner M, Ho D, Robinson J, Sodroski J. Relationship of the human immunodeficiency virus type 1 gp120 third variable loop to a component of the CD4 binding site in the fourth conserved region. J Virol. 1992;66:6997–7004. doi: 10.1128/jvi.66.12.6997-7004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, Ho D D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, KewalRamani V N, Moore J P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]