Abstract
Exosomes are extracellular vesicles well known for facilitating cell-to-cell communication by distributing essential macromolecules like proteins, DNA, mRNA, lipids, and miRNA. These vesicles are abundant in fluids distributed throughout the body, including urine, blood, saliva, and even bile. They are important diagnostic tools for breast, lung, gastrointestinal cancers, etc. However, their application as cancer biomarkers has not yet been implemented in most parts of the world. In this review, we discuss how OMICs profiling of exosomes can be practiced by substituting traditional imaging or biopsy methods for cancer detection. Previous methods like extensive imaging and biopsy used for screening were expensive, mostly invasive, and could not easily provide early detection for various types of cancer. Exosomal biomarkers can be utilized for routine screening by simply collecting body fluids from the individual. We anticipate that the use of exosomes will be brought to light by the success of clinical trials investigating their potential to enhance cancer detection and treatment in the upcoming years.
Graphical Abstract
Keywords: Exosomes, Cancer biomarkers, Clinical signature, Diagnostic tool, Prognostic indicator, Tumor-derived exosomes, Molecular profiling, Cancer diagnosis
Introduction
In the era of precision medicine, identifying and validating reliable cancer biomarkers are paramount for early detection, accurate diagnosis, and effective treatment strategies. Exosomes have gained substantial attention among emerging candidates as promising carriers of valuable information in cancer research. Exosomes are tiny extracellular vesicles (EVs) with a diameter ranging from 30 to 150 nm [1]. They are secreted by cells into bodily fluids such as blood, urine, cerebrospinal fluid (CSF), and saliva. These vesicles transport a collection of nucleic acids, proteins, and lipids. According to the exosome database ExoCarta, 9769 proteins, 3408 mRNAs, 2838 miRNAs, and 1116 lipids have been identified in exosomes from various organisms and bodily fluids [2]. Due to their distinct attributes, including stability, specificity, and capability to traverse biological barriers, exosomes are promising candidates for detecting cancer biomarkers. Exosomes can carry molecules representative of the original cell, including cancer cells. Early cancer detection is made possible by analyzing exosomal content when the cancer cells release the exosomes into the bloodstream or other biofluids. Early detection is essential for treatment to begin at a more manageable and possibly curable stage. Exosome formation entails the fusion and exocytosis of multivesicular bodies (MVBs), releasing them into the extracellular environment [3, 4].
Exosomes facilitate the transfer of exosome-associated RNA to recipient cells, influencing protein functioning and contributing to cellular stress and damage in diseased states. They play a diverse role in various diseases, encompassing neurodegenerative, cancerous, hepatic, and cardiovascular conditions. In the context of cancer, exosomes hold substantial implications for metastasis, drug resistance, and angiogenesis. Specifically, they can modify the extracellular matrix to create a favorable environment for tumor cell migration [5, 6]. Moreover, exosomes influence the migration, invasion, and release of cancer cells by affecting tumor suppressor genes and degrading the extracellular matrix [7, 8].
Most tumors exhibit heterogeneity, comprising different cell types with diverse molecular profiles. In contrast to a single tissue biopsy, exosomes, released by various tumor-resident cells, provide a more comprehensive and representative view of the tumors' heterogeneity. Understanding this complexity is crucial for tailoring treatment plans effectively. The contents of exosomes act as molecular signatures of their cells of origin, making them promising biomarkers. The stability of exosomes, ensured by the lipid bilayer protecting them from external proteases and enzymes, enhances their appeal as diagnostic markers. Consequently, diagnostic tests based on exosomes are gaining momentum for early cancer detection and addressing various ailments. Exosomes can be isolated from easily accessible biofluids such as blood, urine, saliva, and CSF. When comparing liquid biopsy to conventional tissue biopsies, a non-invasive approach, significant improvements become evident. Liquid biopsies offer a less invasive and more dynamic method to monitor the course of cancer and assess the effectiveness of treatment. Exosomes can traverse the blood–brain barrier (BBB) under specific conditions [9], opening up possibilities for therapies involving small molecules, RNA therapy, proteins, and CRISPR gene editing. In RNA therapy, exosomes can deliver RNA molecules, such as mRNA or siRNA, directly to cancer cells, selectively silencing oncogenes or restoring tumor suppressor genes to reduce tumor growth and metastasis [10, 11]. Similarly, exosomes serve as a promising delivery system for CRISPR-Cas9 components, enabling precise genetic editing to correct mutations driving cancer development [12, 13]. They can complement chimeric antigen receptor T (CAR-T) cell therapies targeting cancer cells. CAR exosomes, derived from CAR-T cells, bear CAR on their surface, exhibit elevated levels of cytotoxic molecules, and impede tumor growth [14], thereby enhancing the overall efficacy of CAR-T cell therapy by extending the therapeutic effect beyond the initial infusion site. Cancer cell-derived exosomes, carrying tumor-associated antigens, can modulate the immune response by recruiting and activating dendritic cells (DCs) and other antigen-presenting cells, which stimulate cytotoxic T lymphocytes to recognize and destroy cancer cells [15]. Experimental evidence suggests that circulating exosomes from cancer patients can be utilized for cancer diagnosis and prediction of therapeutic outcomes, potentially reducing the need for invasive biopsies.
This review article aims to comprehensively explore the clinical signature of exosome-based cancer biomarkers, providing an overview of their diverse roles in cancer progression, diagnosis, prognosis, and therapeutic monitoring. This paper uniquely focuses on the dynamic nature of exosomes, which renders them highly suitable for tracking the advancement of the disease, the reaction to treatment, and the emergence of resistance, thereby offering a more sophisticated comprehension of the changing terrain of cancer. We will delve into the biogenesis of exosomes and their specific cargo, including microRNAs (miRNAs), proteins, and metabolites, highlighting their potential as non-invasive biomarkers for various cancer types. This review elucidates the complex exosomal formation and release processes, contributing to our understanding of their roles in cancer progression. Furthermore, we will explore the latest advancements in exosome isolation techniques, analytical methods, and high-throughput technologies that enable the profiling and characterization of exosomal biomarkers, including advanced techniques like OMICS and single exosome profiling. Exosomes can aid in initiating and spreading cancer and are involved in intercellular communication; knowing the precise chemicals that exosomes carry could help identify possible targets for treatment. This discussion also includes ongoing clinical trials to bridge research and clinical practice and a balanced analysis of the advantages and disadvantages of using exosomes as cancer biomarkers, offering insights into their dual roles in cancer promotion and inhibition. Treating cancer in new ways may be possible by focusing on exosomes or their pathways. Additionally, we explore future prospects, such as bioengineering exosomes for enhanced therapeutic capabilities. Ultimately, a deeper understanding of the clinical signature of exosome-based biomarkers will enhance personalized and targeted cancer management approaches, fostering the advancement of precision medicine toward improved patient outcomes and ensuring the review reflects the latest research and innovations in the field.
Biogenesis, secretion, and uptake of exosomes
The endosomal compartment of most eukaryotic cells produces exosomes, which are membrane-bound EVs [16]. These EVs are the intermediate by-products of plasma membrane-derived early- to late endosomes [17, 18]. Processing of early endosomes (EEs) produces a subtype of endosomes carrying several membrane-bound intraluminal vesicles (ILVs) called MVBs. These MVBs consequently fuse with the plasma membrane to release their contents, exosomes, out of cells [19]. There are two distinct mechanisms by which exosomes are produced: ESCRT-dependent (Endosomal sorting complex required for transport)-dependent and ESCRT-independent [19].
The formation of MVBs is regulated by the ESCRT, which consists of four multiprotein complexes: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III. These complexes are recruited to the endosomal membrane to sort selected proteins into ILVs, requiring the ubiquitination of the cytosolic tails of endocytosed receptors [20]. Tsg101, part of ESCRT-I, binds ubiquitinated cargo proteins, activating ESCRT-II, which then initiates the formation of ESCRT-III. ESCRT-III sequesters MVB proteins and recruits a deubiquitinating enzyme to remove ubiquitin tags before sorting proteins into ILVs. An ATPase disassembles the ESCRT-III complex afterward [21].
While ESCRT proteins are essential for targeting membrane proteins for lysosomal degradation, their role in forming ILVs secreted as exosomes is unclear. Proteomic analyses have identified ESCRT complex members such as Alix and Tsg101 in DC exosomes, supporting ESCRT-dependent exosome biogenesis [22]. An ESCRT-0 member has also been implicated in DC exosome secretion [23]. However, targeting MHC class II molecules in activated DCs does not require ubiquitination, and neither Tsg101 nor Alix are involved in proteolipid protein (PLP) sorting into exosomes in oligodendroglial cells [24]. Additionally, the sequestration of the pre-melanosomal protein Pmel17 in ILVs in melanocytes appears independent of ESCRT function [25]. These findings suggest that different MVB subpopulations might use distinct biogenesis mechanisms across various cell types or within the same cell type.
However, the scientific rationale for the ESCRT-independent pathway could be more evident. Numerous studies indicate that some exosomal proteins involving alternative mechanisms are released independently of the ESCRT pathway. Trajkovic et al. demonstrated that while Tsg101 and Alix do not influence the exosomal sorting of PLP, ceramide is essential for the secretion of PLP-containing exosomes [24]. Ceramide's cone-shaped structure may aid membrane invagination of ILVs, and studies have highlighted the role of sphingomyelinases, enzymes converting sphingomyelin to ceramide, in exosome biogenesis. Specifically, acid sphingomyelinase is involved in vesicle release from glial cells, and neutral sphingomyelinase 2 is crucial for miRNA-containing vesicle secretion [26, 27]. Additionally, higher-order oligomerization, or the clustering of protein oligomers, has been implicated in exosome formation, as seen in the exosomal sorting of CD43 in Jurkat T-cells [28] and similar processes involving the transferrin receptor in reticulocytes [29] and the MHC class II complex in lymphocytes [30]. In these instances, antibody-induced oligomerization enhances protein secretion into exosomes. The biogenesis of MVBs is also linked to detergent-resistant domains in exosomal membranes, which include tetraspanin proteins. For example, sorting MHC class II into DC exosomes partially depends on its integration into tetraspanin CD9-containing lipid microdomains [31–33].
One intriguing question is: How does the cell decide what to package into exosomes, and what are the proposed mechanisms for this selective activity? The selective packaging of exosomal content may involve lipid composition, protein and RNA signals, and specific enzymes like sphingomyelinases. Higher-order oligomerization and the presence of tetraspanin-enriched microdomains are also factors influencing what gets packaged into exosomes. Understanding these selective mechanisms further could provide deeper insights into exosome biology.
Additionally, while we have noted differences between exosomes derived from normal and cancer cells, the detailed biogenesis mechanisms in cancer cells require further elucidation. It is conceivable that cancer cells have deregulated or preferential pathways for exosome formation. Identifying these pathways and understanding how they are altered in cancer could uncover potential therapeutic targets. By targeting the unique exosome biogenesis pathways in cancer cells, we might be able to develop treatments that disrupt exosomal communication, contributing to tumor growth and metastasis. Further research could significantly advance our knowledge and therapeutic strategies in oncology.
Exosome secretion and uptake are crucial for maintaining normal cellular activity because they facilitate essential intercellular communication, allowing cells to exchange proteins, lipids, RNA, and other molecules. This exchange supports immune response regulation, tissue repair, and cellular homeostasis. If exosome secretion or uptake is altered, it can disrupt these vital communications, leading to various pathological conditions. For instance, impaired exosome function can contribute to diseases like cancer, where altered exosome-mediated signaling can promote tumor growth, metastasis, and drug resistance [34].
Exosome and cancer
It is well known that exosome secretion and uptake are other important, influential features in normal cellular activity. They play pivotal roles in the intricate landscape of cancer cell communication, contributing to tumor progression and metastasis. Cancer cells release more exosomes than normal cells, using them to exchange information locally and distantly. These exosomes carry bioactive cargo that can promote tumor growth, pre-metastatic niche formation, immune escape, angiogenesis, anti-apoptotic signaling, and drug resistance [35]. In cancer cells, the secretion of exosomes is often dysregulated, leading to the release of a unique cargo that can influence both the local tumor microenvironment and distant organs [36]. Cancer cells release more exosomes than normal cells, using them to exchange information locally and distantly. These exosomes carry bioactive cargo that can promote tumor growth, pre-metastatic niche formation, immune escape, angiogenesis, anti-apoptotic signaling, and drug resistance [37].
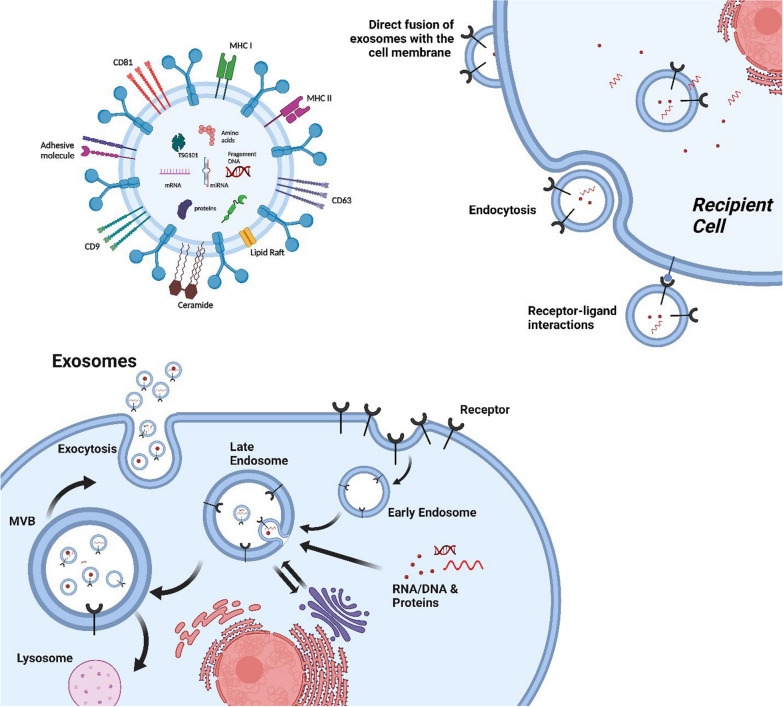
On the receiving end, cancer cells internalize exosomes through various mechanisms, such as endocytosis, phagocytosis, and direct membrane fusion. Once internalized, exosomal cargo can modulate recipient cell behavior, promoting cell proliferation, migration, and evasion of immune surveillance [38]. Figure 1 represents the process simply.
Fig.1.
Biogenesis, Secretion, and Uptake of Exosomes. Exosomes form through the process of inward budding during endocytosis. Specific cargos are sorted into these exosomes within multivesicular bodies (MVBs), where early and late sorting endosomes are assembled. Commonly, Exosomes consist of proteins, lipids, RNAs, and genetic material. The protein content of EVs includes various types, such as transmembrane or lipid-bound proteins found on the cell surface (CD63, CD9, CD81, etc.). Additionally, Exosomes contain lipids like ceramide, different types of RNAs such as messenger RNA (mRNA) and microRNA (miRNA), and DNA fragments. Exosomes are taken up by cells using several mechanisms, including direct fusion of exosomes with the cell membrane of the recipients, receptor-ligand interactions, and endocytosis. EVs transport their contents within the cells comprising proteins, RNAs, and DNAs, releasing them into the cytoplasm or endoplasmic reticulum. MVB: multivesicular body; CD: cluster of differentiation, Created with BioRender.com
Sorting cargo into exosomes is also important in the context of cancer cells compared to normal cells. It is a finely tuned cellular process governed by various molecular mechanisms. One significant aspect is the presence of specific signals and sorting motifs that guide the inclusion of proteins and nucleic acids into these EVs. For instance, certain proteins carry signals that earmark them for exosomal packaging [39].
In the context of cancer, the cell state undergoes substantial changes, leading to alterations in the cargo composition of exosomes. Cancer cells may preferentially sort oncoproteins or mutated nucleic acids into exosomes, disseminating cancer-related information to neighboring or distant cells [40].
The role of exosomes in cancer progression is complex and dual-faceted, depending on their origin and cargo. For instance, exosomes can carry various microRNAs (miRNAs), such as miR-122 [41], which promotes metastasis; mir-9 [42], and miR-135b [43], which enhances angiogenesis; and miR-105, which induces vascular leakiness and promotes metastasis [44]. Other miRNAs like miR-93-5P [45], miR-200 [46], and miR-210 contribute to cancer cell proliferation, metastasis, and tumor progression [47]. Conversely, miR-126 can promote an anti-tumor response, highlighting the dual roles of exosomal miRNAs in cancer regulation [48]. Similarly, long non-coding RNAs (lncRNAs) such as lncRNA-HOTTIP [49]and lncRNA-ZFAS1 [50]enhance drug resistance and cell proliferation. At the same time, proteins like [51] and TGF-β [52] facilitate metastasis and suppress immune responses These bioactive molecules collectively drive cancer progression by enhancing the tumor's ability to invade, resist treatment, and manipulate its surrounding environment.
Consequently, exosomes have become a focal point for developing novel cancer therapies. Strategies include using naturally derived exosomes from immune cells to suppress cancer, inhibiting the release of cancer-derived exosomes, and employing exosomes as carriers for genes or anti-cancer drugs. DC exosomes are particularly promising for cancer therapy due to their antigen-presenting capabilities, which can activate tumor-specific cytotoxic T lymphocytes and natural killer cells [53]. Interfering with cancer cell-derived exosomes by blocking their synthesis, release, or uptake also presents a viable therapeutic approach. For example, inhibiting Rab27a-mediated exosome secretion has been shown to reduce tumor growth and metastasis in mice [54]. Additionally, using exosomes as carriers for miRNAs, proteins, or chemotherapeutic drugs can enhance targeted delivery and therapeutic efficacy while minimizing side effects [55].
Efforts to understand the relationship between cell states, such as cancer transformation and cargo sorting in exosomes, are vital. Researchers seek to unravel how the sorting of goods in cancer cells differs from that of normal cells. This exploration can uncover valuable insights into disease mechanisms and potentially reveal diagnostic markers or therapeutic targets based on the distinctive cargo profiles of cancer cell-derived exosomes [56].
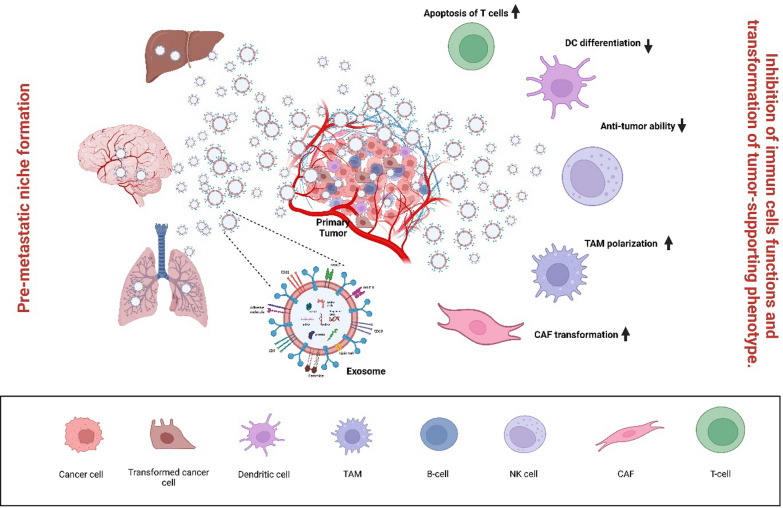
To completely understand the role of exosomes in cancer, their specific physiological functions must be elucidated [57]. The necessity of exosome production for cellular survival is still under debate, and creating mice with complete exosome deletion can help address this question [52, 58, 59]. Furthermore, concluding the functional importance of exosomes in cell-to-cell communication based solely on in vitro experiments in isolated culture systems may not accurately represent physiological conditions. Therefore, there is an increasing need for in vivo studies to explore the functional significance of exosomes in cancer. Figure 2 depicts the relationship between exosomes and cancer progression.
Fig. 2.
Exosomes and Cancer. The role played by exosomes in cancer metastasis and progression. Tumor cells release pathogenic exosomes that inhibit immune cell functions (recognition of cancer cells, cytolytic effects, etc.) and pre-mesenchymal niche formation, Created with BioRender.com
Exosome as source of cancer biomarker
As pointed out earlier, exosomes have been detected in various body fluids such as blood, urine, saliva, amniotic fluid, CSF, ascites, tears, breastfeeding milk, semen, etc. [60, 61]. Cancer exosomes, when collected from a heterogeneous population in body fluids, can aid in diagnosing specific tumor types like glioblastoma, melanoma, pancreatic, breast, and ovarian cancers [62]. Exosome-based biomarker detection is promising in cancer research, offering several advantages over other detection methods [63, 64]. Exosomes can be easily isolated from various bodily fluids, providing convenient accessibility for biomarker analysis [64]. In contrast to invasive tissue biopsies, exosomes offer a non-invasive approach to gathering valuable information. These EVs exhibit remarkable stability in circulation and are defended from enzymatic degradation by a lipid bilayer membrane, ensuring the integrity of their cargo, including nucleic acids and proteins. This stability allows for detecting intact biomarkers, ensuring reliable and accurate results [65]. Furthermore, exosomes actively released by cells carry a selective cargo of biomolecules, reflecting the unique molecular characteristics of the originating cells. This composition provides a more specific and functionally relevant representation of the disease state [65]. Notably, cancer cells release exosomes at the early stages of tumorigenesis, enabling the detection of cancer-specific biomarkers before the onset of clinical symptoms. This early detection potential allows timely interventions and improved patient outcomes [65]. In vitro and preclinical investigations have improved our comprehension of exosome content and its potential use in cancer identification and monitoring [66]. Research also focuses on the role of lipids and metabolites in cancer-derived exosomes, offering new insights into cancer detection and biology [65, 67].
Exosomes may carry unique nucleic acids, such as mutant mRNA like the epidermal growth factor receptor EGFRvIII variant [68], which can serve as accurate biomarkers for glioblastoma. Targeted miRNAs enriched in exosomes can aid in cancer diagnosis and monitoring of cancer progression [62]. Exosome proteins may also contribute to cancer detection and reflect their cellular origin. The transfer of oncoproteins via exosomes between cells may function in carcinogenesis [69, 70].
Exosomes also offer a dynamic snapshot of disease progression and treatment response. Their continuous release and circulation in bodily fluids enable repeated sampling over time, facilitating real-time monitoring of disease status and treatment efficacy [71]. Moreover, exosomes carry various types of biomarkers, including miRNAs, lncRNAs, proteins, and metabolites, providing multiple targets for biomarker analysis. This diversity enhances the chances of identifying robust and reliable biomarkers tailored to specific cancer types [72].
Lately, the "liquid biopsy" technique has emerged as a potential non-invasive method for biomarker detection, utilizing bodily fluids like urine and serum. However, studies have shown that many liquid biomarkers are predominantly located in the lysosomes, limiting their accessibility [73, 74]. In contrast, exosomal biomarkers have demonstrated high diagnostic and prognostic efficiency for cancer detection [75–77]. 4729 individuals from 42 studies were included to check the specificity and sensitivity of exosomes as prognostic biomarkers. From them, 50 prognostic biomarkers were studied. For 13 biomarkers with overall survival present in colon cancer the I2 value (inconsistency index) was 62.94% and P < 0.002;% biomarkers with disease-free survival present in colon cancer showed the I2 value of 0% and P < 0.536, while biomarkers with recurrence-free survival present in colon cancer showed the I2 value of 89.61% and P < 0.0004 biomarkers with overall survival found in gastric cancer, 4 biomarkers with overall survival reported in pancreatic and 5 biomarkers with overall survival reported in liver cancer exhibited the I2 values and P values of 96.71%,81.50%,84.48% and 0.000,0.001,0.000 respectively; again 9 biomarkers showing overall survival in lung cancer had the I2 value of 89.50% and P < 0.000. So, it can be said exosomal biomarkers exhibit specificity and sensitivity, making them valuable tools in cancer diagnosis and monitoring [78].
Given these advantages, exosome-based biomarker detection emerges as a promising avenue in cancer research, offering non-invasive, specific, and dynamic insights into cancer biology, diagnosis, prognosis, and therapeutic monitoring. It can potentially revolutionize the field by providing a more accessible and comprehensive understanding of cancer and its treatment.
Exosomal surface protein as potential biomarkers of cancer
Recent research has shown that exosome-based diagnostics can be successfully developed using quick and high-throughput technology without exosome purification. A microfluidic device called the "ExoChip" was created by Kanwar et al. to capture and stain exosomes with the CD63 antibody and a fluorescent dye [79]. Exosomes may be quantified using a conventional plate reader, and exosomal miRNA analysis can be profiled. To diagnose glioblastoma multiform, a microfluidic chip labeled with a target (CD63, EGFR, or EGFRvIII) specific magnetic nanosensor was used [79]. The "ExoScreen" method, developed by Yoshioka et al. and using photosensitizing beads and CD9 and CD147 antibodies, is extremely quick and analytical. "ExoScreen" may identify EVs enriched in CD9 and CD147 double-positive EVs and grown in tissue culture media for colorectal cancer cells and patient serum [80, 81]. A more specific microfluidic device called the "ExoSearch" chip was created by Zhao et al. to isolate exosomes quantitatively using immunomagnetic beads [82]. Three exosomal tumor protein markers, including CA-125, EpCam, and CD24, were measured by the "ExoSearch" chip during a liquid biopsy of an ovarian cancer patient [56]. In this investigation, we pinpointed biomarkers for diagnosing colon cancer (CC) through proteomic analysis of small EV-derived from CC cell lines. These small-EVs were characterized by western blot analysis, nanoparticle tracking analysis, and transmission electron microscopy, with subsequent examination using mass spectrometry. Western blot analysis revealed the upregulation of five selected proteins in CC. Among these proteins, tetraspanin 1 (TSPAN1) exhibited elevated levels in plasma EVs from CC patients compared to those from healthy controls (HCs), demonstrating a sensitivity of 75.7%. These findings propose TSPAN1 as a robust, non-invasive biomarker for detecting CC [83]. Immune checkpoint inhibitor immunotherapy brings hope for gastric cancer (GC) treatment, but the lack of biomarkers hinders patient selection. Using an EV protein expression array, this study identified four key plasma EV-derived proteins (ARG1/CD3/PD-L1/PD-L2), forming an EV-score that robustly predicted and monitored immunotherapeutic outcomes in 112 GC patients. A high EV score indicates a microenvironment with enhanced antitumor immunity, validated through analysis and experiments. GC patients with EV-score ≥ 1 benefit more from ICIs. At the same time, EV-score < 1 suggests advantages in combining ICIs with HER2-targeted therapies, highlighting the plasma EV-score as a valuable tool for clinical decisions and insights for ICI-regimen improvements [84]. In the subsequent cohort, 96.4% of breast cancer patients exhibited elevated plasma-derived exosomal Del-1 levels at diagnosis. A high postoperative Del-1 level was significantly linked to worse disease-free survival adjusted for clinicopathological characteristics (hazard ratio 24.0; P < 0.0011). This study confirms exosomal Del-1 normalization post-surgery, establishing it as a robust diagnostic biomarker for breast cancer. Moreover, the association between high postoperative Del-1 levels and early relapse suggests its potential as a prognostic biomarker [85].
Exosomal nucleic acid as potential biomarkers of cancer
Ever since Valadi et al. first reported the existence of exosomal miRNAs in 2007, researchers have undertaken pioneering investigations to explore their potential as diagnostic biomarkers for different types of malignancies [86]. Taylor et al. identified eight miRNAs previously recognized as diagnostic indicators for ovarian cancer in circulating tumor exosomes from patients with ovarian cancer [87]. These miRNAs, namely miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205, and miR-214, were detected in the exosomes [88]. In a study by Rabinowits et al., miRNA profiling analysis was conducted on exosomes extracted from lung cancer patients, tumor biopsy samples, and control groups [88]. The results showed that exosomes from lung cancer patients and tumor biopsy samples exhibited similar miRNA patterns, distinct from those observed in the exosomes from the control group. This finding indicated the potential of circulating exosomal miRNAs as liquid biopsy markers (liquid biopsy involves identifying and segregating circulating tumor cells, circulating tumor DNA, and exosomes, which serve as valuable sources of genomic and proteomic insights for individuals diagnosed with cancer) for lung cancer [88]. Additionally, Kahlert et al. discovered large fragments of double-stranded genomic DNA (> 10 kb) in exosomes derived from pancreatic cancer cell lines and patients [89]. They suggested that exosomal DNA sequencing could be utilized to predict treatment options and evaluate therapy resistance. Whole-genome sequencing of exosomes obtained from pancreatic cancer patients revealed mutations in KRAS and p53 [90].
Exosomes from differentiated thyroid carcinoma (DTC) patients' serum showed decreased miR-130a-3p compared to benign cases and healthy controls. This miRNA correlated with DTC characteristics, such as tumor size, lymph node metastasis, and TNM stage. Combining exosomal miR-130a-3p with other markers (antithyroglobulin autoantibodies and thyroglobulin) improved the sensitivity and specificity of diagnostic biomarkers. The study identified insulin-like growth factor (IGF)-1 as a target gene, with a negative correlation between serum IGF-1 and exosomal miR-130a-3p levels. These findings suggest reduced exosomal miR-130a-3p as a sensitive biomarker for DTC diagnosis [91]. Serum exosomal miR-29a levels were significantly reduced in papillary thyroid carcinoma (PTC) cases, showing effective differentiation from normal controls by ROC analysis. Post-surgery, these levels increased significantly at 30 and 90 days. Lower miR-29a expression correlated with higher recurrence risk, worse clinical variables, and shorter survival and was an independent prognostic indicator for overall survival in both univariate and multivariate analyses [92]. A three-miRNA panel (miR-25-3p, miR-296-5p, miR-92a-3p) consistently showed up-regulation in PTC patients versus healthy controls, demonstrating superior diagnostic performance (Area under the ROC Curve: AUCs: 0.727, 0.771, 0.862) in multiple stages, and strong differentiation from benign goiters (AUC: 0.969). Analysis of tissue and exosome samples supported their close association with PTC, suggesting this serum panel is a valuable diagnostic tool [93]. Stable exosomal miRNAs were analyzed for accurate diagnosis of indeterminate thyroid nodules. Exosomes from 13 PTC and 7 nodular goiter (NG) patients were studied, identifying 129 differentially expressed miRNAs, with miR-5189-3p showing optimal performance (AUC: 0.951) in distinguishing PTC from NG. Enriched target genes in cancer pathways suggest the potential use of these plasma exosomal miRNAs as diagnostic biomarkers for thyroid nodules [94]. Eight plasma exosomal miRNA candidates were identified via RNA-seq, with miR-16-2-3p, miR-223-5p, miR-34c-5p, miR-182-5p, miR-223-3p, and miR-146b-5p lower in nodules vs. controls and miR-16-2-3p and miR-223-5p higher in PTC cases than benign nodules. These miRNAs, particularly miR-16-2-3p and miR-223-5p, serve as potent indicators for thyroid nodule detection, and combined panels enhance diagnostic sensitivity and specificity compared to single markers [95]. Small RNA sequencing identified 41 potential exosomal miRNA markers for PTC, with 4 miRNAs (miR-376a-3p, miR-4306, miR-4433a-5p, miR-485-3p) showing significantly increased expression in PTC patients compared to healthy and benign nodules. MiR-485-3p demonstrated the highest AUCs for diagnosing PTC, particularly in patients with high-risk factors like larger tumor size, advanced stage, and lymph node metastasis [96]. The three-miRNA panel in plasma effectively discriminates PTC from healthy control (HC) or nodular goiter (NG) (AUC: 0.877), with miR-346 and miR-34a-5p up-regulated in PTC tissues and consistently elevated in PTC plasma exosomes [97]. Exosomal lncRNAs, particularly RP11-77G23.5 and PHEX-AS1 in EpCAM-specific exosomes, show promise as diagnostic biomarkers for lung cancer, distinguishing malignancy and offering insights into subtype classification and disease progression. Their elevated levels in lung adenocarcinoma and distinct expression patterns related to tumor stages and metastasis underscore their diagnostic potential [98].
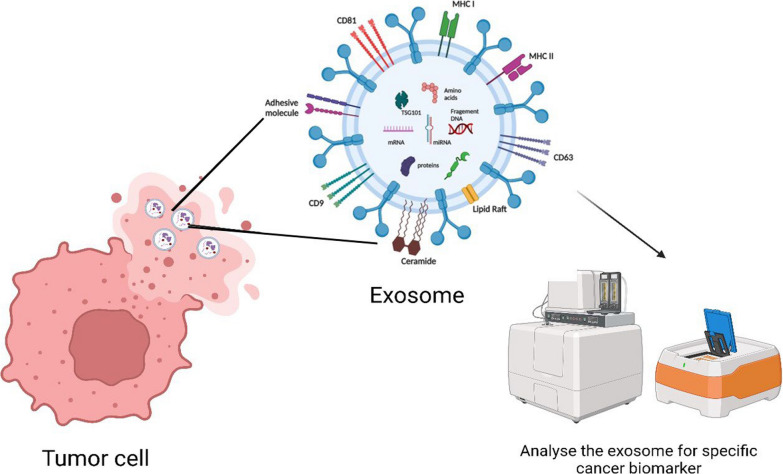
Over the previous three years, additional exosomal miRNAs have been identified employing a combination of RNA sequencing-based miRNA profiling, ExoQuick precipitation, ultracentrifugation, and the commercial Exo-miR kit (Bioo Scientific, Austin, TX, USA). These cancer models encompass glioblastoma, breast, colon, prostate, and pancreatic cancers. ExoQuick is a proprietary polymer, so it has the advantage of gent precipitation of exosomes [99]. A list of exosomal biomarkers is listed in Table 1, and Fig. 3 depicts the utility of exosomes as a cancer biomarker.
Table 1.
List of Exosomal biomarkers
| Exosomal biomarkers | EVs’ source | Application | References | |
|---|---|---|---|---|
| cells | Biopsy | |||
| EGFR, or EGFRvIII | – | Serum from patients with glioblastoma | Diagnosis and prognosis | [80] |
| CD147 or CD9 | Colorectal cancer cell lines (HCT116 cells, HCT15 cells, HT29 cells, COLO201 cells, COLO205 cells, WiDr cells and SW1116 cells) | Serum from patients with colorectal cancer | Diagnosis | [81] |
| CA-125, EpCAM, CD24 | – | Plasma from a patient with ovarian cancer | Diagnosis | [82] |
| Tetraspanin 1 (TSPAN1) | Colon cancer cell lines (HT-29 and HCT-116) | – | Diagnosis | [83] |
| ARG1, CD3, PD-L1, PD-L2 | – | Plasma from a patient with gastric cancer | Diagnosis and Prognosis | [84] |
| Developmental endothelial locus-1 (Del-1) | – | Plasma from a patient with breast cancer | Diagnosis and Prognosis | [85] |
| miR-1246 | Breast cancer cell lines (MCF-7 and MDA-MB-231) | – | Diagnosis | [100] |
| miR-200b, miR-200c, miR-141 and miR-375 | - | Pleural effusions from patients with lung adenocarcinomas | Diagnosis | [101] |
| Mutated KRAS and p53 DNA | - | Serum from patients with pancreatic cancer | Diagnosis | [89] |
| BRAF(V600E) mutation in exoDNA | Melanoma cell lines, wild-type (WT; SK-MEL-146 and SK-MEL-147) or mutated BRAF (SK-MEL-28, SK-MEL-133, SK-MEL-192, and SK-MEL-267) | - | Diagnosis | [90] |
| miR-130a-3p | – | Serum from patients with differentiated thyroid carcinoma (DTC) | Diagnosis | [91] |
| miR-29a | – | Serum from patients with papillary thyroid carcinoma (PTC) | Diagnosis and Prognosis | [92] |
| miR-25-3p, miR-296-5p and miR-92a-3p | – | Serum from patients with PTC | Diagnosis | [93] |
| miR-5189-3p | – | Plasma from patients with PTC | Diagnosis | [94] |
| miR-16–2-3p and miR-223-5p | – | Plasma from patients with PTC | Diagnosis | [95] |
| miR-346 and miR-34a-5p | – | Plasma from patients with PTC | Diagnosis | [96] |
| lncRNAs RP11-77G23.5 and PHEX-AS1 | Lung cancer cell line (NCI-H1299) | Serum from patients with lung cancer | Diagnosis | [98] |
EGFR: Epidermal Growth Factor Receptor; EGFRvIII: Epidermal Growth Factor Receptor Variant III; CD147: Cluster of Differentiation 147; CD9: Cluster of Differentiation 9; CA-125: Cancer Antigen 125; EpCAM: Epithelial Cell Adhesion Molecule; TSPAN1: Tetraspanin 1; ARG1: Arginase 1; CD3: Cluster of Differentiation 3; PD-L1: Programmed Cell Death Ligand 1; PD-L2: Programmed Cell Death Ligand 2; KRAS: Kirsten Rat Sarcoma Viral Oncogene Homolog; p53: Tumor Protein p53; BRAF(V600E): B-Raf Proto-Oncogene, Serine/Threonine Kinase (V600E mutation)
Fig. 3.
Exosome as Cancer Biomarkers. Each tumor cell releases specific exosomes that contain microRNA, DNA, proteins, and metabolites that can be used as biomarkers of specific cancer, Created with BioRender.com
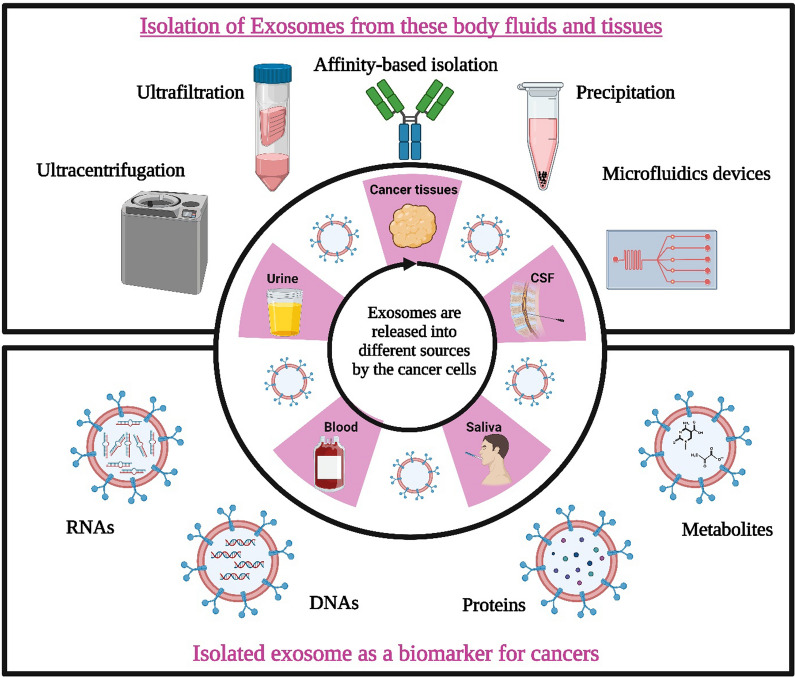
Isolation of exosomes
Exosomes are isolated on a different basis: Firstly, based on size, and secondly, based on its affinity. Ultracentrifugation is considered a gold standard in exosome isolation. However, in recent research, some new techniques for the isolation of exosomes have been practiced, and they are broadly classified based on their mechanism, namely Ultracentrifugation (UC), density gradient (DG) centrifugation, infiltration techniques, immunoaffinity, capture-based techniques, exosome precipitation, and use of acoustic nano-filters [102].
Ultracentrifugation
This method is commonly employed and widely recognized as the standard approach for isolating exosomes [103]. Although it is one of the most widely adopted techniques for exosome isolation, it depends on a few factors, like rotor type, centrifugation type, and sample viscosity. Hence, these parameters are optimized before standardizing the protocols for performing ultracentrifugation [104]. This method is advantageous because it is easy to perform and has a significantly higher exosome purity yield than other methods [105, 106]. However, there are some disadvantages of the above method, which should be counted: the downgrade of the quality of exosomes, which debars it from clinical applications. It happens because, during high-speed centrifugation, exosomes are subjected to a high shear force, which tends to damage them [107, 108].
Size-based techniques
They are categorized into ultrafiltration, sequential filtration, and size exclusion chromatography (SEC). SEC is the most advantageous of all three because of the following features: high yield, low cost-to-benefit ratio, and low destructive outcomes. This procedure also allows the smooth extraction of exosomes from serum and plasma [109]. A recent study found ultrafiltration is superior to UC since it recovers more particles, including exosomes, smaller than 100 nm. The size distributions of exosomes extracted through UC or SEC were identical, as demonstrated by TEM and NanoSight. With less processing time than the traditional UC protocol, ultrafiltration techniques increase exosome yield and isolation efficiency by producing more particles. Despite being widely employed in many sectors, these size-based approaches still have a lengthy running time, limiting their applicability in therapy and research.
Capture-based techniques
This technique produces a purity exosome based on the immunoaffinity principle [110]. Washing in a stationary phase can successfully capture immobilized particular exosomes, depending on the precise immunological interaction between the antibody and antigen. This method successfully separates exosomes that carry particular target membrane proteins. Due to thorough assessments of the effectiveness of recycling exosomes, the conclusion that capture-based strategies incorporating the Ep-CAM biomarker constitute the best strategy for separating exosomes compared to other methods has been primarily recognized through thorough evaluations of the effectiveness of recycling exosomes [111].
Precipitation technique
The most popular polymer employed in exosome isolation, polyethylene glycol (PEG), vigorously encourages enrichment and raises exosome yield. This technique was claimed to be practical for separating numerous biomolecules and viruses from physiological fluids before its application with exosomes [112]. Due to their simplicity, rapidity, lack of exosome destruction, and minimal need for extra equipment, precipitation-based approaches for exosome isolation are the most appealing for clinical research. However, it has been observed that these procedures have a problem with the sample's co-isolation of other contaminants, such as non-exosomal proteins (such as albumin) and other particles [113].
Microfluidic-based techniques
Microfluidics technologies have been successfully integrated with size-based separation, immunoaffinity-based separation, and dynamic separation techniques. Recently, a novel exosome isolation technology called the ExoTIC gadget has been introduced. The ExoTIC gadget has gained significant popularity due to its remarkable advantages, including high yield, purity, and efficiency. It is particularly well-suited for extracting exosomes from serum or other physiological fluids, surpassing conventional methods such as PEG precipitation (including the ExoQuickTM approach) or UC. Despite its numerous benefits, such as high purity, controllability, separation specificity, and efficiency, there are still specific challenges associated with the ExoTIC gadget, such as the requirement for complex isolation devices and limitations due to the need for strong immunoaffinity [113, 114].
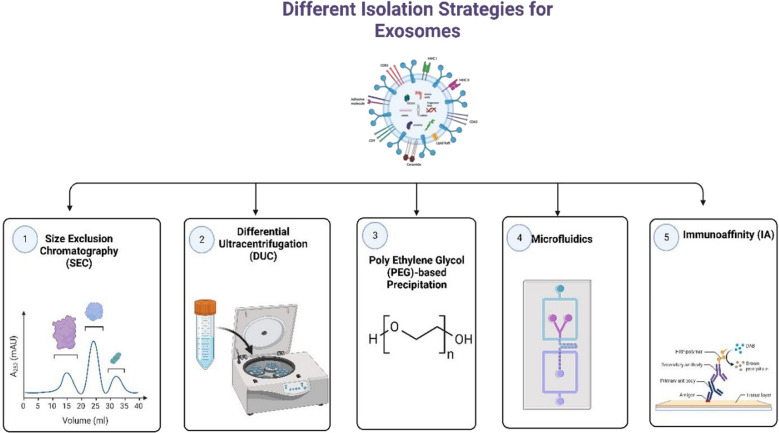
Above all, a good exosome isolation technique should be easy to use, quick, effective, affordable, and scalable. Additionally, it shouldn't harm the exosomes or call for additional tools. Different approaches offer unique benefits and drawbacks regarding effectiveness, repeatability, and influence on functional outcomes. Further advancement of exosome research for both fundamental and clinical applications can be facilitated by optimizing isolation processes and employing combinations of isolation approaches. Figure 4 depicts the different strategies involved in the isolation of exosomes. Table 2 shows how exosomes have already been isolated using different analytical techniques.
Fig. 4.
Different Isolation Strategies for Exosomes. Traditional methods for isolating exosomes include size exclusion chromatography (SEC) and differential ultracentrifugation (DUC). SEC involves using biofluids as a mobile phase against a porous stationary phase to elute molecules based on their size, with larger particles eluting first, followed by smaller exosomes, resulting in a longer elution time due to increased path length. In addition to these conventional methods, more innovative techniques are available for exosome isolation. In addition to these conventional methods, more innovative techniques are available for exosome isolation. One such technique is PEG-based precipitation, which facilitates the aggregation of exosomes in large numbers using a polymer solution. Another approach is immunoaffinity (IA) capture, where antibodies targeted against exosomal surface proteins are used to isolate specific exosome populations. Microfluidics (MF) technology, utilizing chips with specific antibody-mediated binding, enables efficient capture of exosomes, Created with BioRender.com
Table 2.
Different isolation techniques for exosomes in cancer
| Exosome source | Isolation approach | Clinical importance | Reference |
|---|---|---|---|
| Human breast milk | Ultracentrifugation and immuno-isolation on paramagnetic beads | It is crucial for the maturation of the infant’s immune system | [131] |
| Human cancer cells containing activated EGFR (A431, A549, DLD-1) | Ultracentrifugation | Tumor cell-derived microvesicles could act as a unique form of angiogenesis-modulating stimuli | [132] |
|
U373 (human astrocyto ma) cells |
Ultracentrifugation | Membrane microvesicles of cancer cells can contribute to a horizontal propagation of oncogenes | [133] |
| Neuroblastoma | Differential ultracentrifugation (DUC), OptiPrep (Sigma-Aldrich) density gradient centrifugation (ODGC), and SEC | Exosome leads to the production of pro-tumorigenic cytokines and chemokines in mesenchymal stromal cells | [134] |
| Platelet-free plasma | Ultracentrifugation | A better method for the isolation of smaller exosomes from complex biological fluid, that is, Plasma | [135] |
| Human dendritic cells | Centrifugation | Frequently employed exosome markers, including major histocompatibility complex, flotillin, and heat-shock 70-kDa proteins, were detected in all EVs irrespective of their subtypes | [136] |
| Liquid biopsies of patients with pancreatic ductal adenocarcinoma (PDAC) | Serial ultracentrifugation | Exosomes contain distinct tumor DNA, which has different biology than circulating cell-free DNA (cfDNA) shed from dying tissues | [137] |
| Pancreatic ductal adenocarcinoma (PDAC) exosomal ‘surfaceome’ | Centrifugation | Cell surface proteins analysis of exosomes from pancreatic ductal adenocarcinoma (PDAC) enables the capture and molecular profiling of tumor-specific DNA | [138] |
| Dendritic-cell culture | Ultracentrifugation | Exosomes represent a novel source of tumor-rejection antigens for T-cell cross priming, relevant for immune-interventions | [139] |
| Non-small-cell lung cancer cell line, A549 | Ultracentrifugation | Exosomes originating from cancer cells with Rab27a overexpression exhibited enhanced anti-tumor immune effects | [140] |
| CAR-T cells | Ultracentrifugation | Exosomes carrying CAR (chimeric antigen receptor) display elevated levels of cytotoxic molecules and impede tumor growth | [14] |
| Human saliva | Density gradient centrifugation | Diagnostic approaches that specifically target EVs found in saliva | [141] |
| Urine | ME-Kit (microvesicles enrichment kit), Ultracentrifugation | The clinical feasibility of the Vn96-peptide-based EV isolation method enables large-scale protein profiling of urinary EV biomarkers | [142] |
| HeLa cells | PEG-based approach | The potential functions of exosomes in the buildup and transportation of protein degradation intermediates | [112] |
The advanced approach of exosome profiling
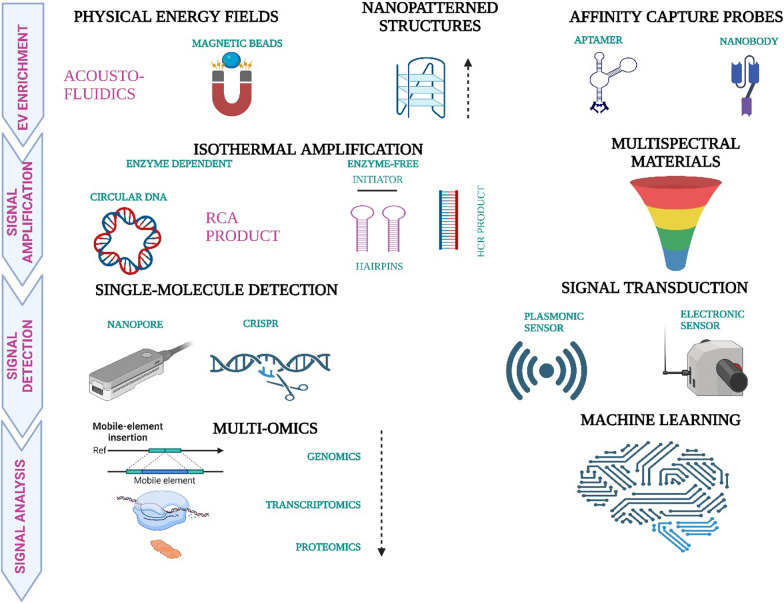
Throughout the years, there has been relatively rapid development in the different methods of exosome profiling. MiRNA profiling through microarray analysis has been a potential screening tool for early detection of cancers, especially ovarian cancer. However, there is a deficit of established and proven molecular markers [115, 116]. Nowadays, nanotechnology has become a vital tool for the efficient profiling of exosomes. Although SERS (Surface-Enhanced Raman Spectroscopy) has the same fundamental principle as Raman spectroscopy, it demands a substrate modification [117, 118]. It is a super-sensitive multiplexing approach that gives authentic results using low-volume/concentration analytes [119]. Recently published in a paper, TPEX (Templated Plasmonics for Exosomes) is a nanotechnology platform that makes space for the analytical study of multiselective molecular profiling of exosomes apart from their prompt in situ assessment of the biomolecular and biophysical compositions. It provides multiplexed and quick inspection of exosomal targets with exceptional results upon administration on a microfluidic smartphone-based sensor [120]. Of many others, advanced iFCM (imaging Flow Cytometry) is an approach to carry out multiparametric and high-throughput vesicle-by-vesicle representation of the exosomes, resulting in efficient recovery of specific vesicle subsets [121]. Lastly, over the years, development in OMICS-based technologies has significantly advanced the studies of markers of proteins and exosomes [122–124], which in turn have led to elaborate research in the field of diagnostic methods [81, 125] related to exosomes evaluation of glycomic profiles with the help of lectin microarray-based technologies and mass-spectrometry are noteworthy mentions [126–129]. Exosomes and artificial intelligence (AI) are indeed emerging as promising tools in cancer diagnosis, and their combination holds great potential for advancing our ability to detect and understand various types of cancers. Integrating exosomes and AI can open new doors in cancer diagnosis. A recent study demonstrates the efficacy of using artificial intelligence to simultaneously detect six early-stage cancers by analyzing exosome profiles via surface-enhanced Raman spectroscopy. In a dataset of 520 test samples, our system achieved a robust cancer identification rate (AUC: 0.970) and proficiently classified tumor organ types in 278 patients (mean AUC: 0.945). The integrated decision model showed a sensitivity of 90.2% and specificity of 94.4%, successfully predicting tumor organs in 72% of positive cases. Notably, our non-specific Raman signature analysis method holds the potential for expanding diagnostic applications to other diseases [130]. Figure 5 depicts advanced exosome profiling approaches.
Fig. 5.
Representation of single exosome profiling methods. Created with BioRender.com
While these advanced technologies, such as SERS, TPEX, and iFCM, offer significant promise in exosome profiling, their potential clinical applications also warrant discussion. Integrating these methods into clinical practice could greatly enhance early cancer detection and monitoring. For example, SERS' high sensitivity and specificity in detecting exosomal markers make it a valuable tool for routine clinical diagnostics. TPEX, with its smartphone-based microfluidic sensor, provides a feasible approach for point-of-care testing, thus facilitating rapid and accurate exosome analysis in a clinical setting. Furthermore, combining AI with exosome profiling technologies can streamline the diagnostic process, offering precise and non-invasive cancer detection. These advancements improve diagnostic accuracy and potentially make exosome-based diagnostics more accessible in clinical environments, thereby bridging the gap between research innovations and practical healthcare applications.
OMICS profiling of exosomes
OMICS profiling tends to concentrate on detecting all the bioinformation of exosomes. This includes its genomics, transcriptomics, proteomics, metabolomics, and lipidomics, which help to diagnose cancer at early stage and, as a result, improve the patient survival rate [143].
Exosomes are seen to act as essential mediators as they transport specific molecules among different populations of cells [144]. It belongs to the class of EVs, including ectosomes, apoptotic bodies, and microvesicles. These are secreted by almost all types of cells, including tumor cells, and involve extracellular communication. Their lipid bilayer nature makes them stable in body fluids and contains many nucleic acids, proteins, and lipids. Various body fluids contain exosomes, providing an alternative approach to detecting tumors [145]. Hence, exosomes and their essential components have a significant potential to serve as biomarkers for diagnosing early-stage cancer. In the case of breast cancer, the tumor mass present is first analyzed by imaging and is then characterized by needle biopsy to check its chance of malignancy. These methods are invasive and quite time-consuming. Nowadays, exosomal miRNAs are known to be useful biomarkers for detecting breast cancer [146]. The presence of miR-1246 and miR21 in plasma was predominantly used as the initial biomarker to detect breast cancer [147]. In 2018, Li et al. reported an exosomal miR106a-363 cluster, a novel diagnostic biomarker for detecting breast cancer [148]. Recently, miR-92b-5p was found in stable breast cancer cell lines as the latest therapeutic strategy for its detection [148].
Diagnosis of cancerous diseases is mainly done through imaging methods and a biopsy, which would confirm the result [149, 150]. However, these traditional methods were invasive, costly, and uncomfortable for the patient. In addition, small-size early cancer may not be detected in imaging studies, and a biopsy of the early cancer may not be possible. On top of it, the accuracy of a biopsy is greatly influenced by the experience of the procedures. So, OMICS profiling of exosomes offers a more convenient way of diagnosing cancer, which is non-invasive, inexpensive, reproducible, procedure-independent, and has the potential for early detection [151].
Exosomes were used to check the dysregulated genes for different cancer types. Hence, the observed tumor heterogeneity character was used as it indicated the presence of a difference in genes and cell behaviors between different types of tumors. Hence, a combination of bioinformatics, machine learning, and DNA sequencing can be done to determine the dysregulated genes and the corresponding type of tumor [152, 153].
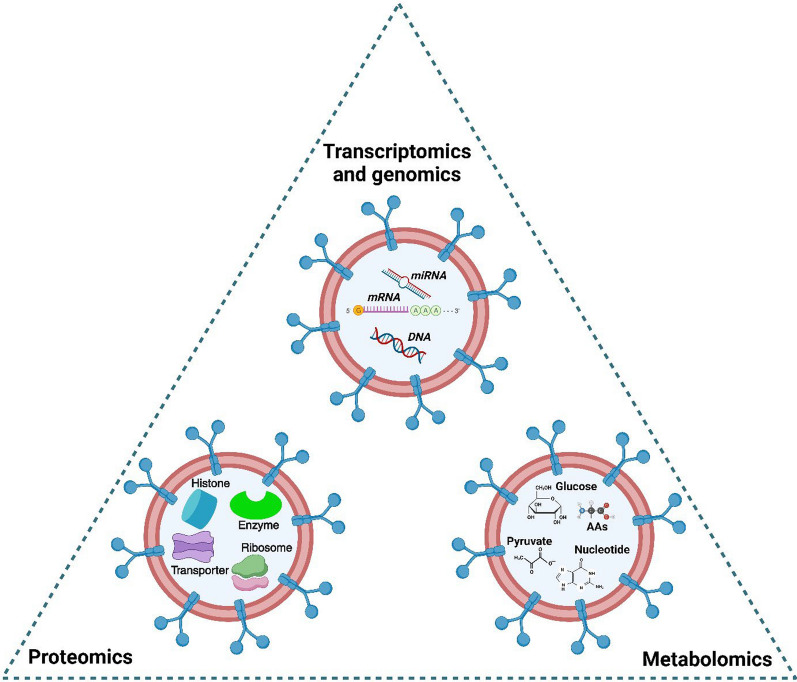
Hence, exosomal markers are established to be useful targets for the detection of cancer. Cancer cells or tumors and other diseased cells release more exosomes than normal healthy cells, indicating the presence of disease. So, the methods that utilize exosomes can be implemented for routine screening and are also useful for tumors for which routine screening is unavailable. Exosome’s OMICS profiling can be used to verify the presence of malignant lesions. Hence, it can distinguish between those and benign lesions. Also, it is observed that acidic tumor environments can lead to an increase in the release of exosomes in body fluids [154]. However, the isolation, detection, and quantification of exosomes are some of its limitations, and hence, this method isn't utilized rapidly for routine screening yet. Figure 6 depicts the major OMICS processes involved in the profiling of exosomes.
Fig. 6.
OMICS Profiling of Exosomes. This profiling 3 ways: Transcriptomics and genomics using the messenger RNA (mRNA), microRNA (miRNA), and DNA present in the cancer-specific exosome; Proteomics using the proteins, histones, transporters present in the exosome; Metabolomics using the metabolites such as glucose, pyruvate, nucleotides, amino acids (AAs) specific for cancer. Created with BioRender.com
OMICS technologies face several challenges and limitations in exosome research. Isolating pure exosome populations from biological fluids is difficult due to other EVs and contaminants, which can affect the accuracy and reproducibility of OMICS analyses [155]. The high heterogeneity of exosomes in size, content, and function complicates the interpretation of OMICS data and the identification of specific biomarkers [156]. Detecting low-abundance molecules requires highly sensitive techniques, which current OMICS technologies may lack [157]. The large and complex datasets generated necessitate sophisticated bioinformatics tools for analysis, but interpreting these data accurately is challenging due to the lack of standardized protocols and reference databases [158]. Variability in experimental procedures and analytical methods can lead to inconsistent results, and the lack of standardization hampers reproducibility across different studies [159]. Although OMICS technologies can identify potential biomarkers or functional molecules, validating their biological relevance is challenging and requires additional, resource-intensive experiments [160]. Both technical and biological variability can influence OMICS data, complicating the identification of true biological signals, and disentangling these sources of variability necessitates robust experimental designs and statistical approaches [102]. Addressing these challenges requires ongoing advancements in isolation techniques, analytical methods, bioinformatics tools, and standardization across the field of exosome research.
Single exosome profiling
Owing to a difference in origins, exosomes have a highly heterogeneous molecular composition [70, 161]. Hence, the study of exosomes should be conducted individually. Otherwise, the heterogeneity would not be properly detected in the bulk analysis. Bulk-level analysis methods like mass spectroscopy or ELISA may give inaccurate results in detecting exosome heterogeneity; hence, these methods are not preferred [162]. Currently, EV analysis is limited to surface proteins only. So, single exosome profiling can be done on the same to get a more concise and accurate diagnosis.
Nowadays, many methods of single exosome profiling are being used, and most employ the basic principles of light scattering, fluorescent sensing, or electron absorption. These are derived both for the isolation and sensing of EVs like exosomes. The purification and analysis methods must be made simpler to utilize these analysis methods for routine screening and cancer detection.
A high-throughput method known as the proximity barcoding assay can be employed for single exosome analysis. This technique allows for profiling over a hundred surface proteins on a single exosome, facilitating the distinction between different exosomes based on their heterogeneous surface protein compositions. By analyzing human body fluids, various exosome sub-populations can be effectively identified [136, 163].
Advances in single exosome profiling techniques have been marked by the development of various methodologies that allow for more detailed and precise analysis [164]. Digital PCR has been adapted for single EV analysis, which can amplify and identify RNA content, targeting miRNA and mRNA, including cancer markers. Recent advancements enable the sequestering of EVs in droplets before lysing, offering improved sensitivity and specificity in mutation detection [165]. Additionally, digital PCR has been leveraged to detect membrane proteins on single EVs, expanding its application beyond nucleic acid analysis. DNA-tagged antibodies in digital PCR provide a powerful approach to identifying multiple coinciding membrane proteins on single EVs, offering richer data and more specific diagnostic information [161].
Digital ELISA, inspired by the digital immunoassay methodology, has been applied to detect membrane proteins on single EVs [166]. This approach involves labeling EVs with antibodies, sequestering them into droplets, and using fluorescence to visualize protein markers. While suitable for single biomarker detection, challenges remain in identifying multiple markers on single EVs [166]. Flow cytometry has been optimized for EV analysis, overcoming limitations in conventional systems using DNA aptamers, nanoparticle tags, or advanced imaging flow cytometry [167]. These enhancements allow for improved detection and characterization of EVs, contributing to a better understanding of EV subpopulations and providing valuable information on surface proteins and biomarkers [168].
Nanoparticle tracking analysis (NTA) has evolved to include fluorescence capabilities, enabling the detection of specific biomarkers through immunolabeling. NTA remains a high-throughput method for single EV analysis, providing information on concentration, size, and surface markers. However, challenges in multiplexing with fluorophores limit its capabilities compared to flow cytometry [169].
Raman spectroscopy and various trapping techniques have been employed for the chemical composition analysis of single EVs [170]. While unable to directly identify macromolecular biomarkers, Raman spectroscopy, combined with electromagnetic trapping, distinguishes EV populations and detects labeled biomarkers with strong Raman scattering cross-sections [171]. Microscopy-based methods, such as fluorescence and total internal reflective fluorescence (TIRF) microscopy, have allowed for direct visualization of single EVs. Microfluidic chip-based systems enable the immobilization and imaging of EVs, offering multiplexed detection of surface proteins and RNA content. However, these methods are generally lower throughput than other techniques [165].
Now, the classification of exosomes based on their proteomic characteristics can be done using a machine-learning algorithm known as FlowSOM [172]. This algorithm helps to generate clusters of exosomes by using a self-organizing map. Then, a t-SNE or t-distributed stochastic neighbor embedding plot is used to visualize the exosome sub-populations. Also, along with the progression of the disease, the alteration of the presence of different sub-populations of exosomes could be monitored through single exosome profiling.
The continuous refinement of single-exosome profiling techniques, including digital PCR, digital ELISA, flow cytometry, NTA, Raman spectroscopy, and microscopy, has created a comprehensive toolkit for understanding exosome heterogeneity and functional diversity at the single-exosome level. These advancements significantly enhance the study of EV biology and biomarker discovery, providing new insights into exosome diagnostic and therapeutic potential. Figure 7 depicts the usage of single exosome profiling in the context of cancer.
Fig. 7.
Application of Single-Exosome Profiling in Cancer. Step 1: A sample containing exosomes (small vesicles) is collected and processed. Step 2: Exosomes are isolated using ultracentrifugation or microfluidics, then captured and trapped using nanoscale traps or microfluidic chambers. Step 3: The surface proteins and biomarkers on the trapped exosome are identified and analyzed using mass spectrometry or fluorescence microscopy techniques. Step 4: The cargo contents (e.g., RNA, DNA, proteins) of the exosome are extracted and analyzed using techniques like qRT-PCR. Step 5: The data from the analysis is integrated to create a comprehensive profile of the individual exosome, including its size, shape, surface markers, and cargo contents. Created with Biorender.com
Advantages and disadvantages of exosome-based cancer biomarker
Exosomes help bring forth a lot of information about the tumor state of the patient and assist in sorting the same into tumor subtypes by undergoing genomic and proteomic scanning and analysis [173]. The former helps design therapeutic treatments considering the genetic makeup and abnormalities of the growing tumor. In contrast, the latter helps curate certain processes that directly target proteins involved in tumor growth. Both of these approaches give insights into the tumor's metastasis rate through specific markers and provide a profile of the heterogenicity and complexity of the tumor studied. They lend out fitting therapeutic strategies, like that of the recently evolved method of liquid biopsies, against cancer treatment [174–177]. As one of the least intrusive and dynamic methods, this detects cancer-specific biomarkers that provide an overview of tumor prognosis, a comprehensive and accurate capturing of different tumor parts' genetic and molecular makeup, and an effective and targeted therapy. One of the crucial advantages of exosomes being used as biomarkers is that they can express MHC molecules on their cell surface, thus presenting antigens through indirect and direct pathways [178, 179]. It is true for some tumor-derived EVs that present MHCs loaded with tumor-processed antigenic peptides and antigenic proteins, which later form complexes with anticancer autoantibodies circulating in the plasma produced by various B cell subpopulations [180]. They bring forth an elaborate area of research on how this process of antigen presentation may be utilized in advancing immunotherapy against cancer. They are also remarkably durable in storage conditions [181, 182], facilitating the preservation of the biomolecules within them without degradation over an extended period, allowing them to withstand various standardized protocols in clinical settings for analysis. Abundant studies indicate that the intensity of GPC1 in patients of pancreatic cancer brings to the fore an exciting insight of using these for early detection of this cancer [183], along with it being an appealing non-invasive screening and diagnostic tool for a wide category of cancers [184]. Several experiments proclaim that serum-derived exosomal DNA can be useful in detecting parental tumor cell mutations [90, 185–187]. It has also come to our knowledge that exosomes fostering non-coding RNAs might help track cancer progression and diagnosis, similar to identifying breast cancer biomarkers [188–193]. It is a super-sensitive approach that gives authentic results using low-volume/concentration analytes [194, 195]. On the contrary, standardized techniques are needed to analyze, estimate, and segregate exosomes for varied clinical implementations. It is exceedingly difficult to achieve optimized efficacy while isolating pure exosomes from multiplex fluids like blood or cell culture supernatants. This induces fluctuating and jumbled exosome reproducibility, causing difficulty interpreting and concluding outcomes. We are yet to reach the desired improvement in the liquid biopsy tools. The current tools often lead to erroneous cancer detection and monitoring.
While exosomes hold great promise as cancer biomarkers due to their ability to reflect the molecular composition of their cells of origin, several disadvantages and challenges limit their clinical application. Firstly, the isolation and purification of exosomes from body fluids are complex and can be contaminated by other EVs and proteins, affecting the accuracy and reproducibility of results [155]. Additionally, the heterogeneous nature of exosomes, arising from different cellular sources and varying physiological states, complicates the identification of specific and reliable biomarkers [156]. The sensitivity of current detection technologies may be insufficient to accurately quantify low-abundance exosomal components, leading to potential misinterpretations [157]. Moreover, the lack of standardized protocols for exosome isolation, characterization, and analysis results in variability across studies, undermining reproducibility and comparability [159]. Finally, while OMICS technologies can identify potential exosomal biomarkers, validating their clinical relevance and biological function requires extensive and resource-intensive follow-up studies [160]. These challenges necessitate ongoing technological advancements and standardization efforts to realize the full potential of exosomes as cancer biomarkers.
Clinical trials of exosome-based cancer biomarker
Clinical trials investigating exosome-based cancer biomarkers have generated significant interest due to the potential of exosomes to provide non-invasive, accurate, and dynamic insights into tumor biology. However, thoroughly discussing these trials requires understanding their real-world applications, limitations, and the translational journey from clinical trials to medical practice.
Clinical trials on exosome-based biomarkers aim to evaluate their effectiveness in early cancer detection, monitoring disease progression, predicting treatment response and prognosis, and identifying therapeutic targets. These trials often involve collecting and analyzing exosomes from various body fluids, such as blood, urine, and saliva, to determine the presence and levels of specific cancer-related molecules, including proteins, lipids, and nucleic acids [136]. One significant scope of these trials is the potential for non-invasive cancer diagnostics. Exosome-based liquid biopsies can offer a less invasive alternative to traditional tissue biopsies, enabling more frequent monitoring of tumor dynamics and potentially improving patient outcomes. For example, detecting and quantifying exosomal mutations, such as EGFRvIII in glioblastoma, can provide critical information about tumor status and treatment efficacy [68]. However, there are notable limitations. The heterogeneity of exosomes, stemming from their diverse cellular origins and the varying physiological states of their parent cells, poses a challenge for standardization and consistency in biomarker discovery [156]. Additionally, the current sensitivity of detection technologies may not be sufficient to accurately measure low-abundance exosomal components, potentially leading to false negatives or positives [157]. Moreover, the lack of standardized protocols for exosome isolation and analysis further complicates the reproducibility of clinical trial results [159].
Translating the results of clinical trials into oncological practice involves several critical steps. To ensure their reliability and clinical relevance, robust validation of identified biomarkers is required through large-scale studies and cross-cohort comparisons. Once validated, the next step is the integration of these biomarkers into diagnostic, prognostic, and therapeutic frameworks. For diagnostic purposes, exosome-based assays must be developed and standardized for clinical use. These assays must demonstrate high sensitivity, specificity, and reproducibility to gain regulatory approval and clinical adoption. For instance, assays detecting exosomal PD-L1 could potentially guide immunotherapy decisions by identifying patients most likely to benefit from such treatments [196]. In therapeutic contexts, exosome-based biomarkers can help personalize treatment plans. By monitoring exosomal content over time, clinicians can assess treatment responses and adjust therapies accordingly, enhancing personalized medicine approaches. Exosome analysis can provide insights into disease progression and recurrence risk, aiding in patient stratification and long-term management. For example, exosomal miRNAs have been investigated for their prognostic potential in various cancers, providing valuable information on patient outcomes [197].
It is crucial to address their current limitations through ongoing research and technological advancements to enhance the clinical utility of exosome-based biomarkers. Improvements in isolation techniques, detection sensitivity, and bioinformatics analysis will be pivotal in overcoming the challenges associated with exosome heterogeneity and low-abundance biomolecules. Moreover, collaborative efforts to establish standardized protocols and consensus guidelines will facilitate more consistent and reproducible findings across studies and clinical settings. Such efforts will also expedite the regulatory approval process, enabling faster translation of research findings into clinical practice.
In conclusion, while exosome-based cancer biomarkers hold great promise, their successful integration into oncological practice requires addressing several key challenges and limitations. Ongoing clinical trials and research efforts are essential to validate these biomarkers and develop reliable diagnostic, prognostic, and therapeutic tools to enhance cancer patient care. Table 3 lists all the clinical trials with exosomes in different types of cancer and their significance.
Table 3.
Clinical trials with exosomes in cancer
| Cancer type | Trail ID | Exosome source | Clinical importance |
|---|---|---|---|
| Early lung cancer | NCT03542253 | Blood | The expression of exosomal miRNA was significantly elevated in early-stage lung cancer tissues compared to adjacent non-cancerous tissues. Moreover, the levels of miRNA-A in the adjacent tissues were notably higher than those observed in peripheral blood exosomes |
| High-grade prostate cancer | NCT02702856 | Non-catheter urine | The objective was to evaluate the correlation between an Exosome Urine Test score and the detection of high-grade (Gleason grade/score ≥ 7) prostate cancer through a prostate needle biopsy |
| Pancreatic cancer | NCT03821909 | Portal venous blood | The aim was to assess the practicality and safety of obtaining portal venous blood samples using endoscopic ultrasound (EUS), as well as to identify portal venous circulating tumor cells (CTCs) and analyze mRNA markers of exosomes through RNA-seq |
| Lung cancer | NCT04529915 | Blood | The objective was to investigate the abundant exosomes in blood samples and conduct clinical studies to assess the feasibility of diagnosing lung cancer |
| Advanced gastric cancer | NCT01779583 | Plasma | The primary objective was to examine the molecular profile of exosomes derived from gastric cancer |
| Early-staged lung cancer | NCT04939324 | Blood | This study aimed to analyze the molecular profiling of exosomes obtained from samples collected from the tumor-draining vein. The goal is to identify molecular characteristics that can be prognostic indicators for cancer recurrence following surgery |
| Pancreatic cancer | NCT02393703 | Blood tissue | The objective was to isolate and examine exosomes, which are small vesicles containing crucial proteins and nucleic acids functioning as messenger systems |
| HER2-positive breast cancer | NCT04288141 | Blood tumor | The assessment of HER2-HER3 dimer expression in tumor samples and blood (exosome) samples was obtained from patients diagnosed with HER2-positive breast cancer undergoing HER2-targeted therapies |
| Breast cancer | NCT05286684 | CSF | Evaluating the practicality of exosome analysis in cerebrospinal fluid as part of the diagnostic evaluation for metastatic meningitis (Exo-LCR) |
| Bladder cancer | NCT05270174 | Urine | Investigate the potential of lncRNA-ElNAT1 in urine exosomes as a novel target for preoperative diagnosis of lymph node metastasis |
| Pancreatic ductal adenocarcinoma (PDAC) | NCT03032913 | Blood | Assessing the diagnostic precision of CTCs and quantification of onco-exosomes in the diagnosis of pancreatic cancer—PANC-CTC study |
Future prospects
Research on exosomes is gaining popularity in the diagnosis and treatment of oncological diseases. It has been demonstrated that exosomes are crucial cell-to-cell communication transmitters. Exosomes are excellent therapeutic targets for cancer and perfect drug delivery vehicles due to several beneficial characteristics. Exosomes generated from cancer cells contain various proteins, lipids, DNA, RNA, and metabolites unique to cancer cells, which can be utilized as biomarkers for various cancers [198]. Exosomes are ideal targets for cancer diagnostics because they offer a high concentration and protected environment for their payload. Recent research has shown that exosomes maintain tissue homeostasis by modulating cell–cell communication through the chemicals they contain. Furthermore, the development of cancer is linked to exosomes that are secreted from cancer cells. Thus, understanding the function of exosomes in cancer will improve the efficacy of novel therapeutic and diagnostic strategies.
Exosomes, in particular, are helpful sources of biomarkers due to their affinity for their parent cells and ability to load cargo selectively [199]. Studies have shown exosomal miRNAs to be useful as molecular diagnostic markers for cancers, and miRNAs can be transported using nanoparticle platforms to provide targeted treatments for cancers [200]. Adipose-derived mesenchymal stem cells can transfer miR-122 via exosomes, making hepatocellular carcinoma cells more susceptible to chemotherapeutic treatments [201]. Exosomes are superior to other nanoparticles due to their remarkable biocompatibility, low immunogenicity, high stability, extended half-life, capacity to pass physical barriers like the BBB, and targetability. Additionally, their propensity for bioengineering and capacity to transport functional biomolecules, such as therapeutic proteins, chemotherapeutics, and nucleic acids, have garnered significant attention lately [202].
Despite the potential advantages, challenges and disadvantages are associated with the clinical use of exosomes. Since both cancer and normal cells produce exosomes, identifying specific markers or marker panels produced exclusively or at high levels in cancer cells is crucial for early cancer detection. Tumor-specific targeting is necessary for therapeutic techniques to reduce off-target effects [203]. Cancer-derived exosomes contribute to immune evasion, tumor formation, progression, angiogenesis, metastasis, anti-apoptotic signalling, and treatment resistance. Conversely, exosomes from healthy cells, including DC, T, and B cells, can significantly slow down tumor formation. Therefore, depending on their cell of origin and bioactive payload, exosomes may have a dual role in promoting, inhibiting, or controlling cancer development [204].
In conclusion, while exosomes offer tremendous promise as cancer biomarkers and therapeutic vehicles, further research and clinical validation are essential to overcome limitations. Their use as cancer biomarkers in the clinic presents both advantages and challenges. The development of standardized isolation techniques, comprehensive profiling methods, and targeted delivery systems will be pivotal in realizing the clinical utility of exosome-based biomarkers and therapies. With continued technological advancements and collaboration between researchers and clinicians, exosome-based cancer biomarkers may soon revolutionize cancer diagnosis and management, ultimately leading to improved patient outcomes.
Conclusions
The clinical signature of exosome-based cancer biomarkers represents a promising avenue for improving cancer diagnosis and prognosis. This review explored various critical exosome-related aspects and their potential as cancer biomarkers. We have provided a comprehensive overview of the significance of exosomes in cancer research, highlighting their role as essential mediators of intercellular communication and potential carriers of diagnostic information.
The biogenesis of exosomes sheds light on the complex process by which these tiny vesicles are formed and released from cells, indicating their involvement in tumor development and progression. Moreover, we have focused on the intricate relationship between exosomes and cancer, showcasing exosomes' multifaceted roles in tumor microenvironment modulation, immune response evasion, and metastasis promotion. The concept of exosomes as a source of cancer biomarkers demonstrates how their cargo of nucleic acids, proteins, and lipids holds immense diagnostic potential for detecting and monitoring cancer.
Methodologies for isolating exosomes are crucial for obtaining pure and reliable samples for biomarker research. OMICS profiling of exosomes highlights the wide array of information that can be gleaned from exosome cargo analysis, paving the way for personalized cancer diagnosis and treatment. Single exosome profiling showcases the sensitivity of this approach, allowing for the detection of subtle changes in the exosomal content with potential diagnostic applications.
We have explored advanced approaches in exosome profiling, indicating the continuous advancements in technology that promise to enhance the precision and clinical utility of exosome-based cancer biomarkers. The advantages and disadvantages of exosome-based cancer biomarkers underscore the need for careful validation and standardization to ensure their successful translation into clinical practice. Exosomes offer several advantages, including specificity, non-invasiveness, and the ability to carry a diverse range of biomolecules. However, challenges such as heterogeneity, complex isolation methods, and high costs must be addressed.
Lastly, we have touched upon the ongoing clinical trials investigating the feasibility and efficacy of exosome-based cancer biomarkers, underscoring this field's growing interest and potential. These trials will provide valuable insights and help bridge the gap between research and clinical application.
The clinical signature of exosome-based cancer biomarkers holds tremendous promise as a non-invasive and sensitive approach for cancer diagnosis, prognosis, and monitoring. However, more research and rigorous clinical validation are needed to fully realize their potential and ensure their successful integration into routine clinical practice. With continued advancements in technology and collaboration between researchers and clinicians, exosome-based cancer biomarkers may soon revolutionize how we diagnose and manage cancer, ultimately leading to improved patient outcomes. The ongoing exploration of the advantages and disadvantages of exosome-based biomarkers and advancements in isolation and profiling technologies will be pivotal in overcoming current challenges and enhancing their clinical utility.
In conclusion, exosomes present a unique opportunity in oncology, offering a novel and promising approach to cancer management. The future of exosome research is bright, with the potential to significantly impact patient care and treatment outcomes through innovative diagnostic and therapeutic strategies.
Acknowledgements
Not applicable.
Abbreviations
- BBB
Blood Brain Barrier
- CAR
Chimeric antigen receptor
- CSF
Cerebrospinal fluid
- CTC
Circulating tumor cells
- CAR-T
Chimeric antigen receptor T
- DTC
Differentiated thyroid carcinoma
- DUC
Differential ultracentrifugation
- EFGR
Epidermal growth factor receptor
- EpCAM
Epithelial cellular adhesion molecule
- ESCRT
Endosomal sorting complexes required for transport
- EUS
Endoscopic ultrasound
- EV
Extracellular vesicle
- Exo-LCR
Exosome analysis as a part of the diagnostic evaluation for metastatic meningitis
- IGF
Insulin-like growth factor
- lncRNA
Long non-coding RNA
- HRS
Hepatocyte growth factor Regulated tyrosine kinase Substrate
- IA
Immunoaffinity
- iFCM
Imaging flow cytometry
- ILV
Intraluminal vesicle
- MF
Microfluidics
- MVB
Multivesicular body
- NG
Nodular goiter
- ODGC
OptiPrep density gradient centrifugation
- PTC
Papillary thyroid carcinoma
- PANC Study
Pancreatitis, A National Cohort Study
- PDAC
Pancreatic ductal adenocarcinoma
- PEG
Poly ethylene glycol
- SEC
Size exclusion chromatography
- STAM
Signal transducing adaptor molecule
- TEM
Transmission electron microscopy
- UC
Ultracentrifugation
- UIM
Ubiquitin interacting motif
- ZFD
Zinc finger domain
Author contributions
Subhrojyoti Ghosh, Anand Krishnan, Prakash Gangadaran and Byeong-Cheol Ahn contributed to the conception, writing, and discussion of this manuscript. Subhrojyoti Ghosh, Ramya Lakshmi Rajendran and Atharva A Mahajan equally contributed and wrote the initial draft of the manuscript. Ankita Chowdhury, Aishi Bera, Sudeepta Guha, Kashmira Chakraborty, Rajanyaa Chowdhury, Aritra Paul, Shreya Jha, Anuvab Dey, Amit Dubey, Sukhamoy Gorai, Purbasha Das and Chae Moon Hong contributed to the initial draft of the manuscript. All authors have approved the final version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2022R1A2C2005057).
Availability of data and material
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Subhrojyoti Ghosh, Ramya Lakshmi Rajendran and Atharva A. Mahajan contributed equally to this work.
Contributor Information
Anand Krishnan, Email: krishnana1@ufs.ac.za.
Prakash Gangadaran, Email: prakashg@knu.ac.kr.
Byeong-Cheol Ahn, Email: abc2000@knu.ac.kr.
References
- 1.Brennan K, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep. 2020;10(1):1039. 10.1038/s41598-020-57497-7. 10.1038/s41598-020-57497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–89. 10.1146/annurev-cellbio-101512-122326. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 3.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5(4):317–23. 10.1038/nrm1360. 10.1038/nrm1360 [DOI] [PubMed] [Google Scholar]
- 4.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23(1):519–47. 10.1146/annurev.cellbio.23.090506.123319. 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weidle UH, Birzele F, Kollmorgen G, Rüger R. The multiple roles of exosomes in metastasis. Cancer Genom Proteom. 2017;14(1):1–16. 10.21873/cgp.20015. 10.21873/cgp.20015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu W, Rana S, Zöller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15(8):875-IN4. 10.1593/neo.13786. 10.1593/neo.13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–4. 10.1038/nature15376. 10.1038/nature15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu D, Deng S, Liu T, Han R, Zhang T, Xu Y. TGF-β-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 2018;7(10):5118–29. 10.1002/cam4.1758. 10.1002/cam4.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CC, et al. Elucidation of exosome migration across the blood-brain barrier model In Vitro. Cell Mol Bioeng. 2016;9(4):509–29. 10.1007/s12195-016-0458-3. 10.1007/s12195-016-0458-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin Biol Ther. 2012;12(sup1):S189–97. 10.1517/14712598.2012.680018. 10.1517/14712598.2012.680018 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J Nanobiotechnology. 2022;20(1):279. 10.1186/s12951-022-01472-z. 10.1186/s12951-022-01472-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SM, Yang Y, Oh SJ, Hong Y, Seo M, Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. 10.1016/j.jconrel.2017.09.013. 10.1016/j.jconrel.2017.09.013 [DOI] [PubMed] [Google Scholar]
- 13.McAndrews KM, Xiao F, Chronopoulos A, LeBleu VS, Kugeratski FG, Kalluri R. Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic Kras G12D in pancreatic cancer. Life Sci Alliance. 2021;4(9):e202000875. 10.26508/lsa.202000875. 10.26508/lsa.202000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu W, et al. CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity. Nat Commun. 2019;10(1):4355. 10.1038/s41467-019-12321-3. 10.1038/s41467-019-12321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimura A, Sawada K, Kimura T. Is the exosome a potential target for cancer immunotherapy? Ann Transl Med. 2017;5(5):117–117. 10.21037/atm.2017.01.47. 10.21037/atm.2017.01.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kar R, et al. Exosome-based smart drug delivery tool for cancer theranostics. ACS Biomater Sci Eng. 2023;9(2):577–94. 10.1021/acsbiomaterials.2c01329. 10.1021/acsbiomaterials.2c01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–500. 10.1038/emboj.2011.286. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118(4):1917–50. 10.1021/acs.chemrev.7b00534. 10.1021/acs.chemrev.7b00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. Challenges and opportunities in exosome research—perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019. 10.1063/1.5087122. 10.1063/1.5087122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3(12):893–905. 10.1038/nrm973. 10.1038/nrm973 [DOI] [PubMed] [Google Scholar]
- 21.Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16(4):415–21. 10.1016/j.ceb.2004.06.003. 10.1016/j.ceb.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 22.Théry C, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–18. 10.4049/jimmunol.166.12.7309. 10.4049/jimmunol.166.12.7309 [DOI] [PubMed] [Google Scholar]
- 23.Tamai K, et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399(3):384–90. 10.1016/j.bbrc.2010.07.083. 10.1016/j.bbrc.2010.07.083 [DOI] [PubMed] [Google Scholar]
- 24.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–7. 10.1126/science.1153124. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T, Gu F, Gruenberg J. Lipids, lipid domains and lipid–protein interactions in endocytic membrane traffic. Semin Cell Dev Biol. 1998;9(5):517–26. 10.1006/scdb.1998.0257. 10.1006/scdb.1998.0257 [DOI] [PubMed] [Google Scholar]
- 26.Bianco F, et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009;28(8):1043–54. 10.1038/emboj.2009.45. 10.1038/emboj.2009.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of MicroRNAs in living cells. J Biol Chem. 2010;285(23):17442–52. 10.1074/jbc.M110.107821. 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5(6):e158. 10.1371/journal.pbio.0050158. 10.1371/journal.pbio.0050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J Cell Sci. 1997;110(16):1867–77. 10.1242/jcs.110.16.1867. 10.1242/jcs.110.16.1867 [DOI] [PubMed] [Google Scholar]
- 30.Muntasell A, Berger AC, Roche PA. T cell-induced secretion of MHC class II–peptide complexes on B cell exosomes. EMBO J. 2007;26(19):4263–72. 10.1038/sj.emboj.7601842. 10.1038/sj.emboj.7601842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buschow SI, et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10(10):1528–42. 10.1111/j.1600-0854.2009.00963.x. 10.1111/j.1600-0854.2009.00963.x [DOI] [PubMed] [Google Scholar]
- 32.Urbanelli L, et al. Signaling pathways in exosomes biogenesis, secretion and fate. Genes (Basel). 2013;4(2):152–70. 10.3390/genes4020152. 10.3390/genes4020152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krylova SV, Feng D. ‘The machinery of exosomes: biogenesis release, and Uptake.’ Int J Mol Sci. 2023;24(2):1337. 10.3390/ijms24021337. 10.3390/ijms24021337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Yang J, Cao D, You Y, Shen K, Peng P. Regulation of exosomes released from normal ovarian epithelial cells and ovarian cancer cells. Tumor Biol. 2016;37(12):15763–71. 10.1007/s13277-016-5394-2. 10.1007/s13277-016-5394-2 [DOI] [PubMed] [Google Scholar]
- 35.Von Schulze A, Deng F. A review on exosome-based cancer therapy. J Cancer Metastasis Treat. 2020. 10.20517/2394-4722.2020.79. 10.20517/2394-4722.2020.79 [DOI] [Google Scholar]
- 36.Dhar R, Mallik S, Devi A. Exosomal microRNAs (exoMIRs): micromolecules with macro impact in oral cancer. 3 Biotech. 2022;12(7):155. 10.1007/s13205-022-03217-z. 10.1007/s13205-022-03217-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinbichler TB, Dudás J, Riechelmann H, Skvortsova I-I. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–81. 10.1016/j.semcancer.2017.02.006. 10.1016/j.semcancer.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Tian L, Lu J, Ng IO-L. Exosomes and cancer—diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis. 2022;11(1):54. 10.1038/s41389-022-00431-5. 10.1038/s41389-022-00431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang WH, Cerione RA, Antonyak MA. Extracellular vesicles and their roles in cancer progression. Methods Mol Biol. 2021;2174:143–70. 10.1007/978-1-0716-0759-6_10. 10.1007/978-1-0716-0759-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A, As SG, Ganesh PS, Saravanan M, Sunny B. Exosome mediated cancer therapeutic approach: present status and future prospectives. Asian Pac J Cancer Prev. 2023;24(2):363. 10.31557/APJCP.2023.24.2.363. 10.31557/APJCP.2023.24.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. 10.1016/j.cmet.2006.01.005. 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 42.Zhuang G, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–23. 10.1038/emboj.2012.183. 10.1038/emboj.2012.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124(25):3748–57. 10.1182/blood-2014-05-576116. 10.1182/blood-2014-05-576116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–15. 10.1016/j.ccr.2014.03.007. 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MengXin L, et al. miR-93–5p transferred by exosomes promotes the proliferation of esophageal cancer cells via intercellular communication by targeting PTEN. Biomed Environ Sci. 2018;31(3):171–85. 10.3967/BES2018.023. 10.3967/BES2018.023 [DOI] [PubMed] [Google Scholar]
- 46.Le MTN, et al. miR-200–containing extracellular vesicles promote breast cancer cell metastasis. J Clin Investig. 2014;124(12):5109–28. 10.1172/JCI75695. 10.1172/JCI75695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui H, et al. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2015;34(28):3640–50. 10.1038/onc.2014.300. 10.1038/onc.2014.300 [DOI] [PubMed] [Google Scholar]
- 48.Monaco F, et al. Exosomal transfer of miR-126 promotes the anti-tumour response in malignant mesothelioma: role of miR-126 in cancer-stroma communication. Cancer Lett. 2019;463:27–36. 10.1016/j.canlet.2019.08.001. 10.1016/j.canlet.2019.08.001 [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Lv B, Su Y, Wang X, Bu J, Yao L. <p>Exosome-mediated Transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 Axis</p>. Onco Targets Ther. 2019;12:11325–38. 10.2147/OTT.S231846. 10.2147/OTT.S231846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan L, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143(6):991–1004. 10.1007/s00432-017-2361-2. 10.1007/s00432-017-2361-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–26. 10.1038/ncb3169. 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rong L, Li R, Li S, Luo R. Immunosuppression of breast cancer cells mediated by transforming growth factor-β in exosomes from cancer cells. Oncol Lett. 2016;11(1):500–4. 10.3892/ol.2015.3841. 10.3892/ol.2015.3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, et al. Dendritic cells loaded with tumor derived exosomes for cancer immunotherapy. Oncotarget. 2018;9(2):2887–94. 10.18632/oncotarget.20812. 10.18632/oncotarget.20812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bobrie A, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72(19):4920–30. 10.1158/0008-5472.CAN-12-0925. 10.1158/0008-5472.CAN-12-0925 [DOI] [PubMed] [Google Scholar]
- 55.Tang K, et al. Delivery of chemotherapeutic drugs in tumour cell-derived microparticles. Nat Commun. 2012;3(1):1282. 10.1038/ncomms2282. 10.1038/ncomms2282 [DOI] [PubMed] [Google Scholar]
- 56.Pitzer CR, Paez HG, Alway SE. The contribution of tumor derived exosomes to cancer cachexia. Cells. 2023. 10.3390/CELLS12020292. 10.3390/CELLS12020292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohammadi M, Zargartalebi H, Salahandish R, Aburashed R, Wey Yong K, Sanati-Nezhad A. Emerging technologies and commercial products in exosome-based cancer diagnosis and prognosis. Biosens Bioelectron. 2021;183:113176. 10.1016/j.bios.2021.113176. 10.1016/j.bios.2021.113176 [DOI] [PubMed] [Google Scholar]
- 58.Ge R, Tan E, Sharghi-Namini S, Asada HH. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 2012;5(3):323–32. 10.1007/s12307-012-0110-2. 10.1007/s12307-012-0110-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim MS, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–64. 10.1016/j.nano.2015.10.012. 10.1016/j.nano.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tickner JA, Urquhart AJ, Stephenson S-A, Richard DJ, Oâ€TMByrne KJ. Functions and therapeutic roles of exosomes in cancer. Front Oncol. 2014. 10.3389/fonc.2014.00127. 10.3389/fonc.2014.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol. 2018. 10.3389/fonc.2018.00066. 10.3389/fonc.2018.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Zheng Y, Zhao M. Exosome-based cancer therapy: implication for targeting cancer stem cells. Front Pharmacol. 2017. 10.3389/fphar.2016.00533. 10.3389/fphar.2016.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Asti E, Garnier D, Lee TH, Montermini L, Meehan B, Rak J. Oncogenic extracellular vesicles in brain tumor progression. Front Physiol. 2012. 10.3389/fphys.2012.00294. 10.3389/fphys.2012.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paskeh MDA, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. 2022;15(1):83. 10.1186/s13045-022-01305-4. 10.1186/s13045-022-01305-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochimica et Biophysica Acta (BBA) Rev Cancer. 2019;1871(2):455–68. 10.1016/j.bbcan.2019.04.004. 10.1016/j.bbcan.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–42. 10.1007/s10555-013-9441-9. 10.1007/s10555-013-9441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:1–11. 10.1155/2011/842849. 10.1155/2011/842849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. 10.1038/ncb1800. 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kosaka N, Yoshioka Y, Tominaga N, Hagiwara K, Katsuda T, Ochiya T. Dark side of the exosome: the role of the exosome in cancer metastasis and targeting the exosome as a strategy for cancer therapy. Future Oncol. 2014;10(4):671–81. 10.2217/fon.13.222. 10.2217/fon.13.222 [DOI] [PubMed] [Google Scholar]
- 70.Kalluri R, Leleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020. 10.1126/science.aau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nam G, Choi Y, Kim GB, Kim S, Kim SA, Kim I. Emerging prospects of exosomes for cancer treatment: from conventional therapy to immunotherapy. Adv Mater. 2020;32(51):2002440. 10.1002/adma.202002440. 10.1002/adma.202002440 [DOI] [PubMed] [Google Scholar]
- 72.Zhu L, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13(1):152. 10.1186/s13045-020-00987-y. 10.1186/s13045-020-00987-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of micrornas detectable in serum and saliva is concentrated in exosomes. PLoS ONE. 2012;7(3): e30679. 10.1371/journal.pone.0030679. 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. 10.1038/cr.2015.82. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22(2):283–99. 10.3233/CBM-171011. 10.3233/CBM-171011 [DOI] [PubMed] [Google Scholar]
- 76.Santasusagna S, et al. Prognostic impact of miR-200 family members in plasma and exosomes from tumor-draining versus peripheral veins of colon cancer patients. Oncology. 2018;95(5):309–18. 10.1159/000490726. 10.1159/000490726 [DOI] [PubMed] [Google Scholar]
- 77.Liu W, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–51. 10.2147/OTT.S140062. 10.2147/OTT.S140062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong C-H, Chen Y-C. Clinical significance of exosomes as potential biomarkers in cancer. World J Clin Cases. 2019;7(2):171–90. 10.12998/wjcc.v7.i2.171. 10.12998/wjcc.v7.i2.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14(11):1891–900. 10.1039/C4LC00136B. 10.1039/C4LC00136B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shao H, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–40. 10.1038/nm.2994. 10.1038/nm.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshioka Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5(1):3591. 10.1038/ncomms4591. 10.1038/ncomms4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16(3):489–96. 10.1039/C5LC01117E. 10.1039/C5LC01117E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee CH, Im EJ, Moon PG, Baek MC. Discovery of a diagnostic biomarker for colon cancer through proteomic profiling of small extracellular vesicles. BMC Cancer. 2018. 10.1186/S12885-018-4952-Y. 10.1186/S12885-018-4952-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang C, et al. Plasma extracellular vesicle derived protein profile predicting and monitoring immunotherapeutic outcomes of gastric cancer. J Extracell Vesicles. 2022. 10.1002/JEV2.12209. 10.1002/JEV2.12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee SJ, et al. Exosomal Del-1 as a potent diagnostic marker for breast cancer: prospective cohort study. Clin Breast Cancer. 2021;21(6):e748–56. 10.1016/J.CLBC.2021.02.002. 10.1016/J.CLBC.2021.02.002 [DOI] [PubMed] [Google Scholar]
- 86.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. 10.1038/ncb1596. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 87.Beach A, Zhang H-G, Ratajczak MZ, Kakar SS. Exosomes: an overview of biogenesis, composition and role in ovarian cancer. J Ovarian Res. 2014;7(1):14. 10.1186/1757-2215-7-14. 10.1186/1757-2215-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. exosomal MicroRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. 10.3816/CLC.2009.n.006. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 89.Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–75. 10.1074/jbc.C113.532267. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–9. 10.1038/cr.2014.44. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin G, et al. [Retracted] exosomal miR-130a-3p promotes the progression of differentiated thyroid cancer by targeting insulin-like growth factor 1. Oncol Lett. 2023;26(4):1–1. 10.3892/OL.2023.14017. 10.3892/OL.2023.14017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Wen Q, Wang Y, Li X, Jin X, Wang G. Decreased serum exosomal miR-29a expression and its clinical significance in papillary thyroid carcinoma. J Clin Lab Anal. 2021;35(1):e23560. 10.1002/JCLA.23560. 10.1002/JCLA.23560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zou X, et al. A three-microRNA panel in serum as novel biomarker for papillary thyroid carcinoma diagnosis. Chin Med J (Engl). 2020;133(21):2543–51. 10.1097/CM9.0000000000001107. 10.1097/CM9.0000000000001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan Q, et al. Exosomal miRNAs are potential diagnostic biomarkers between malignant and benign thyroid nodules based on next-generation sequencing. Carcinogenesis. 2020;41(1):18–24. 10.1093/CARCIN/BGZ160. 10.1093/CARCIN/BGZ160 [DOI] [PubMed] [Google Scholar]
- 95.Liang M, et al. A panel of plasma exosomal miRNAs as potential biomarkers for differential diagnosis of thyroid nodules. Front Genet. 2020;11:539614. 10.3389/FGENE.2020.00449/BIBTEX. 10.3389/FGENE.2020.00449/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dai D, Tan Y, Guo L, Tang A, Zhao Y. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur J Endocrinol. 2020;182(1):111–21. 10.1530/EJE-19-0524. 10.1530/EJE-19-0524 [DOI] [PubMed] [Google Scholar]
- 97.Wang Z, et al. A three plasma microRNA signature for papillary thyroid carcinoma diagnosis in Chinese patients. Gene. 2019;693:37–45. 10.1016/J.GENE.2019.01.016. 10.1016/J.GENE.2019.01.016 [DOI] [PubMed] [Google Scholar]
- 98.Shen X, et al. Evaluation of EpCAM-specific exosomal lncRNAs as potential diagnostic biomarkers for lung cancer using droplet digital PCR. J Mol Med. 2022;100(1):87–100. 10.1007/S00109-021-02145-4/METRICS. 10.1007/S00109-021-02145-4/METRICS [DOI] [PubMed] [Google Scholar]
- 99.Yamanaka R, Abe E, Sato T, Hayano A, Takashima Y. Secondary intracranial tumors following radiotherapy for pituitary adenomas: a systematic review. Cancers (Basel). 2017. 10.3390/CANCERS9080103. 10.3390/CANCERS9080103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hydbring P, et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer. 2018;124:45–52. 10.1016/J.LUNGCAN.2018.07.018. 10.1016/J.LUNGCAN.2018.07.018 [DOI] [PubMed] [Google Scholar]
- 101.Zhai LY, et al. In situ detection of plasma exosomal MicroRNA-1246 for breast cancer diagnostics by a Au nanoflare probe. ACS Appl Mater Interfaces. 2018;10(46):39478–86. 10.1021/ACSAMI.8B12725. 10.1021/ACSAMI.8B12725 [DOI] [PubMed] [Google Scholar]
- 102.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome isolation techniques. Theranostics. 2017;7(3):789–804. 10.7150/thno.18133. 10.7150/thno.18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Helwa I, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS ONE. 2017;12(1):e0170628. 10.1371/journal.pone.0170628. 10.1371/journal.pone.0170628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeppesen DK, et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles. 2014;3(1):25011. 10.3402/jev.v3.25011. 10.3402/jev.v3.25011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Patel GK, et al. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep. 2019;9(1):5335. 10.1038/s41598-019-41800-2. 10.1038/s41598-019-41800-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng Q, Zhang J, Zhou G. Comparison of plasma exosomes by differential ultracentrifugation and solvent precipitation methods. Clin Lab. 2018;64(6):991–8. 10.7754/CLIN.LAB.2018.180104. 10.7754/CLIN.LAB.2018.180104 [DOI] [PubMed] [Google Scholar]
- 107.Mol EA, Goumans M-J, Doevendans PA, Sluijter JPG, Vader P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine. 2017;13(6):2061–5. 10.1016/j.nano.2017.03.011. 10.1016/j.nano.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 108.Konoshenko MYu, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed Res Int. 2018;2018:1–27. 10.1155/2018/8545347. 10.1155/2018/8545347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu R, Greening DW, Rai A, Ji H, Simpson RJ. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods. 2015;87:11–25. 10.1016/j.ymeth.2015.04.008. 10.1016/j.ymeth.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 110.Oksvold MP, Neurauter A, Pedersen KW. Magnetic bead-based isolation of exosomes. New York: Springer New York; 2015. 10.1007/978-1-4939-1538-5_27. [DOI] [PubMed] [Google Scholar]
- 111.Amrollahi P, Rodrigues M, Lyon CJ, Goel A, Han H, Hu TY. Ultra-sensitive automated profiling of EpCAM expression on tumor-derived extracellular vesicles. Front Genet. 2019. 10.3389/fgene.2019.01273. 10.3389/fgene.2019.01273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weng Y, et al. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 2016;141(15):4640–6. 10.1039/C6AN00892E. 10.1039/C6AN00892E [DOI] [PubMed] [Google Scholar]
- 113.Lin S, et al. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16(9):1903916. 10.1002/smll.201903916. 10.1002/smll.201903916 [DOI] [PubMed] [Google Scholar]
- 114.Yang F, Liao X, Tian Y, Li G. Exosome separation using microfluidic systems: size-based, immunoaffinity-based and dynamic methodologies. Biotechnol J. 2017;12(4):1600699. 10.1002/biot.201600699. 10.1002/biot.201600699 [DOI] [PubMed] [Google Scholar]
- 115.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. 10.1038/nature03702. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 116.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–707. 10.1158/0008-5472.CAN-07-1936. 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- 117.Moskovits M. Surface-enhanced Raman spectroscopy: a brief retrospective. J Raman Spectrosc. 2005;36(6–7):485–96. 10.1002/jrs.1362. 10.1002/jrs.1362 [DOI] [Google Scholar]
- 118.Valley N, Greeneltch N, Van Duyne RP, Schatz GC. A look at the origin and magnitude of the chemical contribution to the enhancement mechanism of surface-enhanced Raman spectroscopy (SERS): theory and experiment. J Phys Chem Lett. 2013;4(16):2599–604. 10.1021/jz4012383. 10.1021/jz4012383 [DOI] [Google Scholar]
- 119.Gómez M, Lazzari M. Reliable and cheap SERS active substrates. Mater Today. 2014;17(7):358–9. 10.1016/j.mattod.2014.08.001. 10.1016/j.mattod.2014.08.001 [DOI] [Google Scholar]
- 120.Wu X, et al. Exosome-templated nanoplasmonics for multiparametric molecular profiling. Sci Adv. 2020. 10.1126/sciadv.aba2556. 10.1126/sciadv.aba2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mastoridis S, Bertolino GM, Whitehouse G, Dazzi F, Sanchez-Fueyo A, Martinez-Llordella M. Multiparametric analysis of circulating exosomes and other small extracellular vesicles by advanced imaging flow cytometry. Front Immunol. 2018. 10.3389/fimmu.2018.01583. 10.3389/fimmu.2018.01583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res. 2005;4(4):1104–9. 10.1021/pr050027n. 10.1021/pr050027n [DOI] [PubMed] [Google Scholar]
- 123.Drake RR, et al. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5(10):1957–67. 10.1074/mcp.M600176-MCP200. 10.1074/mcp.M600176-MCP200 [DOI] [PubMed] [Google Scholar]
- 124.Haab BB. Antibody arrays in cancer research. Mol Cell Proteomics. 2005;4(4):377–83. 10.1074/mcp.M500010-MCP200. 10.1074/mcp.M500010-MCP200 [DOI] [PubMed] [Google Scholar]
- 125.Ueda K, Ishikawa N, Tatsuguchi A, Saichi N, Fujii R, Nakagawa H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci Rep. 2014;4(1):6232. 10.1038/srep06232. 10.1038/srep06232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Saraswat M, Joenväära S, Musante L, Peltoniemi H, Holthofer H, Renkonen R. N-linked (N-) Glycoproteomics of Urimary Exosomes*. Mol Cell Proteomics. 2015;14(2):263–76. 10.1074/mcp.M114.040345. 10.1074/mcp.M114.040345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Escrevente C, et al. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS ONE. 2013;8(10):e78631. 10.1371/journal.pone.0078631. 10.1371/journal.pone.0078631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gerlach JQ, et al. Surface glycosylation profiles of urine extracellular vesicles. PLoS ONE. 2013;8(9):e74801. 10.1371/journal.pone.0074801. 10.1371/journal.pone.0074801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saito S, Hiemori K, Kiyoi K, Tateno H. Glycome analysis of extracellular vesicles derived from human induced pluripotent stem cells using lectin microarray. Sci Rep. 2018;8(1):3997. 10.1038/s41598-018-22450-2. 10.1038/s41598-018-22450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martins TS, Catita J, Rosa IM, Da Cruz e Silva OAB, Henriques AG. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE. 2018. 10.1371/JOURNAL.PONE.0198820. 10.1371/JOURNAL.PONE.0198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179(3):1969–78. 10.4049/jimmunol.179.3.1969. 10.4049/jimmunol.179.3.1969 [DOI] [PubMed] [Google Scholar]
- 132.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci. 2009;106(10):3794–9. 10.1073/pnas.0804543106. 10.1073/pnas.0804543106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. 10.1038/ncb1725. 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- 134.Nakata R, et al. Contribution of neuroblastoma-derived exosomes to the production of pro-tumorigenic signals by bone marrow mesenchymal stromal cells. J Extracell Vesicles. 2017;6(1):1332941. 10.1080/20013078.2017.1332941. 10.1080/20013078.2017.1332941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tian Y, et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell Vesicles. 2020;9(1):1697028. 10.1080/20013078.2019.1697028. 10.1080/20013078.2019.1697028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci. 2016. 10.1073/pnas.1521230113. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Allenson K, et al. High prevalence of mutantKRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28(4):741–7. 10.1093/annonc/mdx004. 10.1093/annonc/mdx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Castillo J, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29(1):223–9. 10.1093/annonc/mdx542. 10.1093/annonc/mdx542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wolfers J, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7(3):297–303. 10.1038/85438. 10.1038/85438 [DOI] [PubMed] [Google Scholar]
- 140.Li W, et al. Exosomes derived from Rab27a-overexpressing tumor cells elicit efficient induction of antitumor immunity. Mol Med Rep. 2013;8(6):1876–82. 10.3892/mmr.2013.1738. 10.3892/mmr.2013.1738 [DOI] [PubMed] [Google Scholar]
- 141.Iwai K, Minamisawa T, Suga K, Yajima Y, Shiba K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J Extracell Vesicles. 2016;5(1):30829. 10.3402/jev.v5.30829. 10.3402/jev.v5.30829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bijnsdorp IV, et al. Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method. J Extracell Vesicles. 2017;6(1):1313091. 10.1080/20013078.2017.1313091. 10.1080/20013078.2017.1313091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shaba E, et al. Multi-omics integrative approach of extracellular vesicles: a future challenging milestone. Proteomes. 2022;10(2):12. 10.3390/proteomes10020012. 10.3390/proteomes10020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mostafazadeh M, Samadi N, Kahroba H, Baradaran B, Haiaty S, Nouri M. Potential roles and prognostic significance of exosomes in cancer drug resistance. Cell Biosci. 2021;11(1):1. 10.1186/s13578-020-00515-y. 10.1186/s13578-020-00515-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational Implications of Tumor Heterogeneity. Clin Cancer Res. 2015;21(6):1258–66. 10.1158/1078-0432.CCR-14-1429. 10.1158/1078-0432.CCR-14-1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Makler A, Asghar W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev Mol Diagn. 2020;20(4):387–400. 10.1080/14737159.2020.1731308. 10.1080/14737159.2020.1731308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hannafon BN, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90. 10.1186/s13058-016-0753-x. 10.1186/s13058-016-0753-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee J-K, et al. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS ONE. 2013;8(12):e84256. 10.1371/journal.pone.0084256. 10.1371/journal.pone.0084256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Soung Y, Ford S, Zhang V, Chung J. Exosomes in cancer diagnostics. Cancers (Basel). 2017. 10.3390/cancers9010008. 10.3390/cancers9010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Egan TK. Monitoring patients undergoing cancer therapy. Lab Med. 2000;31(12):666–71. 10.1309/R078-Y40Q-PAJP-1RPP. 10.1309/R078-Y40Q-PAJP-1RPP [DOI] [Google Scholar]
- 151.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. 10.3322/caac.21763. 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 152.Shen Y, Huang J, Jia L, Zhang C, Xu J. Bioinformatics and machine learning driven key genes screening for hepatocellular carcinoma. Biochem Biophys Rep. 2024;37: 101587. 10.1016/j.bbrep.2023.101587. 10.1016/j.bbrep.2023.101587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cassim S, Chepulis L, Keenan R, Kidd J, Firth M, Lawrenson R. Patient and carer perceived barriers to early presentation and diagnosis of lung cancer: a systematic review. BMC Cancer. 2019;19(1):25. 10.1186/s12885-018-5169-9. 10.1186/s12885-018-5169-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Logozzi M, et al. Microenvironmental pH and exosome levels interplay in human cancer cell lines of different histotypes. Cancers (Basel). 2018;10(10):370. 10.3390/cancers10100370. 10.3390/cancers10100370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Théry C, et al. Minimal information for studies of extracellular vesicles (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. 10.1080/20013078.2018.1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mateescu B, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6(1):1286095. 10.1080/20013078.2017.1286095. 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jeppesen DK, et al. Reassessment of exosome composition. Cell. 2019;177(2):428–44518. 10.1016/j.cell.2019.02.029. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tkach M, Théry C. Communication by Extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–32. 10.1016/j.cell.2016.01.043. 10.1016/j.cell.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 159.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013. 10.3402/JEV.V2I0.20360. 10.3402/JEV.V2I0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Van Deun J, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228–32. 10.1038/nmeth.4185. 10.1038/nmeth.4185 [DOI] [PubMed] [Google Scholar]
- 161.Hilton SH, White IM. Advances in the analysis of single extracellular vesicles: a critical review. Sensors Actuators Reports. 2021;3:100052. 10.1016/j.snr.2021.100052. 10.1016/j.snr.2021.100052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ferguson S, Yang KS, Weissleder R. Single extracellular vesicle analysis for early cancer detection. Trends Mol Med. 2022;28(8):681–92. 10.1016/j.molmed.2022.05.003. 10.1016/j.molmed.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bebelman MP, Janssen E, Pegtel DM, Crudden C. The forces driving cancer extracellular vesicle secretion. Neoplasia. 2021;23(1):149–57. 10.1016/j.neo.2020.11.011. 10.1016/j.neo.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Qiu L, Liu X, Zhu L, Luo L, Sun N, Pei R. Current advances in technologies for single extracellular vesicle analysis and its clinical applications in cancer diagnosis. Biosensors (Basel). 2023. 10.3390/BIOS13010129. 10.3390/BIOS13010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee K, et al. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12(1):494–503. 10.1021/ACSNANO.7B07060. 10.1021/ACSNANO.7B07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu C, et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018;18(7):4226–32. 10.1021/ACS.NANOLETT.8B01184. 10.1021/ACS.NANOLETT.8B01184 [DOI] [PubMed] [Google Scholar]
- 167.Ricklefs FL, et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019. 10.1080/20013078.2019.1588555. 10.1080/20013078.2019.1588555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shen W, et al. A single extracellular vesicle (EV) flow cytometry approach to reveal EV heterogeneity. Angew Chem Int Ed Engl. 2018;57(48):15675–80. 10.1002/ANIE.201806901. 10.1002/ANIE.201806901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7(6):780–8. 10.1016/J.NANO.2011.04.003. 10.1016/J.NANO.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Spedalieri C, Kneipp J. Surface enhanced Raman scattering for probing cellular biochemistry. Nanoscale. 2022;14(14):5314–28. 10.1039/D2NR00449F. 10.1039/D2NR00449F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu MY, Ling DX, Ling L, Li W, Li YQ. Stable optical trapping and sensitive characterization of nanostructures using standing-wave Raman tweezers. Sci Reports. 2017;7(1):1–8. 10.1038/srep42930. 10.1038/srep42930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Guo W, et al. Single-exosome profiling identifies ITGB3+ and ITGAM+ exosome subpopulations as promising early diagnostic biomarkers and therapeutic targets for colorectal cancer. Research. 2023. 10.34133/research.0041. 10.34133/research.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hoshino A, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044-1061.e18. 10.1016/j.cell.2020.07.009. 10.1016/j.cell.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fitts CA, Ji N, Li Y, Tan C. Exploiting exosomes in cancer liquid biopsies and drug delivery. Adv Healthc Mater. 2019;8(6):1801268. 10.1002/adhm.201801268. 10.1002/adhm.201801268 [DOI] [PubMed] [Google Scholar]
- 175.Li S, Yi M, Dong B, Tan X, Luo S, Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 2021;148(11):2640–51. 10.1002/ijc.33386. 10.1002/ijc.33386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer. 2022;21(1):54. 10.1186/s12943-022-01525-9. 10.1186/s12943-022-01525-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yu W, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32(4):466–77. 10.1016/j.annonc.2021.01.074. 10.1016/j.annonc.2021.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Clayton A, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247(1–2):163–74. 10.1016/S0022-1759(00)00321-5. 10.1016/S0022-1759(00)00321-5 [DOI] [PubMed] [Google Scholar]
- 179.Mirzakhani M, Shahbazi M, Oliaei F, Mohammadnia-Afrouzi M. Immunological biomarkers of tolerance in human kidney transplantation: an updated literature review. J Cell Physiol. 2019;234(5):5762–74. 10.1002/jcp.27480. 10.1002/jcp.27480 [DOI] [PubMed] [Google Scholar]
- 180.Kato T, Fahrmann JF, Hanash SM, Vykoukal J. Extracellular vesicles mediate B cell immune response and are a potential target for cancer therapy. Cells. 2020;9(6):1518. 10.3390/cells9061518. 10.3390/cells9061518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Stickney Z, Losacco J, McDevitt S, Zhang Z, Lu B. Development of exosome surface display technology in living human cells. Biochem Biophys Res Commun. 2016;472(1):53–9. 10.1016/j.bbrc.2016.02.058. 10.1016/j.bbrc.2016.02.058 [DOI] [PubMed] [Google Scholar]
- 182.Logozzi M, et al. High levels of exosomes expressing cd63 and caveolin-1 in plasma of melanoma patients. PLoS ONE. 2009;4(4):e5219. 10.1371/journal.pone.0005219. 10.1371/journal.pone.0005219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. 10.1038/nature14581. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li J, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. 2017;21(5):838–47. 10.1111/jcmm.12941. 10.1111/jcmm.12941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Castellanos-Rizaldos E, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res. 2018;24(12):2944–50. 10.1158/1078-0432.CCR-17-3369. 10.1158/1078-0432.CCR-17-3369 [DOI] [PubMed] [Google Scholar]
- 186.Krug AK, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29(3):700–6. 10.1093/annonc/mdx765. 10.1093/annonc/mdx765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139(7):1443–8. 10.1002/ijc.30179. 10.1002/ijc.30179 [DOI] [PubMed] [Google Scholar]
- 188.Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5. 10.1111/j.1365-2141.2008.07077.x. 10.1111/j.1365-2141.2008.07077.x [DOI] [PubMed] [Google Scholar]
- 189.Tanaka Y, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119(6):1159–67. 10.1002/cncr.27895. 10.1002/cncr.27895 [DOI] [PubMed] [Google Scholar]
- 190.Lan F, Qing Q, Pan Q, Hu M, Yu H, Yue X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol. 2018;41(1):25–33. 10.1007/s13402-017-0355-3. 10.1007/s13402-017-0355-3 [DOI] [PubMed] [Google Scholar]
- 191.Kanaoka R, et al. Usefulness of plasma exosomal MicroRNA-451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncology. 2018;94(5):311–23. 10.1159/000487006. 10.1159/000487006 [DOI] [PubMed] [Google Scholar]
- 192.Lee YR, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144(6):1444–52. 10.1002/ijc.31931. 10.1002/ijc.31931 [DOI] [PubMed] [Google Scholar]
- 193.De Long J, et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res. 2015;7(11):2500–9. [PMC free article] [PubMed] [Google Scholar]
- 194.Chen G, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–6. 10.1038/s41586-018-0392-8. 10.1038/s41586-018-0392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yuwen D-L, Sheng B-B, Liu J, Wenyu W, Shu Y-Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2650–8. [PubMed] [Google Scholar]
- 196.Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical significance of PD-L1 þ exosomes in plasma of head and neck cancer patients. Clin Cancer Res. 2018;24(4):896–905. 10.1158/1078-0432.CCR-17-2664/87451/AM/CLINICAL-SIGNIFICANCE-OF-PD-L1-EXOSOMES-IN-PLASMA. 10.1158/1078-0432.CCR-17-2664/87451/AM/CLINICAL-SIGNIFICANCE-OF-PD-L1-EXOSOMES-IN-PLASMA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang J, et al. Exosome and exosomal MicroRNA: trafficking, sorting, and function. Genomics Proteomics Bioinform. 2015;13(1):17–24. 10.1016/j.gpb.2015.02.001. 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fang Z, Ding YX, Li F. Exosomes: promising biomarkers and targets for cancer. World J Gastrointest Oncol. 2022;14(8):1594–6. 10.4251/WJGO.V14.I8.1594. 10.4251/WJGO.V14.I8.1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bhat EA, Sajjad N, Thokar FM. Current advancement of exosomes as biomarkers for cancer diagnosis and forecasting. Cancer Treat Res Commun. 2021;28:100417. 10.1016/j.ctarc.2021.100417. 10.1016/j.ctarc.2021.100417 [DOI] [PubMed] [Google Scholar]
- 200.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. 10.1016/j.plipres.2017.03.001. 10.1016/j.plipres.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 201.Lou G, et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8(1):122. 10.1186/s13045-015-0220-7. 10.1186/s13045-015-0220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Samanta S, Rajasingh S, Drosos N, Zhou Z, Dawn B, Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharmacol Sin. 2018;39(4):501–13. 10.1038/aps.2017.162. 10.1038/aps.2017.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sauter ER. Future perspectives for body fluid exosomes and cancer. Transl Cancer Res. 2017;6(S8):S1394–7. 10.21037/tcr.2017.09.28. 10.21037/tcr.2017.09.28 [DOI] [Google Scholar]
- 204.Wu X, et al. The roles of exosomes as future therapeutic agents and diagnostic tools for glioma. Front Oncol. 2021. 10.3389/fonc.2021.733529. 10.3389/fonc.2021.733529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.