Abstract
The baculovirus Bombyx mori nucleopolyhedrovirus (BmNPV) contains five related open reading frames (ORFs). Recent sequence analyses of several other baculovirus genomes reveal that these ORFs belong to a unique multigene family called the baculovirus repeated ORFs (bro) family. Here we have characterized these five genes from BmNPV at the transcriptional and translational levels. Reverse transcription-PCR and primer extension analyses indicated that transcription of all bro genes occurs by 2 to 4 h postinfection (p.i.) and reaches maximal levels between at 8 and 12 h p.i. Transcription of all genes is initiated between 50 and 70 nucleotides upstream of the start codon, at a characteristic C(T)AGT motif. Expression of a cat reporter gene under the control of each bro promoter provides evidence that a viral factor(s) is required for the transcription of all bro genes. Immunoblot analysis indicated that a population of BRO proteins is produced vigorously between at 8 and 14 h p.i. Immunohistochemical analysis by confocal microscopy showed that BRO proteins are localized in both the nucleus and the cytoplasm at 8 h p.i. Four BmNPV mutants, in which the bro-a, bro-b, bro-c, and bro-e genes were individually inactivated, were successfully isolated. However, exhaustive efforts failed to isolate a bro-d-deficient mutant. Similarly, it was not possible to isolate a double-deletion bro-a bro-c mutant. The bro-d gene may play an irreplaceable functional role(s) during viral infection, while bro-a and bro-c may functionally complement each other.
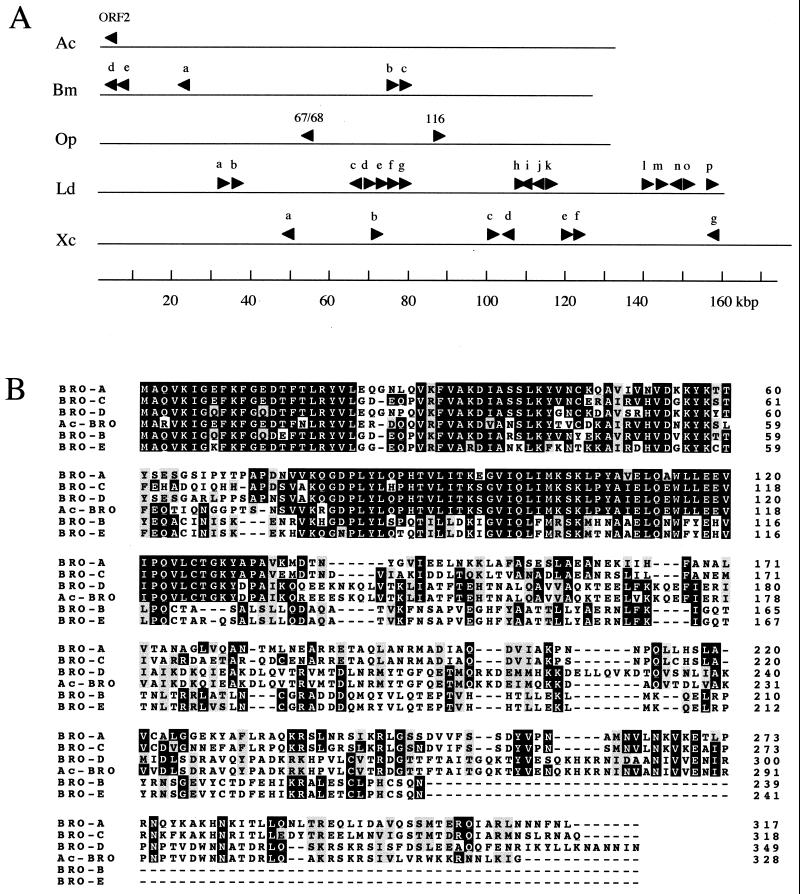
Bombyx mori nucleopolyhedrovirus (BmNPV) is a member of the Baculoviridae, a large family of viruses with circular double-stranded DNA genomes that are pathogenic for invertebrates, particularly insects of the order Lepidoptera. The BmNPV genome is about 128 kb long and is predicted to contain 136 open reading frames (ORFs) (5). Although the genomes of BmNPV and Autographa californica NPV (AcNPV) are closely related in ORF content, ORF homology (on average about 90% amino acid sequence identity), and genome organization, a major distinction is the presence of five ORFs in BmNPV related to AcNPV ORF2 (5). These ORFs (ORFs 22, 80, 81, 131, and 132) are members of the baculovirus repeated ORFs (bro) family (5). In addition, Kuzio et al. (10) reported that there are 16 bro genes in the genome of Lymantria dispar NPV (LdNPV) and that one of these, Ld-bro-n, was the most highly conserved ORF between LdNPV and AcNPV/BmNPV (82% amino acid sequence identity). Whereas AcNPV has only one bro gene, there are three bro genes (one encoding a small truncated 88-amino-acid protein and two ORFs) in Orgyia pseudotsugata NPV (OpNPV) (2) and seven bro genes in Xestia c-nigrum granulovirus (XcGV) (8) (Fig. 1A). According to Kuzio et al. (10), most bro genes have a related sequence in their N-terminal regions but show differing degrees of similarity in other regions. They proposed that the bro gene family can be separated into four groups (groups I to IV) based on the relationships of the variable regions.
FIG. 1.
bro gene family in baculoviruses and alignment of BROs. (A) Presence of the bro family in various baculoviruses. The maps show the genome organization of bro genes in various baculoviruses. The arrowheads denote the direction of transcription. Baculoviruses aligned was as follows. Ac, AcNPV; Bm, BmNPV; Op, OpNPV; Ld, LdNPV; Xc, XcGV. (B) Alignment of amino acid sequences of BmNPV BROs and AcNPV BRO (accession no. L22858). The alignment was conducted with the Clustal X program and modified by using Boxshade. The alignment of three or more identical amino acids is indicated by white letters within black boxes, and three or more similar residues are shown in shaded boxes. Dashes indicate gaps.
Although this gene family appears to be widespread, highly repetitive, and highly conserved, its function has not been determined. In this report, we describe the characterization of BmNPV bro gene expression at the transcriptional and translational level. In addition, we have examined the locations of products of these genes in cells during the infection cycle.
MATERIALS AND METHODS
Cells, insects, and viruses.
The BmN (BmN-4) cell line was maintained in TC-100 with 10% fetal bovine serum as described previously (11). Larvae of the silkworm B. mori were reared on an artificial diet at 27°C (12). The BmNPV T3 isolate and the recombinants were propagated on BmN cells. Transfection of plasmid DNA and cotransfection of BmNPV DNA and plasmids carrying the β-galactosidase gene inserted into each bro gene (Table 1) were performed by following the instructions in the manual provided by the manufacturer of Lipofectin, with no modification (Gibco-BRL). Virus infections and plaque assays were performed as described previously (11).
TABLE 1.
Plasmids used for construction of deletion mutants and transient-transfection assays
| Gene | Plasmids used for construction
|
Isolation of recombinantd | |||
|---|---|---|---|---|---|
| Plasmidab | ORFbc | lacZ inserted atab: | Genomic clone | ||
| bro-a | HindHeh3, HindIII-EcoRI (19720–22514) | 20852←21805 | BstE II - BstE II (21287–21562) | HindH | + |
| bro-b | EcoAph2, HindIII-PstI (74903–77340) | 76776→77495 | Sty I (76870) | EcoA | + |
| bro-c | PstBps20, PstI-SmaI (77341–80428) | 77570→78526 | Bbs I - Bbs I (77606–78431) | PstB | + |
| bro-d | EcoEex9 EcoRI-XbaI (124275–147) | 124934←125983 | Sac II - Sty I (125224–125881) | EcoE | − |
| bro-e | EcoEex9 | 126058←126783 | Not I - Bbs I (126241–126742) | EcoE | + |
Construction of the clones is described in Materials and Methods.
Numbers indicate the locations in the BmNPV genome.
Arrows indicate the direction of transcription of each ORF.
+, recombinant virus was purified; −, recombinant virus was not purified.
DNA manipulations.
To construct plasmids for bro gene deletions, DNA fragments from genomic libraries (13) were first inserted into pTZ19R (Pharmacia Biotech Inc.). Table 1 summarizes all the constructions; for example, HindHeh3 (Table 1) was constructed by subcloning the 2.8-kb HindIII-EcoRI fragment of genomic clone HindH into pTZ19R. Insertion of a β-galactosidase gene cassette containing a Drosophila melanogaster heat shock promoter (hsp70) (designated lacZ) into each bro gene coding region was performed as described previously (6). Viral DNA was purified from culture supernatant and polyhedra of virus-infected BmN cells (11). The reporter plasmid pSK-CAT was constructed as follows. Plasmid pMAMneo-CAT (Clontech) was digested with NheI, filled in with Klenow fragment, and digested with XhoI; then the 0.8-kb DNA fragment containing the cat gene was inserted into pBluescript SK(−). Plasmid pSK-CAT was then used to construct pAp-CAT, pBp-CAT, pCp-CAT, pDp-CAT, and pEp-CAT, containing the upstream regions of bro-a, bro-b, bro-c, bro-d, and bro-e, respectively. These plasmids were constructed as follows. Plasmid HinHeh3 was digested with XhoI, and the 398-bp fragment was inserted into pSK-CAT (pAp-CAT). For pBp-CAT, EcoAph2 was digested with SphI and DraI and the 400-bp fragment was inserted into pSK-CAT. Plasmid PstBps20 was digested with PstI and DraI, and the 255-bp fragment was inserted into pSK-CAT (pCp-CAT). The 381-bp NotI-DraI fragment and the 421-bp NspV-SalI fragment of EcoEex9 were inserted into pSK-CAT to construct pDp-CAT and pEp-CAT, respectively. Each reporter plasmid contains at least 230 nucleotides (nt) upstream of ATG initiation codon. Routine methods for DNA manipulations were as described by Sambrook et al. (18).
RNA manipulations.
Total RNA was isolated as described by Durantel et al. (4) from BmNPV-infected and mock-infected BmN cells. For reverse transcription-PCR (RT-PCR), first-strand cDNA was synthesized from 1 μg of total RNA with oligo(dT) primers and SuperScript II reverse transcriptase (Gibco BRL). PCR amplification was carried out with primer pairs that had been shown to bind to each bro gene specifically. For primer extension, the specific primers pBro-a (5′-TTTGTCAACATTAACTATCACAGCTTG-3′), pBro-b (5′-CATCGACATGAACACGTACTGCCTTTTCAT-3′), pBro-c (5′-CTGCGACGTGGACACGTATTGCCCGTTCAC-3′), pBro-d (5′-TTTATCCACGTGTCTGCTTACAGCGTC-3′), and pBro-e (5′-TCGACGTGATCTCTAATTGCTTTTTTGGTG-3′) were synthesized. A 20-μg portion of total RNA was annealed at 55°C to IRD-41-labeled primers. Extension was performed with SuperScript II reverse transcriptase at 42°C for 1 h. The labeled primers were also used to prime nucleotide sequence reactions with the relevant plasmids (Table 1) as templates. The products of primer extension and sequencing were resolved on an LIC-4000 DNA sequencer (LI-COR).
Polyclonal antibody production and immunodetection.
The BmNPV bro-a gene was amplified by PCR with an upstream primer (5′-CAGAATTCGCTCAAGTTAAAATTGG-3′) incorporating an EcoRI site (underlined) and a downstream primer (5′-CAAAGCTTTTACAAGTTAAAATTGT-3′) with a HindIII site (underlined). The amplified fragment was digested with EcoRI and HindIII and then inserted into pET28a(+) (Novagen). The resulting plasmid was transformed into Escherichia coli BL21(DE3) LysE. Recombinant protein was expressed by following the instructions in the manufacturer’s manual (Novagen) and used to raise polyclonal antibodies in a rabbit. Western blotting, immunohistochemistry, and confocal microscopy were performed as described by Okano et al. (14) with minor modifications. For Western blotting, a buffer containing 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid) (pH 11.0) and 10% methanol was used. For immunohistochemistry, mock- or BmNPV-infected BmN cells were fixed at the indicated time postinfection (p.i.) with 2% formalin for 10 min. The cells were then blocked with 1% fetal bovine serum-phosphate-buffered saline for 1 h, incubated with preabsorbed anti-BRO-A antibodies (1:200 dilution) for 1 h at room temperature, and treated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (1:1,000 dilution). Stained cells were analyzed with a confocal laser microscope (model TCS-SP equipped with an Ar-Kr laser; Leica, Heidelberg, Germany). For biochemical fractionation, 1.2 × 107 BmN cells were infected with BmNPV at a multiplicity of infection of 10 and harvested at 14 h p.i. The cell pellets were suspended in 1 ml of buffer A (20 mM Tris-HCl, 1 mM CaCl2 [pH 8.0]) plus 0.3 M sucrose and homogenized with a Wheaton Dounce homogenizer with a loose-fitting pestle. Aliquots of the resultant homogenate were used as the whole-cell sample. Homogenates were mixed with equal volumes of buffer A plus 1.8 M sucrose, loaded onto sucrose cushions (1.8 M sucrose), and centrifuged at 50,000 × g (SW60Ti rotor; Beckman) for 1 h. The pellet (containing nuclei) was suspended in 0.3 M sucrose.
Transient-expression assays.
Cells were collected 72 h after transfection and used for chloramphenicol acetyltransferase (CAT) assays. A FAST CAT Green (Deoxy) chloramphenicol acetyltransferase assay kit (Molecular Probes) was used.
RESULTS
Characteristics of BmNPV bro ORFs.
The genome of BmNPV contains five ORFs (ORFs 22, 80, 81, 131, and 132) that showed high homology to AcNPV ORF2 (5). We now refer to these ORFs as bro-a, bro-b, bro-c, bro-d, and bro-e, respectively (Fig. 1A). The BmNPV bro genes (bro-a to bro-e) were predicted to encode proteins of 317, 239, 318, 349, and 241 amino acids (aa), respectively (Fig. 1B). The bro-b and bro-c genes are adjacent to each other in a head-to-tail arrangement in the BmNPV genome. The bro-d and bro-e genes are also in a head-to-tail arrangement, whereas the bro-a gene is located alone (Fig. 1A). Although BmNPV BROs fall into group I of the BRO family, three distinct subgroups within group I were noted (10). Multiple alignment of the amino acid sequences of BROs showed that all the sequences aligned colinearly from their well-conserved N termini and that BRO-B and BRO-E had truncated C termini (Fig. 1B). Comparative alignment showed significant levels of homology between the different ORFs (e.g., 88.3% for BRO-B and BRO-E, 69.3% for BRO-A and BRO-C, and 80.2% for BRO-D and AcNPV ORF2, but 30 to 40% for the other pairs). In addition, the predicted molecular masses of each pair showing high homology are similar. Thus, bro-a and bro-c were predicted to encode proteins of 35.4 and 35.9 kDa, respectively; bro-b and bro-e could encode proteins of 27.5 and 27.8 kDa, respectively; and bro-d and ORF2 of AcNPV were predicted to encode proteins of 40.1 and 37.8 kDa, respectively. Therefore, they could be subdivided into three subgroups by the differences in their C termini, such as bro-d and Ac-bro in subgroup A, bro-a and bro-c in subgroup B, and bro-b and bro-e in subgroup C (Fig. 1B).
Transcriptional analysis.
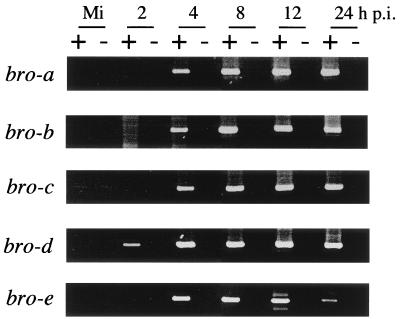
To address whether all the bro genes are expressed, we first performed RT-PCR with total RNAs purified from mock-infected BmN cells or cells at 2, 4, 8, 12, and 24 h p.i. Since their N termini are highly homologous, primers were specific for regions where each bro gene is distinguishable. These experiments showed that bro-d transcription can be detected from 2 h p.i., that transcription of the other bro genes can be detected from 4 h p.i., and that all transcripts persist through 24 h p.i. (Fig. 2).
FIG. 2.
RT-PCR analysis of bro genes. Total RNA was extracted from mock-infected BmN cells (Mi) or cells at 2, 4, 8, 12, and 24 h p.i. RT-PCR was performed with five sets of the specific primers for each bro either with (+) or without (−) reverse transcriptase.
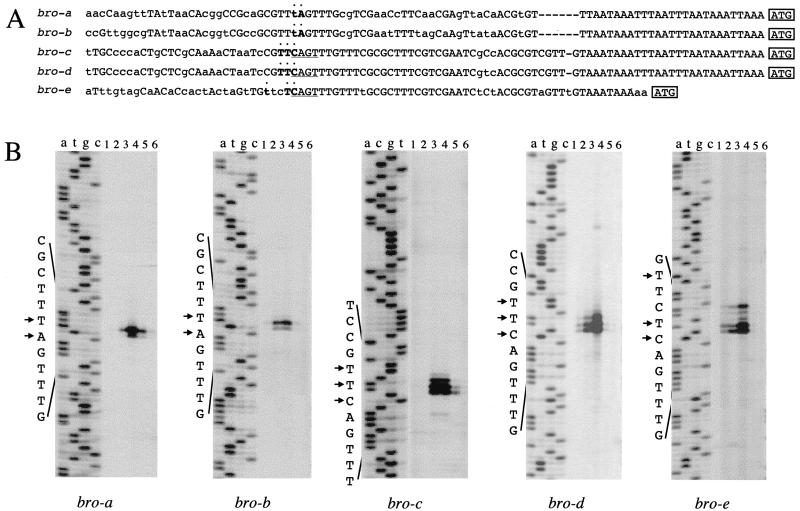
As shown in Fig. 3A, the upstream regions of bro genes are well conserved. Although TATA box-like sequence was not found, a consensus baculovirus early-gene promoter motif (CAGT) was found in the 5′-flanking regions of bro-c, bro-d, and bro-e. This supports the idea that the BmNPV bro genes are expressed in the early stage of infection. To map the initiation sites of bro transcripts, primer extension was performed with total RNA purified from mock- or BmNPV-infected cells at 4, 8, 12, 24, and 48 h p.i. The 5′ ends of the bro-c, bro-d, and bro-e transcripts mapped to the region containing the CAGT motif (Fig. 3). The bro-a and bro-b transcripts initiated at a TAGT motif instead of CAGT. For all the bro genes, the transcription initiation site was found in a similar position relative to the ATG initiation codon. The start sites of bro-a and bro-b transcripts were found 63 nt upstream of the ATG codon, bro-c and bro-d transcripts initiate 68 nt upstream of the ATG codon, and bro-e transcripts start 52 nt upstream from the initiation codon. Transcription of all bro genes reached a maximal level between 8 and 12 h p.i. and decreased quickly. Taken together with the results of the RT-PCR analysis, these results led us to conclude that the bro genes of BmNPV are transcribed actively in the early stage of infection.
FIG. 3.
Transcriptional analysis of bro genes. (A) Comparison of 5′-flanking regions of bro genes. The capital letters indicate three or more identical nucleotides, and the lowercase letters indicate less than three identical nucleotides or nonidentical sequences. The primer extension products are denoted by boldface and dotted sequences, indicating the transcriptional initiation sites. (B) Primer extension mapping of bro transcripts. Total RNA was isolated from BmNPV-infected BmN cells after mock infection or at 4, 8, 12, 24, 48 h p.i. (lanes 1 to 6). The sequencing ladders were generated by using the plasmids shown in Table 1. The small arrows beside the sequence indicate the initiation sites of transcription.
Transcriptional control of bro genes by bacurovirus factors.
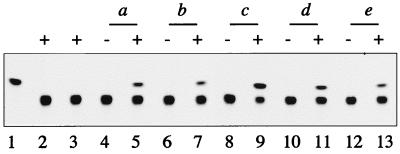
We investigated whether the bro genes could be expressed solely by the host transcription machinery since they are transcribed actively in the early stage of infection. To do this, we constructed reporter plasmids containing the reporter gene cat under the control of each bro gene promoter. These plasmids (pAp-CAT to pEp-CAT) were then cotransfected with or without BmNPV DNA into BmN cells, and CAT was activity assayed. As shown in Fig. 4, CAT activity was observed only when the reporter plasmids were cotransfected with BmNPV DNA. This indicates that activation of each bro promoter is dependent on a viral factor(s) and that all bro genes are delayed-early genes.
FIG. 4.
CAT activity of bro promoter constructs in BmN cells. Reporter plasmids which contain the promoter region of each bro were cotransfected with (+) or without (−) BmNPV DNA into BmN cells. Lane 1 shows a reference standard of the acetylated substrate provided from the manufacturer; lane 2 indicates the activity from the CAT reporter plasmid without a promoter showing no CAT activity in the presence of BmNPV; and lane 3 shows no CAT activity from BmNPV DNA-transfected cells. Cells were collected at 72 h p.i. and used for the CAT assay.
Immunodetection and localization of BRO proteins in infected BmN cells.
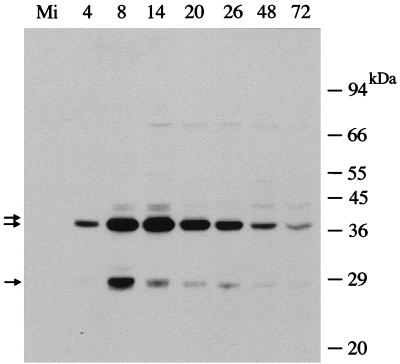
A polyclonal antiserum was raised against His-tagged BRO-A (full-length) expressed in E. coli. The results of Western blot analysis (Fig. 5) suggested that the antibodies recognized all three groups of BmNPV BROs with immunoreactive bands at 28, 38, and 41 kDa, which probably correspond to BRO-B/E, BRO-A/C, and BRO-D, respectively. The expression pattern of the 38-kDa polypeptides was clearly detected at 4 h p.i., increased until 14 h p.i., and then gradually decreased. In contrast, the 28-kDa band reached maximal levels at 8 h p.i. and the levels then decreased quickly. We also found that the 38-kDa polypeptides were associated with budded virus (BV) (data not shown). Even though we detected the production of all three groups of protein, we were not able to distinguish the synthesis of individual BRO proteins because of their similar molecular masses.
FIG. 5.
Western blot analysis of BROs in BmNPV-infected BmN cells. Extracts were prepared from mock- or BmNPV-infected cells collected at the indicated times postinfection. Cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting with anti-BRO antibodies. Arrows show the immunoreactive bands. Size markers (kilodaltons) are indicated are shown on the right.
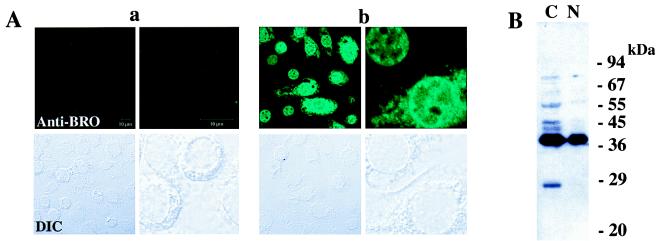
The distribution of BRO proteins in BmNPV-infected BmN cells was then visualized by using anti-BRO antibodies and a confocal laser microscope. Immunoreactive substances were distributed both in the nucleus and cytoplasm (Fig. 6A). To further investigate the distribution of the different BRO polypeptide species, we analyzed total and nuclear subcellular fractions of BmNPV-infected BmN cells by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. As shown in Fig. 6B, the major 38-kDa BRO species was localized both in the cytoplasm and the nucleus whereas the 28-kDa species showed only cytoplasmic localization in BmNPV-infected BmN cells. Several extra protein bands (40 to 68 kDa) were detected in the cellular fraction. These may be ubiquitinated BRO proteins, since the amount of extra proteins increased when the cells were treated with the proteosome inhibitor MG-132 (Calbiochem) (21).
FIG. 6.
Distribution of BRO proteins in BmNPV-infected BmN cells. (A) Intracellular localization of BRO proteins. The top panels show BRO immunofluorescent images, and the bottom panels show differential interface contrast images of the same fields as the top ones. Higher magnifications of specific cells are shown on the right-hand panel in each block. BmN cells were mock infected (a) or infected with BmNPV for 8 h (b) and subjected to immunohistochemistry. (B) Subcellular distribution of BRO proteins. The nuclear fraction (N) and cellular fraction (C) of BmNPV-infected BmN cells (14 h p.i.) were separated and subjected to Western blotting.
Isolation of BmNPV mutants with bro gene deletions.
To determine whether these bro genes are essential for viral growth in BmN cells, we attempted to construct recombinant viruses of BmNPV in which individual bro genes were replaced with a β-galactosidase gene cassette expressed from the Drosophila heat shock promoter. Plasmids containing the β-galactosidase gene inserted into each bro gene (Table 1) were cotransfected with BmNPV DNA into BmN cells. Recombinant viruses were isolated by identification of plaques expressing β-galactosidase. The deletion of each bro gene was confirmed by PCR and Southern hybridization (data not shown). Recombinant viruses lacking bro-a, bro-b, bro-c, or bro-e were successfully isolated. Viral growth curves of these deletion mutants showed that they have the same ability to produce BV as wild-type virus does (data not shown). These results suggest that these genes were not necessary for virus replication in BmN cells. These four mutant viruses were then injected or orally infected into B. mori larvae (fifth instar) to investigate how the deletion of bro-a, bro-b, bro-c, or bro-e affects viral pathology. No detectable differences were observed between the larvae infected with these mutants and those infected with wild-type virus in terms of oral infectivity, time to death, and larval behavior (data not shown).
However, we were not able to obtain recombinant virus lacking bro-d. Several attempts were made to plaque purify this recombinant, but any recombinant virus obtained was always contaminated with wild-type virus. This suggests that bro-d is essential for viral growth in BmN cells. As described above, BmNPV bro genes could be divided into three subgroups according to differences in the C termini and their molecular weights. Therefore, it seemed possible that one member of each group was able to complement the absence of its partner when the other one was disrupted. To investigate this, we attempted to construct a double deletion mutant in which both the bro-a and bro-c genes were deleted. We started with a virus in which the bro-a gene had been replaced with the β-galactosidase gene. We constructed a plasmid with most of bro-a deleted by digestion of HindHeh3 (Table 1) with BstEII and self-ligation, cotransfected this plasmid into BmN cells with a BmNPV mutant carrying the β-galactosidase gene in the bro-a site, and screened for colorless plaques. Deletion of the β-galactosidase and bro-a genes was confirmed by PCR (data not shown). Next, the plasmid used to delete the bro-c gene (Table 1) was cotransfected into BmN cells with BmBroAD to construct a double-deletion mutant. Since the bro-c region would be replaced with the β-galactosidase gene in this plasmid, the final recombinant was selected by the identification of plaques expressing β-galactosidase. Similar to our attempts to isolate a bro-d deletion, we were unable to isolate recombinant virus despite exhaustive efforts. This result supports the possibility that bro-a complements for the absence of bro-c and vice versa and that the presence of at least one member of this bro subgroup is required for virus replication.
DISCUSSION
The bro genes represent the most striking example of multicopy genes with 1 to 16 copies present in different baculovirus genomes identified so far. In BmNPV there are five bro genes representing three subgroups. In this study, we have demonstrated that all five of these genes are transcribed as early genes and that the transcripts of these genes can be detected as early as 4 h p.i. The transcriptional level was maximal between 8 and 12 h p.i. and decreased quickly. The bro-c, bro-d, and bro-e genes have a CAGT early-gene start motif, while bro-a and bro-b have a TAGT sequence instead of CAGT. For all bro genes, transcription is initiated in approximately the same position relative to the ATG codon, and in all cases the start site was mapped to the (C/T)AGT motif. This indicates that TAGT also functions as an early-gene start site for baculovirus transcription. Synthesis of the BRO proteins appeared to correspond to transcription levels. Expression was clearly detected at 4 h p.i. and reached a maximum between at 8 and 14 h p.i. The results of these transcriptional and translational analyses suggest that the BmNPV bro genes are early genes. Interestingly, the AcNPV bro gene (ORF 2) is considered a late gene, since a late-gene promoter motif (ATAAG) is present in its upstream region (3). However, it has not yet been shown that the AcNPV bro gene is expressed in the late stage of infection.
According to Kuzio et al. (10), most bro genes share a related sequence in their N-terminal regions but show differing degrees of similarity in other regions. They proposed that the bro gene family can be separated into four groups (group I to IV) based on the relationship of the variable region and, further, that group I appeared to contain three subgroups of related genes (10). According to this classification, all the bro genes in BmNPV belong to group I. We also suggested that BmNPV bro genes could be divided into three subgroups on the basis of their homology and molecular weight; these are BRO-A and BRO-C, BRO-B and BRO-E, and BRO-D. Sequence comparisons indicated that ORF2 of Spodoptera litura NPV (GenBank accession no. X99073) is closely related to bro-d of BmNPV and that the Leucania separata NPV p20 gene (GenBank accession no. AB009612) is homologous to BmNPV bro-a/c. Although it is not known whether these two NPVs contain multiple copies of the bro gene, since the whole genome sequence is not available, it is clear that at least one copy of the bro family is present in every baculovirus genome analyzed so far.
Other viruses have been reported to contain multigene families in their genome. In particular, the genome sequence of Melanoplus sanguinipes entomopoxvirus (MsEPV) revealed the presence of five multigene families (1). Afonso et al. (1) suggested that one of the multigene families (the ALI motif family) of MsEPV shows homology to AcNPV ORF2 (bro) and putative genes from Chilo iridescent virus (GenBank accession no. AF003534); bacteriophages BK5-T (GenBank accession no. L44593), N15 (GenBank accession no. AF064539), and A2 (GenBank accession no. Y12813); and the bacterium Haemophilus influenzae (GenBank accession no. U32821). Therefore, we searched for homology between these genes and BmNPV bro by a BLAST search. We could find no significant similarity between the MsEPV ALI family and the BmNPV bro family. However, there was slight homology with long gaps between the bro genes and putative genes of bacteriophages BK5-T, r1t, and A2 and ORF 011L of Chilo iridescent virus (data not shown). Therefore, we do not think that the BmNPV bro genes are homologues of the MsEPV ALI motif family. There are other examples of multigene families in other viruses including African swine fever virus (15, 17, 20), molluscum contagiosum virus (19), and human herpesvirus (7). These viruses all contain linear double-stranded DNA genomes. In these viruses, multiple genes clearly show identity within each viral genome, but do not show similarity in comparisons of different viral genomes. The multigene families from African swine fever virus (ASFV) are well known, and there are at least six multigene families in the terminal variable regions of the ASFV genome. Most of them appear to be transcribed during the infection (20). The roles of these multigenes in ASFV infection have not been identified yet. Based on the experiments involving deletion within these terminal regions, Pires et al. suggested that multigene family genes may not be essential for viral growth in cell culture and may be involved in viral host range (15). Afonso et al. also suggested that MsEPV gene families might perform host range functions (1).
It is particularly important to understand the functional significance of the bro family of BmNPV since these genes are heavily expressed in the early stage of infection. The expressions of bro genes required a viral factor(s); therefore, bro genes are delayed-early genes. We have not identified yet which baculovirus factor(s) is required for the expression of bro genes. Many delayed-early genes encode proteins required for viral DNA replication and late-gene expression, and expression of these proteins sets the stage for late-gene expression. However, BRO proteins are listed as being encoded by genes involved in neither DNA replication nor late-gene expression (9, 16). Although the functional role(s) for BmNPV bro genes remains elusive, it is noteworthy that bro-d was found to be essential for viral growth in BmN cells. Furthermore, we provided evidence for potential complementation between bro-a and bro-c. In addition, we found that bro gene products appear to be segregated based on size within the infected cell, with those of 38 kDa localizing to the cytoplasm and nucleus and those of 28 kDa being found only in the cytoplasm. These results may suggest that each subgroup of BmNPV bro genes possesses a different functional role. It is not surprising that members of the same gene family have different functions. Therefore, we think that bro genes of baculovirus may be involved in some other important viral function rather than in host range.
ACKNOWLEDGMENTS
We are grateful to S. Gomi, K. Majima, and T Ohkawa for helping with the isolation of deletion mutants and with the bioassay M. Kurihara and S. Atsuzawa for providing BmN cells and B. mori larvae; and N. Imai for constructing plasmids. We also thank T. Hayakawa for providing the XcGV sequence prior to its publication and G. F. Rohrmann and D. R. O’Reilly for critical reading of the manuscript.
This work was supported by a CREST award from Japan Science and Technology Corporation (S.M.). This work was also supported, in part, by grants from the COE (Center of Excellence) program, Biodesign Research Program of the Science and Technology Agency (S.M. and W.K), and an STA fellowship (E.Z.).
Footnotes
This paper is dedicated to the memory of Susumu Maeda.
REFERENCES
- 1.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrens C H, Russell R L, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 3.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 4.Durantel D, Croizier G, Ravallec M, Lopez-Ferber M. Temporal expression of the AcMNPV lef-4 gene and subcellular localization of the protein. Virology. 1998;241:276–284. doi: 10.1006/viro.1997.8971. [DOI] [PubMed] [Google Scholar]
- 5.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 6.Gomi S, Zhou C E, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230:35–47. doi: 10.1006/viro.1997.8457. [DOI] [PubMed] [Google Scholar]
- 7.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 8.Hayakawa, T., R. Ko, K. Okano, S.-i. Seong, C. Goto, and S. Maeda. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology, in press. [DOI] [PubMed]
- 9.Kool M, Ahrens C H, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 11.Maeda S. Gene transfer vectors of a baculovirus, Bombyx mori, and their use for expression of foreign genes in insect cells. In: Mitsuhashi J, editor. Invertebrate cell system applications. I. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 167–181. [Google Scholar]
- 12.Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Sato Y, Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature. 1985;315:592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- 13.Maeda S, Majima K. Molecular cloning and physical mapping of the genome of Bombyx mori nuclear polyhedrosis virus. J Gen Virol. 1990;71:1851–1855. doi: 10.1099/0022-1317-71-8-1851. [DOI] [PubMed] [Google Scholar]
- 14.Okano K, Mikhailov V S, Maeda S. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J Virol. 1999;73:110–119. doi: 10.1128/jvi.73.1.110-119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pires S, Ribeiro G, Costa J V. Sequence and organization of the left multigene family 110 region of the vero-adapted L60V strain of African swine fever virus. Virus Genes. 1997;15:271–274. doi: 10.1023/a:1007992806818. [DOI] [PubMed] [Google Scholar]
- 16.Rapp J C, Wilson J A, Miller L K. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol. 1998;72:10197–10206. doi: 10.1128/jvi.72.12.10197-10206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez J M, Yanez R J, Pan R, Rodriguez J F, Salas M L, Vinuela E. Multigene families in African swine fever virus: family 505. J Virol. 1994;68:2746–2751. doi: 10.1128/jvi.68.4.2746-2751.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 20.Yozawa T, Kutish G F, Afonso C L, Lu Z, Rock D L. Two novel multigene families, 530 and 300, in the terminal variable regions of African swine fever virus genome. Virology. 1994;202:997–1002. doi: 10.1006/viro.1994.1426. [DOI] [PubMed] [Google Scholar]
- 21.Zemskov, E., W. Kang, and S. Maeda. 1999. Unpublished data.