Abstract
Most people living with HIV-1 experience rapid viral rebound once antiretroviral therapy is interrupted; however, a small fraction remain in viral remission for an extended duration. Understanding the factors that determine whether viral rebound is likely after treatment interruption can enable the development of optimal treatment regimens and therapeutic interventions to potentially achieve a functional cure for HIV-1. We built upon the theoretical framework proposed by Conway and Perelson to construct dynamic models of virus-immune interactions to study factors that influence viral rebound dynamics. We evaluated these models using viral load data from 24 individuals following antiretroviral therapy interruption. The best-performing model accurately captures the heterogeneity of viral dynamics and highlights the importance of the effector cell expansion rate. Our results show that post-treatment controllers and non-controllers can be distinguished based on the effector cell expansion rate in our models. Furthermore, these results demonstrate the potential of using dynamic models incorporating an effector cell response to understand early viral rebound dynamics post-antiretroviral therapy interruption.
Author summary
Most people living with HIV-1 experience rapid viral rebound once antiretroviral therapy is interrupted; however, a small fraction remain in viral remission for an extended duration. The factors that determine viral rebound dynamics after treatment interruption are not well understood. In this study, we built upon a previous theoretical framework to construct dynamic models of virus-immune interactions to study factors that influence viral rebound dynamics. We evaluated these models using viral load data from 24 individuals following antiretroviral therapy interruption. The best-performing model accurately captures the heterogeneity of viral dynamics and robustly shows that people who remain in viral remission for an extended duration after treatment interruption have a higher effector cell expansion rate compared to those who rapidly rebound. These results demonstrate the potential of using viral dynamic models incorporating an effector cell response to understand early viral rebound dynamics post-antiretroviral therapy interruption.
Introduction
Antiretroviral therapy (ART) is successful at suppressing human immunodeficiency virus-1 (HIV-1) below the limit of detection in people with HIV-1 (PWH). However, ART does not completely eliminate HIV-1 due to the presence of latently infected cells, which have low or no HIV-1 gene expression, making it difficult for the immune system to recognize and eliminate them [1]. While continual use of ART robustly controls HIV-1, there are a number of issues with long-term ART, ranging from how well PWH adhere to treatment [2–5] to the potential side effects of taking ART long term [6], such as chronic inflammation and HIV-associated neurocognitive disorders [7–10]. Thus, durable ART-free virological control remains a major goal.
Due to the persistence of the latent reservoir [11–18], if ART is interrupted, latently infected cells that reactivate can lead to viral rebound. In fact, following analytical treatment interruption (ATI) of ART, most persons with HIV-1, including those with an extremely small latent reservoir, experience viral rebound typically within weeks [19–23]. But in rare cases, some individuals maintained low viral loads for an extended duration of months to years [16,22,24,25]. Individuals, who experience rapid viral rebound, are generally referred to as non-controllers (NC), while those who control are termed post-treatment controllers (PTC). The generation of post-treatment control appears to be affected by external factors such as the timing of ART initiation [17,22,25–28], and in human and nonhuman primate models by viral suppression mediated by CD8+ cells [29–34]. Thus, understanding the factors associated with PTC can suggest a clinical path towards durable control in PWH.
Several models have been proposed to explain the mechanisms behind viral rebound following ATI. The studies by Hill et al. [35,36], Pinkevych et al. [37,38], Fennessey et al. [39], and van Dorp et al. [40] hypothesized rebound as a stochastic process due to the reactivation of latently infected cells that release virus and initiate a chain of other successful infection events. Yet, in PTC, viral load is kept at a low level despite a large reservoir size in some people. This suggests that other factors, such as the immune response, also play important roles in determining rebound dynamics of HIV-1 [41–48]. For this reason, Conway and Perelson [49] proposed a model of viral rebound considering both the latent reservoir and immune response dynamics. Their model demonstrated the combined impact of the immune response and the size of the latent reservoir on HIV-1 dynamics post-ATI and lays the foundation for this study.
Some modeling studies have also estimated the distribution of time to rebound. For instance, Conway et al. [50,51] utilized personal biomarkers including HIV-1 reservoir quantification and cell associated HIV-1 RNA to look at the distribution of time to rebound and fit a model to viral rebound data from a collection of ACTG (Advancing Clinical Therapeutics Globally for HIV/AIDS, formerly known as the AIDS Clinical Trials Group) ATI studies [16]. Others studied mathematical and statistical models to fit rebound data of HIV-1 and simian immunodeficiency virus (SIV) [50,52–54], including studies in which additional treatments with monoclonal antibodies or immune stimulants were given prior to ATI [31,55,56]. While these attempts provide valuable insights into the rebound dynamics post-ATI, they either did not aim to establish the biological factors that distinguish PTC and NC or did not fit to individual viral rebound data. In this work, our objective is to examine whether mechanistic models with virus-immune interactions can accurately capture the heterogeneity in viral rebound dynamics and identify potential mechanisms that distinguish the outcomes post-ATI.
Methods
Ethics statement
This research was approved by the Los Alamos National Laboratory Human Subjects Research Review Board (HSRRB). Patients’ consent was not required because this is a retrospective study using data from clinical trials reported by Sharaf et al. [57].
Data
We analyze the data of 24 PWH (9 PTC and 15 NC) from Sharaf et al. [57] for whom we have longitudinal viral load data. The PTC participants were identified from several clinical studies in the ACTG [58–62]. In Sharaf et al., PTCs were defined as individuals who maintained a viral load of less than 400 HIV-1 RNA copies/mL for at least 24 weeks post ATI and where short-term increases (≤ 2 viral load measurements) of over 400 HIV-1 RNA copies/mL were not exclusionary. Non-controllers were defined as individuals who did not meet the PTC definition. We chose to analyze the data between 0- and 52-weeks post ATI to focus on the period during which the viral load data is often used to classify PTC and NC in clinical settings. Note that when non-controllers rebounded, they were often put back on ART, which terminated their data sets.
Mathematical models
Conway and Perelson [49] introduced the following model of viral dynamics to study the phenomenon of PTC:
| (1) |
This model (Eq 1) assumes that target cells (T) are produced at constant rate λT and die at per capita rate dT, respectively. Virus (V) infects target cells with rate constant β and is cleared at per capita rate c. The infection leads to productively infected cells (I) at a rate 1-fL βVT, where fL and (1-fL) are the fractions of infections that lead to latently and productively infected cells, respectively. Infected cells release viruses at per capita rate p and die at per capita rate δ by viral cytopathic effects. Latently infected cells (L) proliferate and decay at per capita rates ρ and dL, respectively, and can reactivate at per capita rate a to become productively infected cells. In the absence of viral infection, effector cells (E), such as CD8+ T-cells and natural killer (NK) cells, are produced at rate λE and die at per capita rate dE. During viral infection, the effector cells recognize infected cells, which stimulates its population to expand. Conway and Perelson modeled this process using the infected-cell dependent growth term , where b is the maximal growth rate and KB is the density of infected cells required for the effector cell growth rate to be at its half-maximal rate. Note if KB is very large, then a large number of infected cells will be needed to activate the expansion of effector cells, and this will lead to a less effective effector cell response.
The effector cells kill productively infected cells with rate constant m, or more precisely, a second order reaction rate constant m, and can become exhausted at maximal rate d. Effector exhaustion is induced by the upregulation of inhibitory receptors during effector expansion that ultimately abrogate the expansion and other functional responses. Chronic infections and cancers typically lead to effector cell exhaustion, which occurs through metabolic and epigenetic reprogramming of the effector cells that incrementally make them worse at proliferating, surviving, and performing effector functions, such as killing and releasing diffusible molecules [63–65]. Conway and Perelson modeled the rate of exhaustion with a similar infected-cell dependent functional response term . These functional response terms had previously been suggested by Bonhoeffer et al. [66].
The model formulation assumes a fraction, fL, of infections lead to the production of latently infected cells. The homeostatic proliferation of latently infected cells at per capita rate ρ follows from prior work [67–70] and is supported by experimental evidence showing that latent cells can proliferate without activating (or expressing viral signals) [18,71–74].
A Simplified Model. We formulated variations of the Conway and Perelson model by altering the model assumptions on the dynamics of latent cells and the effector cells (Section A in S1 Text). In particular, without effector cell exhaustion, we have Simplified Model 1:
| (2) |
For further simplification, we assume that the number of viruses is in quasi-steady state with the number of infected cells in all models including the Conway and Perelson model, so that , as in prior work [75,76]. The initial time (t = 0) is when the individuals are taken off ART. We let , which are approximations of the steady states during ART assuming a small latent reservoir at the time of ATI, that ART was highly effective and suppressed all new infections, and thus that productively infected cells were only generated via activation of latently infected cells. We also studied other models given in Section A in S1 Text. In models without explicit latently infected cell dynamics, we assume the reservoir size does not vary significantly during the first year post-ATI or during the off-ART duration for NCs, the time period we study. This implies L(t) = L(0). Whether the viral load rebounds or not post-ATI is determined by both the immune response and the latent reservoir size. The influence of the reservoir size at the time of ATI has been studied before [36,49], thus we choose to examine whether the immune response—without variation in the latent reservoir size—can capture the viral rebound dynamics for this cohort of participants.
The parameters that we fit, and the fixed parameters are given in Table 1. fL is fixed to 10−6, meaning only 0.0001% of infections result in latent cell production [77]; however, increasing fL up to 10-fold, has little effect on the model dynamics. The assumed size of the latent reservoir was chosen as 1 cell/ml based on estimates of the number of replication competent latently infected cells per million resting CD4 T cells determined by the quantitative viral outgrowth assay [78,79] and assuming patients on long-term ART have close to a million CD4+ T cells per ml. The net rate of change of the latent cell population dL − a + ρ is such that the expected half-life (t1/2) of the reservoir is 44 months [14,15]. We set dL = 0.004 per day [68], and a = 0.001 [49], which gives ρ ≈ 0.0045 per day. We remark that the stability of the latent reservoir is due to the combined effect of proliferation, death, and activation; however, these parameter values are only educated guesses. Furthermore, recent data has suggested that due to proliferation, the reservoir might stop decaying [18].
Table 1. Model parameters.
The double dashed line separates parameters that we fit (above) and fixed (below). References for the fitting parameters are provided for comparison purposes.
| Parameter | Meaning | Unit | |
|---|---|---|---|
| β | Infection rate | mL (HIV RNA copies)-1 day-1 | [80,81] |
| λ E | Production rate of effector cells | cells mL-1 day-1 | [49] |
| m | Effector cell killing rate constant | mL cells-1 day-1 | [49] |
| b | Maximum effector cell growth rate | day -1 | [82] |
| K B | Density of infected cells needed to reach a half-maximal effector cell growth rate | cells mL -1 | [66] |
| p | Viral production rate | (HIV RNA copies) day-1 | [49] |
| d | Maximum effector cell exhaustion rate | day -1 | [83] |
| K D | Density of infected cells needed to reach a half-maximal effector cell exhaustion rate | cells mL -1 | [66] |
| λ T | Production rate of target cells | 104 cells mL-1 day-1 | [84] |
| d T | Death rate of target cells | 0.01 day-1 | [68] |
| f L | Fraction of latently infected cells produced per infection | 10−6 | [49] |
| a | Reactivation rate of latently infected cells | 0.001 day-1 | [49] |
| δ | Death rate of infected cells | 1 day-1 | [85] |
| d E | Death rate of effector cells | 2 day-1 | [83] |
| L 0 | Steady state size of the latent reservoir | 1 cell mL-1 | [78,79] |
| d L | Decay rate of the latent reservoir | 0.004 day-1 | [14,68] |
| ρ | Latently infected cell proliferation rate, | 0.0045 day-1 | [49,86] |
| c | Clearance rate of virus | 23 day-1 | [87] |
Data fitting
We used a nonlinear mixed effects modeling approach (software Monolix 2023R1, Lixoft, SA, Antony, France) to fit Simplified Model 1 and other variants presented in Section A in S1 Text to the viral load data for all individuals simultaneously. We applied left censoring to data points under the limit of detection.
We assumed the fitting parameters follow a logit-normal distribution as we constrained the parameters to be in a fixed range. λE, b, m, and d were constrained between 0 and 10 times the reference values used in Conway and Perelson (Table 1). Imposing bounds on λE, b, m, and d is not necessary; however, this allows us to get the most consistent results across all models tested. We fit β and constrained its range between 10−7.5 and 10−12 mL (HIV RNA copies)-1 day-1 to avoid an unrealistically high viral surge within 24 hours post-ATI due to high values of β. Parameter p was constrained between 0 and 5000 per day. Parameters KB and KD were constrained between 0 and 100,000 cells per mL. Without this upper bound, KD values often tended to infinity. Note that although KD does not appear in Simplified Model 1, it does appear in other model variations that we examined (see Section A in S1 Text). No covariate was used during the initial fitting and comparison of all models. A covariate based on whether a participant is classified as PTC or NC was used later with the best fit model to determine the factors that can distinguish these two groups. Model comparison was done using the corrected Bayesian Information Criterion (BICc) [88] as reported by Monolix.
Result
Model fit and comparison
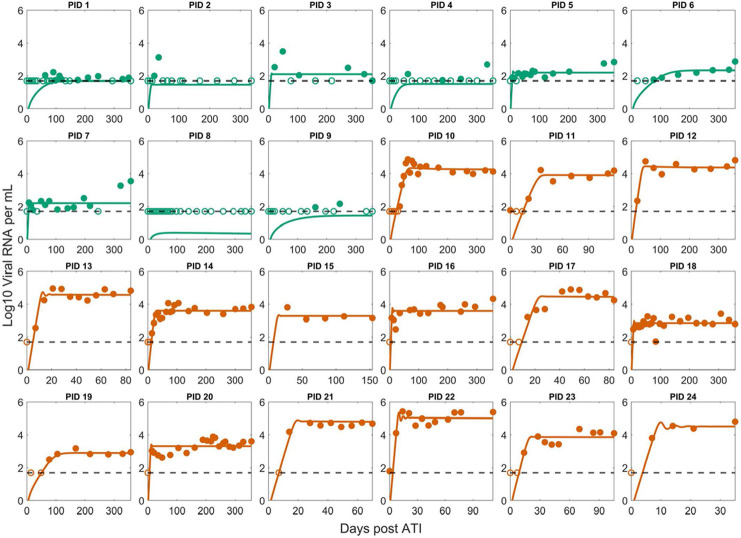
We fit the Conway and Perelson model and variations of it with different assumptions on the dynamics of latent and effector cells (see Section A in S1 Text). The best fit model was Simplified Model 1 described in Eq (2), where effector cell exhaustion is disregarded. Its best-fit to the data is shown in Fig 1. Comparisons of best fits for other models along with population and individual parameters are shown in Figs A-F and Tables A-M in S1 Text. The only difference between Simplified Model 1 and the Conway & Perelson model is the lack of an exhaustion term, which suggests exhaustion is not necessary to explain early viral rebound dynamics (within the first 52 weeks) seen in this set of PWH. This is further supported by the negligible effect of exhaustion estimated from the best fits for the Conway & Perelson model and Simplified Model 3, where the values for d and KD result in a much smaller exhaustion effect compared to the reference values (Tables B and D in S1 Text). This does not imply that effector exhaustion is not relevant to HIV rebound dynamics as it is potentially important to explain the eventual loss of control as occurred in the subset of PTCs who have a truncated x-axis in Fig 1 and in the CHAMP study [25]. In addition, it is an important mechanism to explain the phenomenon where a PTC can have a high viral set point pre-ART and a lower viral set point post-ATI [49], and a variety of other dynamical phenomena [56,64,89].
Fig 1. Best fit of Simplified Model 1 to the post-ATI data from Sharaf et al. [57].
Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Note the x-axis scale varies for the NCs as many of them were put back on ART terminating the data set.
Interestingly, the best fit parameter values for Simplified Model 1 with the exception of KB are close to the parameter values used in Conway and Perelson (compare Tables 1 and 2). This is also true for Simplified Model 1 even when we do not impose bounds on λE, b, and m. All models with effector cell dynamics fit the data relatively well (Table A in S1 Text), which is indicative of the importance of including in the model the dynamics of a population of effector cells that can kill productively infected cells.
Table 2. Best fit population parameters (Simplified Model 1) vs. reference values from Conway and Perelson [49].
| Parameter | Best fit values (population estimate) | Reference values |
|---|---|---|
| β | 1.51×10–8 mL day-1 | 1.5×10–8 mL day-1 |
| λ E | 1.45 mL-1 day-1 | 1 mL-1 day-1 |
| m | 0.83 mL day-1 | 0.42 mL day-1 |
| b | 4.68 day-1 | 1 day-1 |
| K B | 52.62 mL-1 | 0.1 mL-1 |
| p | 3477 day-1 | 2000 day-1 |
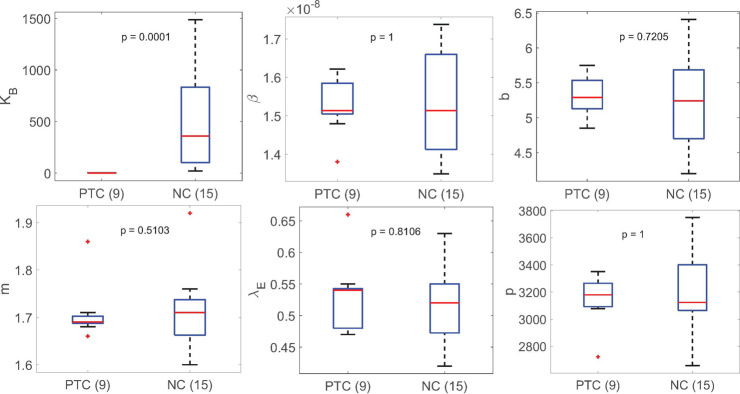
We stratified the best fit parameters (Simplified Model 1) for each person by whether they were PTC or NC (Fig H in S1 Text). The most statistically significant difference using the Mann-Whitney test between the PTC and NC group is the parameter KB (p-value: 0.0005), followed by the immune cell killing rate m (p-value: 0.0169). All model parameters differ at least somewhat between these two groups; however, the degree of significance of the difference for each parameter varies among models. In contrast, the statistically significant difference of KB between PTC and NC is consistently observed across all models tested (Figs G-J in S1 Text).
This observation prompted us to fit Simplified Model 1 using a covariate on KB between PTC and NC. This modification significantly improves the model by 26 BICc points (Fig B and Table A in S1 Text); however, further inclusion of other covariates improves the fit slightly but worsens the BICc score (Table A in S1 Text). The best fit population value of KB for PTC 1.19 cell mL-1 is consistent with the value in Conway and Perelson [49], which indicates a fast expansion of effector cells. The best fit population value of KB for NC is 230-fold higher, indicating a slower expansion of effector cells (Table 3).
Table 3. Best fit population parameters (Simplified Model 1 with a covariate on KB) vs. reference values from Conway and Perelson [49].
| Parameter | Best fit values (population estimate) | Reference values |
|---|---|---|
| β | 1.58×10−8 mL (HIV RNA copies)-1 day-1 |
1.5×10−8 mL (HIV RNA copies)-1 day-1 |
| λ E | 0.50 cells mL-1 day-1 | 1 cells mL-1 day-1 |
| m | 1.72 mL cell-1 day-1 | 0.42 mL cell-1 day-1 |
| b | 5.40 day-1 | 1 day-1 |
| KB(PTC) | 1.19 cells mL-1 | 0.1 cells mL-1 |
| KB(NC) | 277 cells mL-1 | — |
| p | 3220 (HIV RNA copies) day-1 | 2000 (HIV RNA copies) day-1 |
Stratification of the best fit parameter values for Simplified Model 1 with a covariate on KB shows that the inclusion of a covariate on KB is sufficient to explain the variation between PTC and NC (Fig 2). Indeed, comparing the stratified individual parameters estimated using Simplified Model 1 without a covariate (Fig H in S1 Text) and with a covariate on KB (Fig 2), we find that the statistically significant difference between the PTC and NC observed for all other model parameters vanishes. For completeness, we also tested whether having a covariate on other parameters can improve the fit and remove the statistically significant variation in all other parameters including KB. We find that adding a covariate on other parameters (one by one) not only does not improve the fitting compared to the fit with a covariate on KB (Table A in S1 Text), but it also has no effect on the statistically significant difference in KB between PTC and NC. To see whether the model can fit the data well without accounting for individual variation in KB, we tested Simplified Model 1 without random effect for KB, which results in worse fits (Table A in S1 Text). Altogether, these results support the inclusion of a covariate on KB as well as including individual variation in KB.
Fig 2. Summary of best fit parameters in Simplified Model 1 with a covariate on KB stratified based on PTC or NC.
The median value of KB for PTC is 0.94 cell mL-1. The difference in the values of KB is the most significant between PTC and NC. All other parameters are not statistically different between PTC and NC.
Analytical approximation of the rebound dynamics
To better explain why KB separates the NC and PTC participants, we approximated analytically the viral set point Vss after rebound (see Section E in S1 Text) and found
| (3) |
Comparing Vss with the predicted viral load set-point obtained with the full model dynamics (Fig K in S1 Text) demonstrates the accuracy of this approximation.
The clinical classification of PTC individuals by Sharaf et al. is centered on the viral set point and how long it takes to rebound above the threshold of 400 viral RNA copies/mL (i.e., 24 weeks or longer of viral load below 400 copies/mL) [57]. The viral set point approximation Vss shows that the rebound classification is influenced by many factors in highly nonlinear ways. Yet, Vss is simply proportional to KB, which may explain why the estimated value of KB for each individual separates PTC and NC in the model fits (Table H in S1 Text).
Distinct immune response between PTC and NC
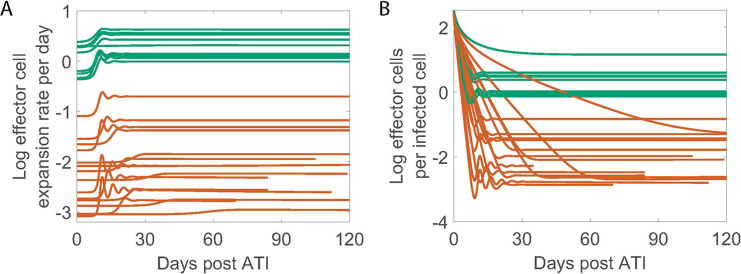
We calculated the effector cell expansion rate per infected cell and the ratio of effector cells to infected cells (E/I) using the best fit parameters for each individual given in Table I in S1 Text. There is a clear distinction in both quantities between PTC and NC (Fig 3). The effector cell expansion rate for individuals in the PTC group is several times higher than for individuals in the NC group (Fig 3A). This result is not surprising given that KB is the only statistically significant difference between the two groups, where higher KB for the NC group leads to a slower expansion rate of the effector cells. Additionally, in PTC individuals, the model predicts at least one effector cell per infected cell throughout the studied duration, whereas in NC individuals, the model predicts about one effector cell for hundreds of infected cells on average (Fig 3B).
Fig 3. Model predicted viral and immune dynamics using Simplified Model 1 with a covariate on KB.
Green is PTC and dark orange is NC. (A) The effector cell expansion rate () on a log10 scale. (B) The ratio of E vs. I plotted on a log10 scale.
Discussion
Differences in the size of the latent reservoir at the time of ART interruption and virus-immune interactions have been recognized as key factors in achieving post-treatment control [49]. Here, our modeling results robustly demonstrate that differences in the expansion rate of the effector cells differentiate PTC and NC in this cohort of 24 PWH.
The size of the latent reservoir determines the overall rate of reactivation of latent cells, which can lead to HIV-1 rebound post-ATI, as pointed out by Hill et al. [35]. However, to do so requires the reactivated cells to start a chain of infections that leads to exponential growth. If the immune system detects these newly reactivated cells and reacts fast enough and strongly enough, it can interrupt this chain of infection events and lead to PTC rather than non-control. Thus, in addition to the size of the reservoir, which Sharaf et al. [57] found to be 7-fold lower in PTCs than in the NCs, other factors, such as virus-immune interactions, may also affect the viral dynamics post-ATI. Esmaeilzadeh et al. [47] showed that autologous neutralizing antibodies apply selection pressure on rebounding variants and may play a role in mediating HIV suppression after ATI. Conway and Perelson [49] suggested that when the immune response is strong, PTC can be achieved. Our modeling results illustrate this concept by accurately capturing the early viral rebound dynamics in this cohort of 24 PWH.
We found that KB the density of infected cells needed to stimulate effectors cells into growth at their half-maximal rate, is significantly lower in PTC compared to NC. Since the clinical definition of PTC depends on the post-ATI set point viral load, and we showed there is an approximately linear relationship between this viral set point and KB in our model (Eq 3), this explains the distinct estimates of KB for PTCs and NCs. These results established an analytical relationship between an early and suppressive immune response and the viral rebound dynamics, echoing the recent results in the SIV system by Vemparala et al. [90]. In practice, it is unlikely that all model parameters including KB for an individual can be estimated prior to an ATI study. However, if an estimate of the value of KB can be obtained, perhaps via correlation with individual’s immunological markers, then it may be possible to determine a threshold value of KB that separates PTCs and NCs a priori.
A plausible biological interpretation of the variation in KB comes from the studies of T cell expansion after contact with an antigen presenting cell (APC) [91,92]. Derivation of KB based on the binding kinetics of a T cell to an APC results in , where kb, kd and kp are the rate constants for T cells binding, dissociation and first order activation/proliferation, respectively. This means that a higher binding affinity, i.e., higher kb and/or lower kd, would result in a lower KB, which gives the interpretation that effector cells in PTC individuals have a higher binding affinity to the antigens presented on the surface of HIV-1 infected cells than the effector cells in NC individuals. This implies that the observed variation in KB may relate to different class I HLA alleles that present the HIV-1 peptides to CD8+ T cells carried by PTC and NC [93–98]. Alternatively, KB could also vary with the effector cell proliferation rate and cytotoxicity. For example, antigenic stimulation in vitro and ex vivo experiments have shown that CD8+ T cells from controllers can proliferate and increase their cytolytic capability more efficiently than those from progressors [99–101]. The effector cell population also includes other cell types, such as NK cells, and there is some evidence that NK cells may play an earlier role than CD8+ T-cells in the control of HIV-1 [42,48]. However, these interpretations remain speculations as KB is modeled as the number of infected cells needed to stimulate robust effector cell expansion, so the precise mechanism associated with KB is difficult to infer without a more detailed immunological model and data. Delineating the exact biological mechanisms that contribute to the differentiating values of KB, perhaps via ex-vivo stimulation, characterization, and TCR-sequencing of CD8+ effector cells in addition to viral sequencing from individuals participating in ATI trials, may aid in finding the right targets for an effective design of vaccines and treatments for HIV-1.
Our results demonstrate that a variation of the Conway-Perelson model without effector cell exhaustion can characterize the viral rebound dynamics in the cohort of 24 individuals from Sharaf et al. [57]. However, there are limitations to the current study that should be noted for future investigation. First, although there is considerable heterogeneity in viral rebound dynamics in the 24 studied participants, this data set is still limited and may not be representative of the broader population of PWH. For instance, the CHAMP study [25] contains individuals with more complex viral rebound dynamics and sometimes a very high initial viral peak before viral remission, which our model fits miss. In addition, since we employed a population fitting approach to fit a data set with mostly non-oscillatory individual data, the best fit model fails to capture the oscillatory features exhibited by several individuals (13, 20, 22, and 23 –Figs 1 and B in S1 Text). This suggests further examination of our modeling framework using data with more heterogeneous viral rebound dynamics is necessary to generalize our results.
Second, while our parameter estimates indicate limited exhaustion of effector cells (Tables B and D in S1 Text), this could be due to several factors such as the duration of ART and the one-year time constraint we put on the data. ART has been shown to restore the functionality of HIV-1 specific CD8+ T-cells and NK cells [102,103], hence lowering the exhaustion level of effector cells. Therefore, exhaustion at the start of ATI may potentially be very limited and unobservable to our model within the one-year time constraint. This limitation is evident when considers participants such as PTC number 7, who showed signs of viral rebound (~2 log increase in viral load) toward the end of year one, which is not captured by the best fit models. Thus, it is likely that effector cell exhaustion plays a role in PTCs who rebound later. Future studies should look further into the interaction of the immune system and virus over longer time scales to examine the possibility of rebound after the initial short-term viral remission. It is also worth pointing out that Simplified Model 2, which does not consider the dynamics of latently infected cells, can fit the data almost as well as Simplified Model 1. This is also likely due to the maximum one-year time constraint in the data and the slow dynamics of the latent reservoir. When considering a time scale on the order of years, some of the assumptions we made in order to derive the approximation of the viral set point are unlikely to hold such as that target cells remain at their disease-free equilibrium value of λ/dT−see the derivation in S5. Thus, this necessitates more intricate mathematical analyses to ascertain the impact of immune parameters, such as KB, on the viral set point [104].
A third ‐ and intriguing–observation, which we did not consider, is that in some PWH, the viral set point post-ATI is orders of magnitude lower than before ART [22]. Qualitatively, this relates to the bistability of the Conway-Perelson model with respect to different initial conditions such as the size of the latent reservoir and the immune response at the time of ATI. If ART can sufficiently reduce the viral burden, it may allow the immune response to catch up. This may happen via the restoration of functionality for HIV-1 specific CD8+ T-cells and NK cells during ART [102,103]; however, the restoration effect may not be sufficient to improve the immune response in NCs to the level of PTCs in most cases [105].
Lastly, by choosing to focus on estimating the immune response parameters in our study, we assumed that the initial conditions of the model for all 24 individuals at the time of ATI were the same. While this assumption is not ideal, when fitting both the initial conditions and the immune parameters, the resulting parameter estimates vary significantly across all participants and models, likely due to parameter identifiability issues, which makes determining the key parameters an unrealistic task. Nevertheless, even with these limitations, our modeling results robustly demonstrate that variation in the effector cell expansion rate sufficiently captures the heterogeneity in viral rebound dynamics post-ATI.
Supporting information
Table A. Model fit comparison (Monolix 2023R1, Lixoft, SA, Antony, France). There are four categories separated by double-lines. Bolded values are the lowest values in the category. The first category contains the Conway and Perelson model with five variations without covariates. The second category tests single covariates for the best fit model in the first category. The third category shows two examples of using two covariates that produce the lowest fitting error. The last category tests the Simplified Model 1 without random effect for KB. Without a random effect, the population estimate for KB is 16.88 (S.E. 0.12)cells mL-1. Table B. Best-fit population parameters for the Conway & Perelson Model vs. reference values from Conway and Perelson [49]. Note the estimated values of KD and d result in a negligible effect of exhaustion. Table C. Best-fit population parameters for the Simplified Model 2 vs. reference values from Conway and Perelson [49]. Table D. Best-fit population parameters for the Simplified Model 3 vs. reference values from Conway and Perelson [49]. Note that the estimated values of KD and d together imply the exhaustion effect is very small, which may be because of this particular set of participants or that it cannot be observed from the limited data. Table E. Best-fit population parameters for the Simplified Model 4 vs. reference values from Conway and Perelson [49]. Note that m* is not the same as m. Table F. Best-fit population parameters for the Simplified Model 5 vs. reference values from Conway and Perelson [49]. Note that m* is not the same as m. Table G. Individual best-fit parameters–Conway & Perelson model. Table H. Individual best-fit parameters–Simplified Model 1. Table I. Individual best-fit parameters–Simplified Model 1 with a covariate on KB (1 is PTC and 2 is NC). For KB, the mean and SD (%) are reported for individual group PTC/NC. Table J. Individual best-fit parameters–Simplified Model 2. Table K. Individual best-fit parameters–Simplified Model 3. Table L. Individual best-fit parameters–Simplified Model 4. Table M. Individual best-fit parameters–Simplified Model 5. Fig A. Best fit of the Conway & Perelson model to the post-ATI data from Sharaf et al. [57]. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Fig B. Best fit of Simplified Model 1 with a covariate on KB to the post-ATI data from Sharaf et al. [57]. Lines show the model predictions using the best fit parameter values from Simplified Model 1 with a covariate on KB. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection (50 viral RNA copies/mL). Open circles are data points below the limit of detection. Filled circles are data points above the limit of detection. Fig C. Best fit of the Simplified Model 2 to the post-ATI data from Sharaf et al. [57]. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Fig D. Best fit of the Simplified Model 3 to the post-ATI data from Sharaf et al. [57]. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Fig E. Best fit of the Simplified Model 4 to the post-ATI data from Sharaf et al. [57]. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Fig F. Best fit of the Simplified Model 5 to the post-ATI data from Sharaf et al. [57]. Green indicates PTC. Dark orange indicates NC. The horizontal dashed line is the limit of detection. Open circles are data points below the limit of detection (50 viral RNA copies/mL). Filled circles are data points above the limit of detection. Fig G. Summary of best-fit parameters in the Conway & Perelson Model stratified based on PTC or NC. The difference in the values of KB is the most significant between PTC and NC. Fig H. Summary of best fit parameters in Simplified Model 1 stratified based on PTC or NC. The difference in the values of KB is the most significant between PTC and NC. Fig I. Summary of best-fit parameters in the Simplified Model 2 stratified based on PTC or NC. The difference in the values of KB is the most significant between PTC and NC. Fig J. Summary of best-fit parameters in the Simplified Model 3 stratified based on PTC or NC. The difference in the values of KB is the most significant between PTC and NC. Fig K. Approximation of the set point viral load Vss using the best fit parameters for each participant compared with the numerical solution of Simplified Model 1 (Eq 2). The horizontal black dotted line is the approximation Vss (Eq 3). Green and dark orange curves correspond to the viral load of the PTC and NC participants, respectively, as predicted by the Simplified Model 1. The horizontal pink dashed line is the 400 viral RNA copies/mL threshold used in the classification of PTC. For participant number 8, Vss is at 0.61 viral RNA copies/mL, which is the full model predicted viral set point after day ~12000.
(DOCX)
(CSV)
Acknowledgments
We would like to thank Christiaan van Dorp for his insightful comments. We thank the participants, staff, and investigators of the ACTG studies.
Disclaimer
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Data Availability
The de-identified viral load data is available in the Supplementary Information.
Funding Statement
This work was performed under the auspices of the US Dept. of Energy under contract 89233218CNA000001 and supported by NIH grants R01-AI152703 (RK), R01-AI028433 (ASP), R01-OD011095 (ASP, JMC), P01-AI169615 (ASP), UM1-AI164561 (RMR), R21-AI143443 (JMC), R01-AI150396 (JZL), P30-AI060354 (JZL), and Los Alamos National Laboratory LDRD 20220791PRD2 (TP), LDRD 20230853PRD2 (JK), and 20210959PRD3 (NS). JMC also acknowledges the support of the National Science Foundation (grant no. DMS-1714654). NP is supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Department of Energy Computational Science Graduate Fellowship under Award Number DE-SC0022158. Research reported in this publication was also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1 AI068634, UM1 AI068636, and UM1 AI106701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hermankova M, Siliciano JD, Zhou Y, Monie D, Chadwick K, Margolick JB, et al. Analysis of Human Immunodeficiency Virus Type 1 Gene Expression in Latently Infected Resting CD4 + T Lymphocytes In Vivo. J Virol. 2003;77:7383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrigan PR, Hogg RS, Dong WWY, Yip B, Wynhoven B, Woodward J, et al. Predictors of HIV Drug-Resistance Mutations in a Large Antiretroviral-Naive Cohort Initiating Triple Antiretroviral Therapy. J Infect Dis. 2005;191:339–47. doi: 10.1086/427192 [DOI] [PubMed] [Google Scholar]
- 3.Maggiolo F, Airoldi M, Kleinloog HD, Callegaro A, Ravasio V, Arici C, et al. Effect of Adherence to HAART on Virologic Outcome and on the Selection of Resistance-Conferring Mutations in NNRTI- or PI-Treated Patients. HIV Clin Trials. 2007;8:282–92. doi: 10.1310/hct0805-282 [DOI] [PubMed] [Google Scholar]
- 4.Gardner EM, Burman WJ, Steiner JF, Anderson PL, Bangsberg DR. Antiretroviral medication adherence and the development of class-specific antiretroviral resistance. AIDS. 2009;23:1035–46. doi: 10.1097/QAD.0b013e32832ba8ec [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom DIS, Hill AL, Rabi SA, Siliciano RF, Nowak MA. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med. 2012;18:1378–85. doi: 10.1038/nm.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla A, Wang C, Patton C, Murray M, Punekar Y, De Ruiter A, et al. A Review of Long-Term Toxicity of Antiretroviral Treatment Regimens and Implications for an Aging Population. Infect Dis Ther. 2018;7:183–95. doi: 10.1007/s40121-018-0201-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sacktor N, Skolasky RL, Seaberg E, Munro C, Becker JT, Martin E, et al. Prevalence of HIV-associated neurocognitive disorders in the Multicenter AIDS Cohort Study. Neurology. 2016;86:334–40. doi: 10.1212/WNL.0000000000002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojha C, Lapierre J, Rodriguez M, Dever S, Zadeh M, DeMarino C, et al. Interplay between Autophagy, Exosomes and HIV-1 Associated Neurological Disorders: New Insights for Diagnosis and Therapeutic Applications. Viruses. 2017;9:176. doi: 10.3390/v9070176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandera A, Taramasso L, Bozzi G, Muscatello A, Robinson JA, Burdo TH, et al. HIV-Associated Neurocognitive Impairment in the Modern ART Era: Are We Close to Discovering Reliable Biomarkers in the Setting of Virological Suppression? Front Aging Neurosci. 2019;11:187. doi: 10.3389/fnagi.2019.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chand S, DeMarino C, Gowen A, Cowen M, Al-Sharif S, Kashanchi F, et al. Methamphetamine Induces the Release of Proadhesive Extracellular Vesicles and Promotes Syncytia Formation: A Potential Role in HIV-1 Neuropathogenesis. Viruses. 2022;14:550. doi: 10.3390/v14030550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci. 1997;94:13193–7. doi: 10.1073/pnas.94.24.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–7. doi: 10.1038/8394 [DOI] [PubMed] [Google Scholar]
- 13.Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, et al. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat Med. 2000;6:82–5. doi: 10.1038/71577 [DOI] [PubMed] [Google Scholar]
- 14.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–8. doi: 10.1038/nm880 [DOI] [PubMed] [Google Scholar]
- 15.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015;212:1361–5. doi: 10.1093/infdis/jiv218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption: AIDS. 2015;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JZ, Aga E, Bosch RJ, Pilkinton M, Kroon E, MacLaren L, et al. Time to Viral Rebound After Interruption of Modern Antiretroviral Therapies. Clin Infect Dis. 2022;74:865–70. doi: 10.1093/cid/ciab541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMyn NF, Varriale J, Fray EJ, Zitzmann C, MacLeod H, Lai J, et al. The latent reservoir of inducible, infectious HIV-1 does not decrease despite decades of antiretroviral therapy. J Clin Invest. 2023;133:e171554. doi: 10.1172/JCI171554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey RT, Bhat N, Yoder C, Chun T-W, Metcalf JA, Dewar R, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci. 1999;96:15109–14. doi: 10.1073/pnas.96.26.15109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz L, Martinez-Picado J, Romeu J, Paredes R, Zayat MK, Marfil S, et al. Structured treatment interruption in chronically HIV-1 infected patients after long-term viral suppression: AIDS. 2000;14:397–403. [DOI] [PubMed] [Google Scholar]
- 21.Chun T-W, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24:2803–8. doi: 10.1097/QAD.0b013e328340a239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sáez-Cirión A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, et al. Post-Treatment HIV-1 Controllers with a Long-Term Virological Remission after the Interruption of Early Initiated Antiretroviral Therapy ANRS VISCONTI Study. Lifson J, editor. PLoS Pathog. 2013;9:e1003211. doi: 10.1371/journal.ppat.1003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee T-H, et al. Antiretroviral-Free HIV-1 Remission and Viral Rebound After Allogeneic Stem Cell Transplantation: Report of 2 Cases. Ann Intern Med. 2014;161:319. doi: 10.7326/M14-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frange P, Faye A, Avettand-Fenoël V, Bellaton E, Descamps D, Angin M, et al. HIV-1 virological remission lasting more than 12 years after interruption of early antiretroviral therapy in a perinatally infected teenager enrolled in the French ANRS EPF-CO10 paediatric cohort: a case report. Lancet HIV. 2016;3:e49–54. doi: 10.1016/S2352-3018(15)00232-5 [DOI] [PubMed] [Google Scholar]
- 25.Namazi G, Fajnzylber JM, Aga E, Bosch RJ, Acosta EP, Sharaf R, et al. The Control of HIV After Antiretroviral Medication Pause (CHAMP) Study: Posttreatment Controllers Identified From 14 Clinical Studies. J Infect Dis. 2018;218:1954–63. doi: 10.1093/infdis/jiy479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etemad B, Esmaeilzadeh E, Li JZ. Learning From the Exceptions: HIV Remission in Post-treatment Controllers. Front Immunol. 2019;10:1749. doi: 10.3389/fimmu.2019.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinkevych M, Docken SS, Okoye AA, Fennessey CM, Del Prete GQ, Pino M, et al. Timing of initiation of anti-retroviral therapy predicts post-treatment control of SIV replication. Siliciano RF, editor. PLOS Pathog. 2023;19:e1011660. doi: 10.1371/journal.ppat.1011660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JZ, Melberg M, Kittilson A, Abdel-Mohsen M, Li Y, Aga E, et al. Predictors of HIV rebound differ by timing of antiretroviral therapy initiation. JCI Insight [Internet]. 2024. [cited 2024 Apr 9];9. Available from: https://insight.jci.org/articles/view/173864?utm_source=TrendMD&utm_medium=cpc&utm_campaign=JCI_Insight_TrendMD_0. doi: 10.1172/jci.insight.173864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harwood OE, Matschke LM, Moriarty RV, Balgeman AJ, Weaver AJ, Ellis-Connell AL, et al. CD8+ cells and small viral reservoirs facilitate post-ART control of SIV replication in M3+ Mauritian cynomolgus macaques initiated on ART two weeks post-infection. Silvestri G, editor. PLOS Pathog. 2023;19:e1011676. doi: 10.1371/journal.ppat.1011676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passaes C, Desjardins D, Chapel A, Monceaux V, Lemaitre J, Mélard A, et al. Early antiretroviral therapy favors post-treatment SIV control associated with the expansion of enhanced memory CD8+ T-cells. Nat Commun. 2024;15:178. doi: 10.1038/s41467-023-44389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim S-Y, Lee J, Osuna CE, Vikhe P, Schalk DR, Chen E, et al. Induction of durable remission by dual immunotherapy in SHIV-infected ART-suppressed macaques. Science. 2024;383:1104–11. doi: 10.1126/science.adf7966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandel D. Post-Intervention HIV Control Linked to Early In Vivo CD8+ T-Cell Proliferative Response to Rebound. Denver, Colorado; 2024. [Google Scholar]
- 33.Crain C. HIV Post-Treatment Controllers Show Enhanced CD8+ T Cell Proliferative Capacity After ART Cessation. Denver, Colorado; 2024. [Google Scholar]
- 34.Blazkova J, Gao F, Marichannegowda MH, Justement JS, Shi V, Whitehead EJ, et al. Distinct mechanisms of long-term virologic control in two HIV-infected individuals after treatment interruption of anti-retroviral therapy. Nat Med. 2021;27:1893–8. doi: 10.1038/s41591-021-01503-6 [DOI] [PubMed] [Google Scholar]
- 35.Hill AL, Rosenbloom DIS, Fu F, Nowak MA, Siliciano RF. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci. 2014;111:13475–80. doi: 10.1073/pnas.1406663111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill AL, Rosenbloom DIS, Goldstein E, Hanhauser E, Kuritzkes DR, Siliciano RF, et al. Real-Time Predictions of Reservoir Size and Rebound Time during Antiretroviral Therapy Interruption Trials for HIV. Weinberger L, editor. PLOS Pathog. 2016;12:e1005535. doi: 10.1371/journal.ppat.1005535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinkevych M, Cromer D, Tolstrup M, Grimm AJ, Cooper DA, Lewin SR, et al. HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5–8 Days—Implications for HIV Remission. Swanstrom R, editor. PLOS Pathog. 2015;11:e1005000. doi: 10.1371/journal.ppat.1005000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinkevych M, Kent SJ, Tolstrup M, Lewin SR, Cooper DA, Søgaard OS, et al. Modeling of Experimental Data Supports HIV Reactivation from Latency after Treatment Interruption on Average Once Every 5–8 Days. Swanstrom R, editor. PLOS Pathog. 2016;12:e1005740. doi: 10.1371/journal.ppat.1005740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fennessey CM, Pinkevych M, Immonen TT, Reynaldi A, Venturi V, Nadella P, et al. Genetically-barcoded SIV facilitates enumeration of rebound variants and estimation of reactivation rates in nonhuman primates following interruption of suppressive antiretroviral therapy. Evans DT, editor. PLOS Pathog. 2017;13:e1006359. doi: 10.1371/journal.ppat.1006359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Dorp CH, Conway JM, Barouch DH, Whitney JB, Perelson AS. Models of SIV rebound after treatment interruption that involve multiple reactivation events. Davenport MP, editor. PLOS Comput Biol. 2020;16:e1008241. doi: 10.1371/journal.pcbi.1008241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, et al. Control of Viremia in Simian Immunodeficiency Virus Infection by CD8 + Lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857 [DOI] [PubMed] [Google Scholar]
- 42.Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med. 2009;265:29–42. doi: 10.1111/j.1365-2796.2008.02045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, et al. CD8+ Lymphocytes Control Viral Replication in SIVmac239-Infected Rhesus Macaques without Decreasing the Lifespan of Productively Infected Cells. Douek DC, editor. PLoS Pathog. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury A, Hayes TL, Bosinger SE, Lawson BO, Vanderford T, Schmitz JE, et al. Differential Impact of In Vivo CD8 + T Lymphocyte Depletion in Controller versus Progressor Simian Immunodeficiency Virus-Infected Macaques. Kirchhoff F, editor. J Virol. 2015;89:8677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y, Cartwright EK, Silvestri G, Perelson AS. CD8+ lymphocyte control of SIV infection during antiretroviral therapy. Douek DC, editor. PLOS Pathog. 2018;14:e1007350. doi: 10.1371/journal.ppat.1007350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Policicchio BB, Cardozo-Ojeda EF, Xu C, Ma D, He T, Raehtz KD, et al. CD8+ T cells control SIV infection using both cytolytic effects and non-cytolytic suppression of virus production. Nat Commun. 2023;14:6657. doi: 10.1038/s41467-023-42435-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esmaeilzadeh E, Etemad B, Lavine CL, Garneau L, Li Y, Regan J, et al. Autologous neutralizing antibodies increase with early antiretroviral therapy and shape HIV rebound after treatment interruption. Sci Transl Med. 2023;15:eabq4490. doi: 10.1126/scitranslmed.abq4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Etemad B, Sun X, Li Y, Melberg M, Moisi D, Gottlieb R, et al. HIV post-treatment controllers have distinct immunological and virological features. Proc Natl Acad Sci. 2023;120:e2218960120. doi: 10.1073/pnas.2218960120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conway JM, Perelson AS. Post-treatment control of HIV infection. Proc Natl Acad Sci. 2015;112:5467–72. doi: 10.1073/pnas.1419162112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway JM, Perelson AS, Li JZ. Predictions of time to HIV viral rebound following ART suspension that incorporate personal biomarkers. Antia R, editor. PLOS Comput Biol. 2019;15:e1007229. doi: 10.1371/journal.pcbi.1007229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conway JM, Meily P, Li JZ, Perelson AS. Unified model of short- and long-term HIV viral rebound for clinical trial planning. J R Soc Interface. 2021;18:rsif.2020.1015, 20201015. doi: 10.1098/rsif.2020.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bing A, Hu Y, Prague M, Hill AL, Li JZ, Bosch RJ, et al. Comparison of empirical and dynamic models for HIV viral load rebound after treatment interruption. Stat Commun Infect Dis. 2020;12:20190021. doi: 10.1515/scid-2019-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang R, Bing A, Wang C, Hu Y, Bosch RJ, DeGruttola V. A flexible nonlinear mixed effects model for HIV viral load rebound after treatment interruption. Stat Med. 2020;39:2051–66. doi: 10.1002/sim.8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao S, Wu L, Yu T, Kouyos R, Günthard HF, Wang R. Nonlinear mixed-effects models for HIV viral load trajectories before and after antiretroviral therapy interruption, incorporating left censoring. Stat Commun Infect Dis. 2022;14:20210001. doi: 10.1515/scid-2021-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyd MA, Dixit NM, Siangphoe U, Buss NE, Salgo MP, Lange JMA, et al. Viral Decay Dynamics in HIV-Infected Patients Receiving Ritonavir-Boosted Saquinavir and Efavirenz With or Without Enfuvirtide: A Randomized, Controlled Trial (HIV-NAT 012). J Infect Dis. 2006;194:1319–22. doi: 10.1086/508291 [DOI] [PubMed] [Google Scholar]
- 56.Wells CR, Cao Y, Durham DP, Byrareddy SN, Ansari AA, Ruddle NH, et al. Mechanistic basis of post-treatment control of SIV after anti-α4β7 antibody therapy. Regoes RR, editor. PLOS Comput Biol. 2021;17:e1009031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharaf R, Lee GQ, Sun X, Etemad B, Aboukhater LM, Hu Z, et al. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest. 2018;128:4074–85. doi: 10.1172/JCI120549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobson JM, Pat Bucy R, Spritzler J, Saag MS, Eron JJ, Coombs RW, et al. Evidence That Intermittent Structured Treatment Interruption, but Not Immunization with ALVAC-HIV vCP1452, Promotes Host Control of HIV Replication: The Results of AIDS Clinical Trials Group 5068. J Infect Dis. 2006;194:623–32. doi: 10.1086/506364 [DOI] [PubMed] [Google Scholar]
- 59.Kilby JM, Bucy RP, Mildvan D, Fischl M, Santana-Bagur J, Lennox J, et al. A Randomized, Partially Blinded Phase 2 Trial of Antiretroviral Therapy, HIV-Specific Immunizations, and Interleukin-2 Cycles to Promote Efficient Control of Viral Replication (ACTG A5024). J Infect Dis. 2006;194:1672–6. doi: 10.1086/509508 [DOI] [PubMed] [Google Scholar]
- 60.Skiest DJ, Brown K, Cooper TW, Hoffman-Roberts H, Mussa HR, Elliott AC. Prospective comparison of methicillin-susceptible and methicillin-resistant community-associated Staphylococcus aureus infections in hospitalized patients. J Infect. 2007;54:427–34. doi: 10.1016/j.jinf.2006.09.012 [DOI] [PubMed] [Google Scholar]
- 61.Volberding P, Demeter L, Bosch RJ, Aga E, Pettinelli C, Hirsch M, et al. Antiretroviral therapy in acute and recent HIV infection: a prospective multicenter stratified trial of intentionally interrupted treatment. AIDS. 2009;23:1987–95. doi: 10.1097/QAD.0b013e32832eb285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schooley RT, Spritzler J, Wang H, Lederman MM, Havlir D, Kuritzkes DR, et al. AIDS Clinical Trials Group 5197: A Placebo-Controlled Trial of Immunization of HIV-1–Infected Persons with a Replication-Deficient Adenovirus Type 5 Vaccine Expressing the HIV-1 Core Protein. J Infect Dis. 2010;202:705–16. doi: 10.1086/655468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99. doi: 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baral S, Antia R, Dixit NM. A dynamical motif comprising the interactions between antigens and CD8 T cells may underlie the outcomes of viral infections. Proc Natl Acad Sci. 2019;116:17393–8. doi: 10.1073/pnas.1902178116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining ‘T cell exhaustion.’ Nat Rev Immunol. 2019;19:665–74. doi: 10.1038/s41577-019-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonhoeffer S, Rembiszewski M, Ortiz GM, Nixon DF. Risks and benefits of structured antiretroviral drug therapy interruptions in HIV-1 infection: AIDS. 2000;14:2313–22. [DOI] [PubMed] [Google Scholar]
- 67.Kim H, Perelson AS. Viral and Latent Reservoir Persistence in HIV-1–Infected Patients on Therapy. De Boer R, editor. PLoS Comput Biol. 2006;2:e135. doi: 10.1371/journal.pcbi.0020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rong L, Perelson AS. Asymmetric division of activated latently infected cells may explain the decay kinetics of the HIV-1 latent reservoir and intermittent viral blips. Math Biosci. 2009;217:77–87. doi: 10.1016/j.mbs.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rong L, Perelson AS. Modeling Latently Infected Cell Activation: Viral and Latent Reservoir Persistence, and Viral Blips in HIV-infected Patients on Potent Therapy. Antia R, editor. PLoS Comput Biol. 2009;5:e1000533. doi: 10.1371/journal.pcbi.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rong L, Perelson AS. Modeling HIV persistence, the latent reservoir, and viral blips. J Theor Biol. 2009;260:308–31. doi: 10.1016/j.jtbi.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, et al. HIV-1 Integration Landscape during Latent and Active Infection. Cell. 2015;160:420–32. doi: 10.1016/j.cell.2015.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, et al. Clonally expanded CD4 + T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci. 2016;113:1883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Callaway D. HIV-1 Infection and Low Steady State Viral Loads. Bull Math Biol. 2002;64:29–64. doi: 10.1006/bulm.2001.0266 [DOI] [PubMed] [Google Scholar]
- 76.Murray JM, Perelson AS. Human Immunodeficiency Virus: Quasi-Species and Drug Resistance. Multiscale Model Simul. 2005;3:300–11. [Google Scholar]
- 77.Conway JM, Perelson AS. Residual Viremia in Treated HIV+ Individuals. Antia R, editor. PLOS Comput Biol. 2016;12:e1004677. doi: 10.1371/journal.pcbi.1004677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. Douek DC, editor. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, et al. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell. 2013;155:540–51. doi: 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stafford MA, Corey L, Cao Y, Daar ES, Ho DD, Perelson AS. Modeling Plasma Virus Concentration during Primary HIV Infection. J Theor Biol. 2000;203:285–301. doi: 10.1006/jtbi.2000.1076 [DOI] [PubMed] [Google Scholar]
- 81.Vaidya NK, Rong L, Marconi VC, Kuritzkes DR, Deeks SG, Perelson AS. Treatment-Mediated Alterations in HIV Fitness Preserve CD4+ T Cell Counts but Have Minimal Effects on Viral Load. Fraser C, editor. PLoS Comput Biol. 2010;6:e1001012. doi: 10.1371/journal.pcbi.1001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davenport MP, Ribeiro RM, Perelson AS. Kinetics of Virus-Specific CD8 + T Cells and the Control of Human Immunodeficiency Virus Infection. J Virol. 2004;78:10096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson PLF, Kochin BF, McAfee MS, Stromnes IM, Regoes RR, Ahmed R, et al. Vaccination Alters the Balance between Protective Immunity, Exhaustion, Escape, and Death in Chronic Infections. J Virol. 2011;85:5565–70. doi: 10.1128/JVI.00166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bofill M, Janossy G, Lee CA, Macdonald-Burns D, Phillips AN, Sabin C, et al. Laboratory control values for CD4 and CD8 T lymphocytes. Implications for HIV-1 diagnosis. Clin Exp Immunol. 2008;88:243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Markowitz M, Louie M, Hurley A, Sun E, Di Mascio M, Perelson AS, et al. A Novel Antiviral Intervention Results in More Accurate Assessment of Human Immunodeficiency Virus Type 1 Replication Dynamics and T-Cell Decay In Vivo. J Virol. 2003;77:5037–8. doi: 10.1128/jvi.77.8.5037-5038.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conway JM, Coombs D. A Stochastic Model of Latently Infected Cell Reactivation and Viral Blip Generation in Treated HIV Patients. Fraser C, editor. PLoS Comput Biol. 2011;7:e1002033. doi: 10.1371/journal.pcbi.1002033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramratnam B, Bonhoeffer S, Binley J, Hurley A, Zhang L, Mittler JE, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. The Lancet. 1999;354:1782–5. doi: 10.1016/S0140-6736(99)02035-8 [DOI] [PubMed] [Google Scholar]
- 88.Burnham KP, Anderson DR. Practical Use of the Information-Theoretic Approach. Model Sel Inference [Internet]. New York, NY: Springer New York; 1998. [cited 2024 Mar 7]. p. 75–117. Available from: http://link.springer.com/10.1007/978-1-4757-2917-7_3. [Google Scholar]
- 89.Desikan R, Raja R, Dixit NM. Early exposure to broadly neutralizing antibodies may trigger a dynamical switch from progressive disease to lasting control of SHIV infection. Yates AJ, editor. PLOS Comput Biol. 2020;16:e1008064. doi: 10.1371/journal.pcbi.1008064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vemparala B, Madelain V, Passaes C, Millet A, Avettand-Fenoel V, Djidjou-Demasse R, et al. Antiviral capacity of the early CD8 T-cell response is predictive of natural control of SIV infection [Internet]. bioRxiv; 2023. [cited 2024 Apr 9]. p. 2023.10.13.562306. Available from: https://www.biorxiv.org/content/10.1101/2023.10.13.562306v1. [Google Scholar]
- 91.De Boer RJ, Perelson AS. Towards a general function describing t cell proliferation. J Theor Biol. 1995;175:567–76. doi: 10.1006/jtbi.1995.0165 [DOI] [PubMed] [Google Scholar]
- 92.Mayer A, Zhang Y, Perelson AS, Wingreen NS. Regulation of T cell expansion by antigen presentation dynamics. Proc Natl Acad Sci. 2019;116:5914–9. doi: 10.1073/pnas.1812800116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.den Uyl D, van der Horst-Bruinsma IE, van Agtmael M. Progression of HIV to AIDS: a protective role for HLA-B27? AIDS Rev. 2004;6:89–96. [PubMed] [Google Scholar]
- 94.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8+ T Cell Response against HIV-1. Watkins D, editor. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ndhlovu ZM, Stampouloglou E, Cesa K, Mavrothalassitis O, Alvino DM, Li JZ, et al. The Breadth of Expandable Memory CD8 + T Cells Inversely Correlates with Residual Viral Loads in HIV Elite Controllers. Silvestri G, editor. J Virol. 2015;89:10735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takata H, Buranapraditkun S, Kessing C, Fletcher JLK, Muir R, Tardif V, et al. Delayed differentiation of potent effector CD8 + T cells reducing viremia and reservoir seeding in acute HIV infection. Sci Transl Med. 2017;9:eaag1809. doi: 10.1126/scitranslmed.aag1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gebara NY, El Kamari V, Rizk N. HIV-1 elite controllers: an immunovirological review and clinical perspectives. J Virus Erad. 2019;5:163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bansal A, Gehre MN, Qin K, Sterrett S, Ali A, Dang Y, et al. HLA-E–restricted HIV-1–specific CD8+ T cell responses in natural infection. J Clin Invest. 2021;131:e148979. doi: 10.1172/JCI148979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–8. doi: 10.1038/ni845 [DOI] [PubMed] [Google Scholar]
- 100.Gaiha GD, McKim KJ, Woods M, Pertel T, Rohrbach J, Barteneva N, et al. Dysfunctional HIV-Specific CD8+ T Cell Proliferation Is Associated with Increased Caspase-8 Activity and Mediated by Necroptosis. Immunity. 2014;41:1001–12. doi: 10.1016/j.immuni.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ndhlovu ZM, Kamya P, Mewalal N, Kløverpris HN, Nkosi T, Pretorius K, et al. Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity. 2015;43:591–604. doi: 10.1016/j.immuni.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rehr M, Cahenzli J, Haas A, Price DA, Gostick E, Huber M, et al. Emergence of Polyfunctional CD8 + T Cells after Prolonged Suppression of Human Immunodeficiency Virus Replication by Antiretroviral Therapy. J Virol. 2008;82:3391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mikulak J, Oriolo F, Zaghi E, Di Vito C, Mavilio D. Natural killer cells in HIV-1 infection and therapy. AIDS. 2017;31:2317–30. doi: 10.1097/QAD.0000000000001645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang W, Ellingson LA. Detecting and Resetting Tipping Points to Create More HIV Post-Treatment Controllers with Bifurcation and Sensitivity Analysis. SIAM J Appl Math. 2023;S493–514. [Google Scholar]
- 105.Migueles SA, Weeks KA, Nou E, Berkley AM, Rood JE, Osborne CM, et al. Defective Human Immunodeficiency Virus-Specific CD8 + T-Cell Polyfunctionality, Proliferation, and Cytotoxicity Are Not Restored by Antiretroviral Therapy. J Virol. 2009;83:11876–89. [DOI] [PMC free article] [PubMed] [Google Scholar]