Abstract
Recent epidemiological research suggests a possible negative correlation between Helicobacter pylori infection and inflammatory bowel disease (IBD). However, conflicting studies have provided unclear evidence regarding these causal relationships. Therefore, recommending specific prevention and treatment strategies for H. pylori infection and IBD is challenging. We used various antibodies (anti-H. pylori IgG, VacA, and GroEl) related to H. pylori infection as indicators. We acquired relevant genetic variants from public databases within the Genome-wide Association Studies (GWAS) dataset using IBDs tool variables from 2 different GWAS datasets. We thoroughly examined the data and screened for IVs that fulfilled these criteria. Subsequently, Bidirectional Mendelian randomization (MR) was conducted to predict the potential causality between the 2. To ensure the accuracy and robustness of our results, we conducted a series of sensitivity analyses. Based on our comprehensive MR analysis, no potential causal relationship was observed between H. pylori infection and IBD. Across various methodologies, including IVW, MR-Egger, and weighted median, our findings showed P values > .05. The only exception was observed in the reverse MR analysis using the MR-Egger method, which yielded a P value of < .05. However, because the IVW method is considered the most statistically significant method for MR, and its P value was > .05, we do not believe that a potential causal relationship exists between them. Our sensitivity analysis did not suggest significant horizontal pleiotropism. Although heterogeneity was detected in the analysis of IBD (IIBDGC source) versus H. pylori GroEL antibody levels (MR-Egger, Qp = 0.038; IVW, Qp = 0.043), the results remained reliable because we selected IVW as a random-effects model in our MR analysis method. Based on our MR research, no direct correlation was observed between H. pylori infection and IBD risk. This implies that eradicating H. pylori may not provide substantial benefits in preventing or treating regional IBD, and vice versa. Nevertheless, the use of H. pylori serological index substitution has limitations, and further research using histological diagnosis and additional MR studies is required to comprehensively assess the link between H. pylori infection and IBD.
Keywords: causal inference, Helicobacter pylori infection, inflammatory bowel disease, Mendelian randomization, null association
1. Introduction
Inflammatory bowel disease (IBD) is a group of idiopathic chronic IBDs, mainly ulcerative colitis (UC), and Crohns disease (CD).[1] IBD seriously affects patients’ quality of life and work capacity, and substantially burdens social development and healthcare systems.[2] IBD affects more than 2.5 million people in Europe and has an increasing incidence in newly industrialized countries in Asia, South America, and the Middle East.[3] Although the pathogenesis of IBD has not yet been fully elucidated, studies have shown that a combination of external environmental factors, intestinal microflora, individual genetic susceptibility, and immune responses influences the pathogenesis of IBD.[4–6]
Helicobacter pylori, a gram-negative bacterium, is a widely prevalent bacterial infection that has infected nearly 60% of the global population. Moreover, it is an important public health concern requiring attention and effective management.[7,8] A recent meta-analysis showed that the prevalence of H. pylori infection in children worldwide is approximately 32%.[9] H. pylori has been associated with various diseases, including peptic ulcer (10%), noncardia cancer (1%–3%), and gastric mucosa-associated lymphoid tissue lymphoma (<0.1%).[10] H. pylori, which colonizes the digestive system, can not only cause localized inflammatory responses but can influence the patient’s immune system.[11–14] This link has led researchers to explore the potential association between H. pylori and the pathogenesis of IBD.
As societies with inadequate living conditions improve sanitation, the prevalence of H. pylori infection tends to decrease. Conversely, the incidence of IBD increases in communities that adopt a Western lifestyle.[15] Several other epidemiologic studies have demonstrated similar results,[16–19] suggesting a negative causal link; that is, the widespread eradication of H. pylori is detrimental to the prevention and control of IBD. However, contrasting findings have been reported in other studies.[20–22] Additionally, Pokrotnieks et al[23] hypothesized that there was no direct cause-and-effect relationship between H. pylori infection and IBD. Instead, the negative correlation established by epidemiological studies may be associated with the transient state of host fucosylated glycans in the gastrointestinal tract. Evidence regarding the relationship between H. pylori infection and IBD is based solely on observational studies, which have certain limitations such as confounding factors, reverse causality, and other sources of bias. Currently, a direct causal relationship between H. pylori infection and IBD remains unclear. This lack of evidence poses challenges in recommending specific prevention or treatment strategies for H. pylori infection and IBD.
Mendelian randomization (MR) is an advanced epidemiological analysis method that facilitates the prediction of causal relationships between factors. MR designs employ genetic variation as an instrumental variable (IV) to capture exposure factors. Genetic variations, typically represented as single-nucleotide polymorphisms (SNPs), are randomly distributed and remain impervious to environmental stimuli and other potential confounding variables. SNPs are genetic variations that occur spontaneously and are entirely independent of environmental factors and other potential confounding variables.[24] With the development of large-scale Genome-wide Association Studies (GWAS) data, MR analyses have become more common, allowing for a rigorous explanation of the causal relationships between complex diseases.
2. Materials and methods
2.1. Mendelian randomized design
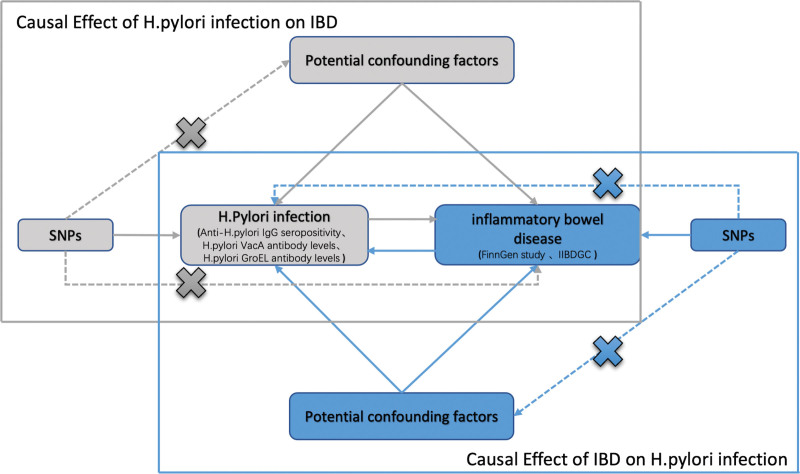
The methodology of this MR study is outlined in detail in Figure 1. Genetic variants were employed as the IV for MR analyses, and the validity of our study relied on 3 fundamental assumptions. The correlation assumption, which asserts that genetic variants are highly correlated with exposure. Second, the independence assumption, which maintains that genetic variants are not influenced by any confounding factors that may mediate the pathway from exposure to outcome. Lastly, the exclusion-restriction assumption, which posits that genetic variants are only expected to impact outcome through exposure.[25]
Figure 1.
Schematic representation of the bidirectional MR study of the causal relationship between H. pylori infection and IBD. SNPs, single-nucleotide polymorphisms. H. pylori, Helicobacter pylori.
2.2. Selection of diagnostic indicators of H. pylori infection
The diagnosis of H. pylori infection is mainly categorized into invasive (endoscopy-based) and noninvasive methods, each with unique advantages and disadvantages, making the selection of an appropriate diagnostic index for H. pylori infection substantial in research. As a noninvasive method, the accuracy of serologic testing is not affected by ulcer bleeding, gastric atrophy, and PPIs or antibiotics, which can lead to false-negative results in other invasive or noninvasive tests.[26] In 1993, a study showed that the sensitivity and specificity of serum anti-H. pylori IgG antibody assay for the diagnosis of HP infection were 96.2% and 60.0%, respectively.[27] The lower specificity of anti-H. pylori IgG antibodies is mainly due to their persistence in the body even after H. pylori eradication. This means that a positive result indicates a current or past infection. In a study conducted among adolescents in 2014, the sensitivity and specificity of IgG antibody testing were 91.2% and 97.4%, respectively.[28] Other H. pylori-associated serological markers may provide stronger indications of H. pylori infection. Epplein et al identified the 4 serologic antibodies with the highest sensitivities associated with H. pylori infection as: the VacA antibody (sensitivity: 100%; 95% CI: 84%–100%), followed by GroEl (sensitivity: 95%; 95% CI: 76%–100%), HcpC (sensitivity: 80%; 95% CI: 57%–82%), and HP1564 (sensitivity: 75%; 95% CI: 53%–59%).[29] VacA is a major virulence factor produced by H. pylori that facilitates gastric colonization by influencing human epithelial cells.[30] GroEL, the molecular chaperone of H. pylori, is a homolog of heat shock protein (HSP60) in prokaryotes and is an essential pathogenesis-associated antigenic component whose primary function is to facilitate protein transport, complete correct folding, and restore the natural conformation. Previous studies have shown that more than 80% of the H. pylori-infected population produces antibodies against GroEL.[31,32] Unfortunately, we did not find any GWAS data related to HcpC and HP564 antibodies. In summary, we used H. pylori-related antibody indicators of H. pylori infection, including serum anti-H. pylori IgG, VacA, and GroEl antibodies.
2.3. Description of data sources
This study identified 2 distinct genetic associations for IBD using independent GWAS datasets. The first dataset included 5673 individuals of European descent with IBD and 213119 European controls from the FinnGen database, ensuring an accurate and consistent diagnosis of IBD through electronic medical records. The second IBD-related GWAS dataset comprised 12,882 European patients and 34,652 European controls from the International Inflammatory Bowel Disease Genetics Consortium. Additionally, the GWAS summary statistics for anti-H. pylori IgG antibody positivity and H. pylori VacA and GroEL antibody levels were obtained from public data in the EBI database, including 8735 European patients. Each GWAS was approved by an appropriate ethics committee. For more detailed information, please see Table 1.
Table 1.
Details of the studies included in the Mendelian randomization analyses
| Phenotype | Consortium | Ethnicity | Sample size | Year | Number of SNPs | Web source |
|---|---|---|---|---|---|---|
| IBD | FinnGen study | European | 5673 | 2021 | 16,380,466 | https://gwas.mrcieu.ac.uk/datasets/finn-b-K11_IBD/ |
| IBD | IIBDGC | European | 12,882 | 2015 | 12,716,084 | https://gwas.mrcieu.ac.uk/datasets/ieu-a-31/ |
| Anti-H. pylori IgG seropositivity | UK biobank | European | 8735 | 2020 | 9170,312 | https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90006910/ |
| H. pylori VacA antibody levels | UK biobank | European | 1571 | 2020 | 9178,635 | https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90006916/ |
| H. pylori GroEL antibody levels | UK biobank | European | 2716 | 2020 | 9172,299 | https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90006913/ |
IBD = inflammatory bowel disease, IIBDGC = International Inflammatory Bowel Disease Genetics Consortium
2.4. Selection of genetic instrumental variable (IV)s for Helicobacter pylori-related antibodies
The IVs of the 3 H. pylori-associated antibodies were obtained separately by GWAS pooling statistics, and a series of quality control measures were used to screen the IV genotypes that met the MR hypothesis. On 1 hand, achieving genome-wide significance for the screened IVs was necessary. On the other hand, obtaining an adequate number of IVs after screening was crucial to enhance statistical power. Hence, the P-value threshold for H. pylori-associated antibody IVs was < 5 × 10−6. Subsequently, a chain imbalance (LI) with a P-value < 5 × 10−8 was established using an R2 of < 0.001, a window size of 10,000 kb, and a P-value < 5 × 10−8 for the linkage disequilibrium (LD) clustering algorithm. To confirm that the affected alleles were of the same type, the exposure and resulting datasets were harmonized. This process involved the removal of SNPs with intermediate allele frequencies and ambiguous SNPs with conflicting alleles. Additionally, the F-statistic for each SNP was calculated using the following formula to assess the strength of the IV:
Where N is the sample size, K is the number of SNPs, and R2 represents the proportion of variation explained by the independent variable.[33] The F-statistics for all SNPs were > 10. Therefore, SNPs that have been rigorously screened were used for subsequent analyses.
2.5. Selection of genetic instrumental variable (IV)s for IBD
Similarly, we performed GWAS pooling statistics to obtain gene IVs for IBD. First, we set relevant parameters to achieve quality control, including P < 5 × 10−8 to reach genome-wide significance of the SNPs and established the same chained disequilibrium (LD) clustering algorithm with an R2 < 0.001, a window size of = 10,000 kb, and P < 5 × 10, balanced (LD) clustering algorithm, and finally harmonized the exposure and resultant datasets. The same formula was used to calculate the F-statistic for each SNP. The SNPs screened after the above steps were used as IV for subsequent analyses.
2.6. Statistical analysis and data visualization
Statistical analyses and data visualization were performed using R programming software, version R4.2.3. About 3 complementary methods, inverse-variance weighted, MR-Egger, and weighted median, were utilized for the MR analysis to ensure robustness. The IVW method is the primary causal estimation method, providing the most accurate results when all selected SNPs are valid IVs.[34] The MR-Egger method utilizes a weighted linear regression to obtain consistent causal estimates, assuming that instrumental strength is independent of the direct effect (InSIDE), even when genetic IVs are not valid. However, this method has low precision and is vulnerable to peripheral genetic variation.[35] The weighted median method is a statistical technique that calculates the weighted median of the Wald ratio estimates and does not require the InSIDE assumption. This method is resistant to horizontal pleiotropic biases. Compared to the MR-Egger method, the weighted median process has a lower type I error and is more effective for causal estimation.[36] These methods are provided by the “TwoSampleMR” R package (Version 0.5.7).[36] Significance was attributed to 2-sided P values of < 0.05. The generation of forest plots was accomplished through utilization of the “forestploter” R package, with version 2.0.1 implemented for this purpose.
3. Results
3.1. The causal effect of Helicobacter pylori infection on IBD
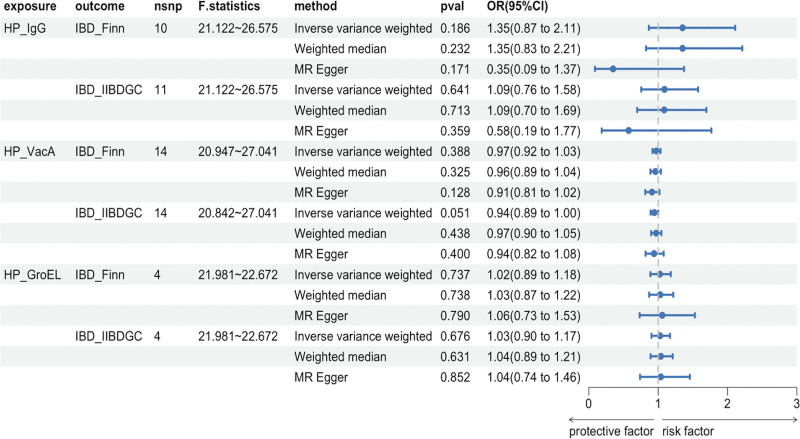
MR analysis was performed to investigate the causal relationship between H. pylori infection and IBD. Various methods including the IVW, MR-Egger, and weighted median were used to validate the results (Fig. 2). All results showed a P-value > 0.05. Figure 3 shows the scatter plots used to visualize the data. The findings of the MR analysis showed that H. pylori infection does not have a causal effect on IBD.
Figure 2.
Forest plot of Mendelian randomization analyses. OR, odds ratio. 95%CI, 95% confidence interval. nsnp, number of single-nucleotide polymorphisms. HP_IgG, Anti-H. pylori IgG seropositivity. HP_VacA, H. pylori VacA antibody levels. HP_GroEL, H. pylori GroEL antibody levels. IBD_Finn, GWAS data source of inflammatory bowel disease in the FinnGen database. IBD_IIBDGG, GWAS data source of inflammatory bowel disease from the International Inflammatory Bowel Disease Genetics Consortium.
Figure 3.
Scatterplot of significant causal relationships between H. pylori infection-related antibody markers and IBD. IBD (Finn), GWAS data source of inflammatory bowel disease in the FinnGen database. IBD (IIBDGG), GWAS data source of inflammatory bowel disease from the International Inflammatory Bowel Disease Genetics Consortium.
3.2. IBD on Helicobacter pylori infection causality
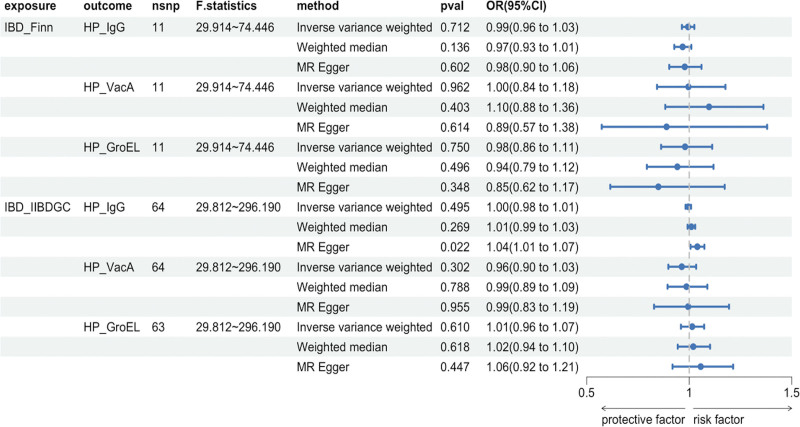
Furthermore, MR analysis was used to examine the causal relationship between IBD development and H. pylori infection. The antibody-mediated cross-reactivity of HPs GroEL, which has a high homology with human HSP60, can contribute to the development of atherosclerosis. Therefore, atherosclerosis-associated Coronary artery disease may be a potential confounding factor. To exclude the effects of horizontal pleiotropy, we excluded rs56062135, which can cause coronary artery disease, from the 2-sample analysis of GroEL antibodies against IBD (IIBDGC source) and HP.[37] Figure 4 shows the results of this study. We utilized the MR-Egger method to analyze the IgG antibody-positive samples for IBD (IIBDGC source) compared to anti-HP. The IVW method indicated a P-value of not more than 0.05, implying that no statistically significant causal relationship existed between the 2. The weighted median method yielded results that were consistent with those of the IVW method. However, MR-Egger resulted in a P-value < 0.05, with the beta direction being the same for several methods. As the IVW method is the most statistically significant method for MR, we do not believe that a potential causal relationship exists between the 2. The analysis did not establish causality between IBD and H. pylori infection.
Figure 4.
Forest plot of reverse Mendelian randomization analyses. OR, odds ratio. 95%CI, 95% confidence interval. HP_IgG, Anti-H. pylori IgG seropositivity. HP_VacA, H. pylori VacA antibody levels. HP_GroEL, H. pylori GroEL antibody levels. IBD_Finn, GWAS data source of inflammatory bowel disease in the FinnGen database. IBD_IIBDGG, GWAS data source of inflammatory bowel disease from the International Inflammatory Bowel Disease Genetics Consortium.
3.3. Sensitivity analysis
We first analyzed all data for heterogeneity, mainly using IVW methods and MR-Egger regression, quantified using Cochrans Q-statistics. Although heterogeneity was detected in the analysis of IBD (IIBDGC source) versus H. pylori GroEL antibody levels (MR-Egger, Qp = 0.038; IVW, Qp = 0.043), because we chose the IVW of the random-effects model for the MR analysis method, the results of the study are still reliable.[38] We utilized the MR-PRESSO method to identify the horizontal polytropy. This method can detect horizontal polytropy anisotropy, correct horizontal polytropy by removing outliers, and determine whether there is a significant change in the causal effects before and after removing outliers.[39] The results for MR-PRESSO are shown in Table 2, with P < .05, which suggests substantial pleiotropy.
Table 2.
Results of sensitivity analysis for bidirectional Mendelian randomization
| Exposure | Outcome | MR-Egger | IVW | MR-PRESSO | ||||
|---|---|---|---|---|---|---|---|---|
| Q | Q_df | Q_pval | Q | Q_df | Q_pval | pval | ||
| HP_IgG | IBD_Finn | 9.216 | 8 | 0.324 | 13.912 | 9 | 0.126 | 0.162 |
| IBD_IIBDGC | 13.443 | 9 | 0.144 | 15.525 | 10 | 0.114 | 0.144 | |
| HP_VacA | IBD_Finn | 11.915 | 12 | 0.453 | 13.808 | 13 | 0.387 | 0.410 |
| IBD_IIBDGC | 4.594 | 12 | 0.970 | 4.599 | 13 | 0.983 | 0.986 | |
| HP_GroEL | IBD_Finn | 0.359 | 2 | 0.836 | 0.394 | 3 | 0.942 | 0.979 |
| IBD_IIBDGC | 1.900 | 2 | 0.387 | 1.903 | 3 | 0.593 | 0.681 | |
| IBD_Finn | HP_IgG | 9.827 | 9 | 0.365 | 10.035 | 10 | 0.437 | 0.288 |
| HP_VacA | 10.420 | 9 | 0.318 | 10.769 | 10 | 0.376 | 0.457 | |
| HP_GroEL | 9.924 | 9 | 0.357 | 10.903 | 10 | 0.365 | 0.421 | |
| IBD_IIBDGC | HP_IgG | 70.143 | 62 | 0.223 | 79.426 | 63 | 0.079 | 0.078 |
| HP_VacA | 76.858 | 62 | 0.097 | 77.027 | 63 | 0.110 | 0.102 | |
| HP_GroEL | 81.901 | 61 | 0.038 | 82.408 | 62 | 0.043 | 0.125 | |
IVW, Inverse-variance weighted
3.4. Meta-merging after MR analysis
In summary, we analyzed the results of different sources of IBD-related GWAS data separately. To further validate the robustness of the results, we performed a meta-analysis of the IVW method results to explore whether the data before and after merging remained consistent. The merged data supported the absence of a causal relationship between H. pylori infection and IBD, as shown in Table 3.
Table 3.
Pooled results were analyzed using meta-analysis. Q_pval, the pval of Cochrans Q-statistics measure
| Exposure | Outcome | OR (95%CI) | Pval | Q_pval |
|---|---|---|---|---|
| HP_IgG | IBD | 1.190 (0.896–1.579) | 0.230 | 0.468 |
| HP_VacA | IBD | 0.975 (0.928–1.024) | 0.309 | 0.121 |
| HP_GroEL | IBD | 1.016 (0.917–1.125) | 0.763 | 0.769 |
| IBD | HP_IgG | 0.995 (0.982–1.009) | 0.476 | 0.807 |
| IBD | HP_VacA | 0.989 (0.962–1.017) | 0.454 | 0.424 |
| IBD | HP_GroEL | 1.013 (0.961–1.068) | 0.640 | 0.836 |
4. Discussion
Evidence from previous studies on the causal relationship between HP infection and IBD has been inconsistent. H. pylori belongs to the genus Helicobacter and is usually observed on the surface epithelium of the stomach; however, H. pylori DNA has been observed in the colon.[40] H. pylori has the potential to induce the production of inflammatory factors in the gastric mucosa. The localized expression of these factors may elicit an immune response at extra-gastric sites, suggesting that HP infection could be implicated in certain autoimmune diseases.[41–43] In IBD, dysregulation of the immune response to commensal bacteria is a crucial pathogenic mechanism. The pathogenesis of IBD, particularly CD, is associated with the TH1 immune response and secretion of pro-inflammatory cytokines.[44] A few patients diagnosed with IBD may suffer from concomitant autoimmune diseases and autoimmune extraintestinal manifestations. Previous studies have indicated that the risk of autoimmune diseases is higher in patients with IBD than in those without.[45] The immunobiological similarity between IBD and H. pylori infection provides the context for a possible causal relationship.
Various observational studies have linked Helicobacter spp., including H. pylori, to the development of IBD. About 1 study identified the presence of H. pylori in the intestinal mucosa of patients with UC-like CD and UC. These findings suggest an association between H. pylori infection and IBD pathogenesis. Further research is warranted to elucidate the mechanisms underlying this relationship and determine the clinical implications of these observations.[46,47] In another study, H. pylori was detected in fecal specimens of most children with CD.[48] Similarly, Laharie et al reported that CD was significantly associated with Helicobacter pullorum or Helicobacter canadensis infection in intestinal biopsies (OR = 2.58; 95% CI: 1.04–6.67).[49] However, other studies yielded contradictory results. In an analysis of intestinal mucosal specimens from patients with IBD, 2 Helicobacter genus-specific PCR assays, 2 H. pylori-specific assays, and a PCR assay designed to amplify fragments of H. heilmannii-like organisms did not reveal any Helicobacter species, which may suggest that Helicobacter is not involved in IBD. A few experts have suggested that H. heilmannii does not play a role in the pathogenesis of IBD.[50] As per the findings of another analogous research study, the presence of Helicobacter species was not identified in the colon biopsies of patients diagnosed with IBD.[51]
Moreover, epidemiological evidence indicates a potential inverse correlation between the prevalence of IBD and H. pylori infection. Regions with a low occurrence of H. pylori infection have experienced a gradual increase in the incidence of IBD following the widespread implementation of treatment plans aimed at eradicating H. pylori.[52] According to a recent meta-analysis, a consistent negative correlation appears to exist between gastric H. pylori infection and IBD, with a statistically significant P-value < 1e−10. This correlation appeared to be more pronounced in patients with CD and IBDU (P-value = 0.38 and 0.43) than in patients with UC (P-value = 0.53) and (P < 1e−10).[53] Several other studies have yielded similar results,[54–58] suggesting that H. pylori infection may play a protective role against IBD. Studies have suggested that H. pylori infection may help in the treatment of IBD, while others disagree. Biopsies revealed no difference in Helicobacter detection between patients with IBD and those with other diseases.[59] In another PCR sequencing study in a Canadian population, no significant difference was observed in the proportion of Helicobacter DNA detected in ileal or colonic samples between patients with IBD and controls. All positive models yielded helicobacter DNA sequences identical to H. pylori 16S rDNA, which was similar to Helicobacter. cinaedi (AF348745), Helicobacter fennelliae (AF348746), Helicobacter heilmannii (AF058768), Helicobacter bilis (AF047843), and Helicobacter hepaticus (AF302103) all differed in 2.5%, 2.5%, 3.6%, 3.9%, and 4.2% of the corresponding sequences, respectively.[60] This finding suggests that no correlation exists between H. pylori infection and IBD.
The controversy surrounding whether H. pylori infection causes IBD arises primarily because of the limitations of observational studies. These studies did not include randomization, prospective analysis, or blinding to account for confounding factors. In contrast, MR selects SNPs as IVs that remain unaffected by environmental and other exposure factors owing to their random distribution.[24] Therefore, we chose MR to analyze H. pylori infection and IBD, which could strictly explain the causal relationship between them.
We used various MR techniques to investigate the causal relationship between H. pylori infection and IBD. However, our extensive bidirectional MR analysis did not provide evidence of a genetically predicted causal association between these 2 conditions.
Despite our efforts, this study has limitations that are worth noting. H. pylori-related antibody indicators were used to assess H. pylori infection. However, these indicators have certain drawbacks that could potentially skew our results, even though we carefully selected several indicators with sensitivity levels exceeding 90%. Furthermore, we acknowledge that there may have been an overlap between participants in the exposure and outcome samples. This overlap may have impacted the accuracy of the MR analysis. However, we attempted to minimize any potential bias by utilizing IVs with strong effects, all of which boasted F-statistics in excess of 10. Our dataset included only European populations. While this choice helped reduce population stratification bias, it is possible that our findings may not be generalizable to other populations.
5. Conclusion
In our study using MR, no direct link was observed between H. pylori infection and the risk of IBD. This suggests that there may not be significant benefits in eradicating H. pylori to prevent or treat regional IBD, and vice versa. However, the limitations of H. pylori serologic index substitution indicate that additional GWAS research based on histologic diagnosis, and further MR studies, are necessary to fully evaluate the relationship between H. pylori infection and IBD.
Author Contributions
Data curation: Kaiqi Yang.
Methodology: Kaiqi Yang.
Software: Kaiqi Yang.
Writing—original draft: Kaiqi Yang.
Visualization: Yuchen Ding.
Resources: Jinlong Chen.
Writing—review & editing: Xiujing Sun.
Abbreviations:
- CD
- Crohns disease
- GWAS
- Genome-wide Association Studies
- H. pylori =
- Helicobacter pylori
- IBD
- inflammatory bowel disease
- IV
- instrumental variable
- LD
- linkage disequilibrium
- MR
- Mendelian randomization
- SNPs
- Single-nucleotide polymorphisms
- UC
- ulcerative colitis.
The authors have no funding and conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
How to cite this article: Yang K, Ding Y, Chen J, Sun X. No potential causal link between HP infection and IBD: A 2way Mendelian randomization study. Medicine 2024;103:8(e37175).
Contributor Information
Kaiqi Yang, Email: yangkaiqi@mail.ccmu.edu.cn.
Yuchen Ding, Email: RichardDing99@163.com.
Jinlong Chen, Email: chenjinlong@mail.ccmu.edu.cn.
References
- [1].Bernstein CN, Fried M, Krabshuis J, et al. Inflammatory bowel disease: a global perspective. 2009.
- [2].Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–7. [DOI] [PubMed] [Google Scholar]
- [3].Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- [4].Danese S, Fiocchi C. Etiopathogenesis of inflammatory bowel diseases. World J Gastroenterol. 2006;12:4807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kugathasan S, Fiocchi C. Progress in basic inflammatory bowel disease research. Semin Pediatr Surg. 2007;16:146–53. [DOI] [PubMed] [Google Scholar]
- [6].Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. [DOI] [PubMed] [Google Scholar]
- [7].Hooi JKY, Lai WY, Ng WK, et al. Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–9. [DOI] [PubMed] [Google Scholar]
- [8].FitzGerald R, Smith SM. An overview of helicobacter pylori infection. Methods Mol Biol. 2021;2283:1–14. [DOI] [PubMed] [Google Scholar]
- [9].Yuan C, Adeloye D, Luk TT, et al.; Global Health Epidemiology Research Group. The global prevalence of and factors associated with Helicobacter pylori infection in children: a systematic review and meta-analysis. Lancet Child Adolesc Health. 2022;6:185–94. [DOI] [PubMed] [Google Scholar]
- [10].Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86. [DOI] [PubMed] [Google Scholar]
- [11].Patel MK, Trombly MI, Kurt-Jones EA. Innate immune responses to Helicobacter pylori infection: an overview. Methods Mol Biol. 2012;921:205–7. [DOI] [PubMed] [Google Scholar]
- [12].Sasaran MO, Melit LE, Dobru ED. MicroRNA modulation of host immune response and inflammation triggered by helicobacter pylori. Int J Mol Sci . 2021;22:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang F, Meng W, Wang B, et al. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. [DOI] [PubMed] [Google Scholar]
- [14].Suarez G, Reyes VE, Beswick EJ. Immune response to H. pylori. World J Gastroenterol. 2006;12:5593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Koloski NA, Bret L, Radford-Smith G. Hygiene hypothesis in inflammatory bowel disease: a critical review of the literature. World J Gastroenterol. 2008;14:165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17. [DOI] [PubMed] [Google Scholar]
- [17].O’Donnell B, Puri P. Long-term results of simple removal of pigment gallstones in childhood. Prog Pediatr Surg. 1977;10:121–7. [PubMed] [Google Scholar]
- [18].Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vare PO, Heikius B, Silvennoinen JA, et al. Seroprevalence of Helicobacter pylori infection in inflammatory bowel disease: is Helicobacter pylori infection a protective factor? Scand J Gastroenterol. 2001;36:1295–300. [DOI] [PubMed] [Google Scholar]
- [20].Rosania R, Von Arnim U, Link A, et al. Helicobacter pylori eradication therapy is not associated with the onset of inflammatory bowel diseases. A case-control study. J Gastrointestin Liver Dis. 2018;27:119–25. [DOI] [PubMed] [Google Scholar]
- [21].Parlak E, Ulker A, Disibeyaz S, et al. There is no significant increase in the incidence of Helicobacter pylori infection in patients with inflammatory bowel disease in Turkey. J Clin Gastroenterol. 2001;33:87–8. [DOI] [PubMed] [Google Scholar]
- [22].Pellicano R, Bresso F, Demarchi B, et al. Prevalence of Helicobacter pylori infection in patients with inflammatory bowel disease: pilot study. Rev Esp Enferm Dig. 2010;102:675–6; author reply 676. [DOI] [PubMed] [Google Scholar]
- [23].Pokrotnieks J, Sitkin S. Helicobacter pylori and inflammatory bowel disease: an unresolved enigma. Inflamm Bowel Dis. 2023;29:e5–6. [DOI] [PubMed] [Google Scholar]
- [24].Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–6. [DOI] [PubMed] [Google Scholar]
- [26].Wang YK, Kuo FC, Liu CJ, et al. Diagnosis of Helicobacter pylori infection: current options and developments. World J Gastroenterol. 2015;21:11221–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yamakawa O, Takemori Y, Noda Y, et al. [Availability of determination of serum anti-Helicobacter pylori IgG antibody in diagnosis of Helicobacter pylori infection]. Nihon Shokakibyo Gakkai Zasshi. 1993;90:2090–6. [PubMed] [Google Scholar]
- [28].Ueda J, Okuda M, Nishiyama T, et al. Diagnostic accuracy of the E-plate serum antibody test kit in detecting Helicobacter pylori infection among Japanese children. J Epidemiol. 2014;24:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Butt J, Blot WJ, Shrubsole MJ, et al. Performance of multiplex serology in discriminating active vs past Helicobacter pylori infection in a primarily African American population in the southeastern United States. Helicobacter. 2020;25:e12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chauhan N, Tay ACY, Marshall BJ, et al. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: an overview. Helicobacter. 2019;24:e12544. [DOI] [PubMed] [Google Scholar]
- [31].Fernandez-de-Larrea N, Michel A, Romero B, et al. Antibody reactivity against Helicobacter pylori proteins in a sample of the Spanish adult population in 2008-2013. Helicobacter. 2017;22. [DOI] [PubMed] [Google Scholar]
- [32].Michel A, Pawlita M, Boeing H, et al. Helicobacter pylori antibody patterns in Germany: a cross-sectional population study. Gut Pathog. 2014;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755–64. [DOI] [PubMed] [Google Scholar]
- [34].Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Papadimitriou N, Dimou N, Tsilidis KK, et al. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020;11:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Keenan JI, Beaugie CR, Jasmann B, et al. Helicobacter species in the human colon. Colorectal Dis. 2010;12:48–53. [DOI] [PubMed] [Google Scholar]
- [41].Meyer F, Wilson KT, James SP. Modulation of innate cytokine responses by products of Helicobacter pylori. Infect Immun. 2000;68:6265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Valle J, Kekki M, Sipponen P, et al. Long-term course and consequences of Helicobacter pylori gastritis. Results of a 32-year follow-up study. Scand J Gastroenterol. 1996;31:546–50. [DOI] [PubMed] [Google Scholar]
- [43].Ram M, Barzilai O, Shapira Y, et al. Helicobacter pylori serology in autoimmune diseases - fact or fiction? Clin Chem Lab Med. 2013;51:1075–82. [DOI] [PubMed] [Google Scholar]
- [44].MacDonald TT, Murch SH. Aetiology and pathogenesis of chronic inflammatory bowel disease. Baillieres Clin Gastroenterol. 1994;8:1–34. [DOI] [PubMed] [Google Scholar]
- [45].Wilson JC, Furlano RI, Jick SS, et al. Inflammatory bowel disease and the risk of autoimmune diseases. J Crohns Colitis. 2016;10:186–93. [DOI] [PubMed] [Google Scholar]
- [46].Oliveira AG, das Gracas Pimenta Sanna M, Rocha GA, et al. Helicobacter species in the intestinal mucosa of patients with ulcerative colitis. J Clin Microbiol. 2004;42:384–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Oliveira AG, Rocha GA, Rocha AM, et al. Isolation of Helicobacter pylori from the intestinal mucosa of patients with Crohn’s disease. Helicobacter. 2006;11:2–9. [DOI] [PubMed] [Google Scholar]
- [48].Man SM, Zhang L, Day AS, et al. Detection of enterohepatic and gastric helicobacter species in fecal specimens of children with Crohn’s disease. Helicobacter. 2008;13:234–8. [DOI] [PubMed] [Google Scholar]
- [49].Laharie D, Asencio C, Asselineau J, et al. Association between entero-hepatic Helicobacter species and Crohn’s disease: a prospective cross-sectional study. Aliment Pharmacol Ther. 2009;30:283–93. [DOI] [PubMed] [Google Scholar]
- [50].Bell SJ, Chisholm SA, Owen RJ, et al. Evaluation of Helicobacter species in inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18:481–6. [DOI] [PubMed] [Google Scholar]
- [51].Huijsdens XW, Linskens RK, Koppes J, et al. Detection of Helicobacter species DNA by quantitative PCR in the gastrointestinal tract of healthy individuals and of patients with inflammatory bowel disease. FEMS Immunol Med Microbiol. 2004;41:79–84. [DOI] [PubMed] [Google Scholar]
- [52].Thia KT, Loftus EV, Jr, Sandborn WJ, et al. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008;103:3167–82. [DOI] [PubMed] [Google Scholar]
- [53].Castano-Rodriguez N, Kaakoush NO, Lee WS, et al. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. 2017;66:235–49. [DOI] [PubMed] [Google Scholar]
- [54].Duggan AE, Usmani I, Neal KR, et al. Appendicectomy, childhood hygiene, Helicobacter pylori status, and risk of inflammatory bowel disease: a case control study. Gut. 1998;43:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].el-Omar E, Penman I, Cruikshank G, et al. Low prevalence of Helicobacter pylori in inflammatory bowel disease: association with sulphasalazine. Gut. 1994;35:1385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Parente F, Molteni P, Bollani S, et al. Prevalence of Helicobacter pylori infection and related upper gastrointestinal lesions in patients with inflammatory bowel diseases. A cross-sectional study with matching. Scand J Gastroenterol. 1997;32:1140–6. [DOI] [PubMed] [Google Scholar]
- [57].Pearce CB, Duncan HD, Timmis L, et al. Assessment of the prevalence of infection with Helicobacter pylori in patients with inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2000;12:439–43. [DOI] [PubMed] [Google Scholar]
- [58].Piodi LP, Bardella M, Rocchia C, et al. Possible protective effect of 5-aminosalicylic acid on Helicobacter pylori infection in patients with inflammatory bowel disease. J Clin Gastroenterol. 2003;36:22–5. [DOI] [PubMed] [Google Scholar]
- [59].Sturegard E, Hertervig E, Sjunnesson H, et al. Helicobacter species in human colon biopsies. Aliment Pharmacol Ther. 2004;19:613–4. [DOI] [PubMed] [Google Scholar]
- [60].Grehan M, Danon S, Lee A, et al. Absence of mucosa-associated colonic Helicobacters in an Australian urban population. J Clin Microbiol. 2004;42:874–6. [DOI] [PMC free article] [PubMed] [Google Scholar]