Abstract
Objective
Testing for the Janus activating kinase 2 (JAK2) V617F mutation is important for diagnosing and treating myeloproliferative neoplasms (MPNs). Recently, urine cell-free DNA (ucfDNA) was reported to be useful for detecting tumor-specific gene mutations in several solid tumors. However, its utility in detecting such mutations in hematological malignancies has not yet been assessed. In this study, we assessed whether or not the JAK2 V617F mutation could be detected in ucfDNA and whether or not its positivity rate in ucfDNA was associated with the JAK2 V617F allele ratio of peripheral blood cells in patients with MPN.
Methods
The JAK2 V617F allele ratio of genomic DNA from peripheral blood cells was determined using quantitative polymerase chain reaction (qPCR) or droplet digital PCR (ddPCR). ucfDNA was subjected to ddPCR. The correlation between the JAK2 V617F mutation positivity rates of blood-derived DNA and those of ucfDNA was assessed.
Materials
Twelve patients with polycythemia vera and 12 patients with essential thrombocythemia were enrolled. Ethylenediaminetetraacetic acid-treated peripheral blood (100 mL) and 15-30 mL of fresh urine were used.
Results
The JAK2 V617F mutation was detected in the ucfDNA from all 20 JAK2 V617F mutation-positive patients. In addition, the JAK2 V617F mutation positivity rate of ucfDNA was correlated with the JAK2 V617F allele ratio of blood-derived DNA, including in both estimated glomerular filtration rate (eGFR) groups (patients with an eGFR ≥50 or <50 mL/min/1.73 m2).
Conclusion
Our results indicate that ucfDNA is a valuable tool for diagnosing and monitoring MPN. Given these findings, other disease-specific gene mutations in hematological malignancies may also be detectable in ucfDNA.
Keywords: JAK2 V617F mutation, urinary cell-free DNA, myeloproliferative neoplasm, digital PCR, liquid biopsy
Introduction
Polycythemia vera (PV) and essential thrombocythemia (ET) are types of myeloproliferative neoplasms (MPNs), which are hematological malignancies resulting from acquired genetic alterations in hematopoietic precursor cells in the bone marrow (1,2). Over the past decade, many causative genetic mutations have been identified in MPN (1). Among these genetic mutations, the Janus activating kinase 2 (JAK2) V617F mutation is found in 92% of PV patients and 55% of ET patients (1). Positivity for this genetic mutation is a diagnostic criterion for these conditions (3). In addition, a high JAK2 V617F allele burden in peripheral blood cells has been reported to be associated with an increased risk of thrombosis in PV patients (4). Thus, testing for the JAK2 V617F mutation provides clinically important information for therapeutic intervention and diagnoses.
Recent advances in nucleotide technology have allowed the detection of low levels of mutation-containing tumor-derived DNA in the blood, cerebrospinal fluid, and urine (5,6). The detection of genetic mutations using bodily fluids (i.e. in a “liquid biopsy”) has been widely applied to cancer diagnostics and monitoring. In solid tumors, it has been reported that tumor-derived genetic mutations can be detected using urinary cell-free DNA (ucfDNA), including lung, uroepithelial, pancreatic, and colon cancers (7-10). However, little is known about the diagnostic applications of ucfDNA in hematopoietic tumors (11).
In the present study, we detected the JAK2 V617F mutation in urine samples obtained from patients with PV or ET using the droplet digital polymerase chain reaction (ddPCR) method to develop non-invasive diagnostic or monitoring tests for these diseases. We compared the results of this technique with the mutated allele burden in the peripheral blood cells.
Materials and Methods
Patients
This study was approved by the Wakayama Medical University Ethics Committee (No. 3817) and conducted in accordance with the Declaration of Helsinki. Twenty patients positive for the JAK2 V617F mutation were enrolled in this study after providing their written informed consent. Four patients negative for the JAK2 V617F mutation were also enrolled. All patients were diagnosed with PV or ET at Wakayama Medical University Hospital and visited our hospital between March and May 2023 for routine follow-up. Urine samples from five healthy volunteers were used as controls.
DNA preparation
Genomic DNA was isolated from 100 μL of EDTA-treated peripheral blood using a DNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Urine was collected from five healthy volunteers, in addition to PV and ET patients. Urine was collected during outpatient visits (the time of which varied), but samples were not collected from the first urine in the morning. A total of 15-30 mL of fresh urine was then centrifuged at 1,600×g for 10 min at room temperature within 3 h of collection. After centrifugation, the supernatant was separated into 10-mL samples and stored at -80°C. When fresh urine could not be processed within 3 h, 500 μL of urine preservation solution (StreckⓇ Urine Preserve; Streck, La Vista, USA) was added to 10 mL of urine at room temperature before storage. Within five days of the StreckⓇ Urine Preserve being added to the urine sample, the urine was centrifuged, and the supernatant was stored.
After thawing at 37°C, the supernatant was centrifuged at 16,000×g at 4°C. Subsequently, ucfDNA was extracted from 5 mL of the supernatant using a QIAampⓇ MinEluteⓇ ccfDNA Midi kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Finally, ucfDNA was eluted in 25 μL TE buffer. The amount of ucfDNA was quantified using a Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's protocol. The ucfDNA concentration was calculated as the ucfDNA content in nanograms per milliliter of urine.
ddPCR
ucfDNA was analyzed for the JAK2 V617F mutation using ddPCR (QX200 system; Bio-Rad Laboratories, Hercules, USA). The ddPCR primers used for detecting JAK2 V617F were purchased from Bio-Rad (Unique Assay ID: dHsaMDV27944642). The ddPCR assays were conducted according to the manufacturer's protocol. In brief, 20 μL of ddPCR reaction solution was prepared by adding 10 μL of ddPCR Supermix and 1 μL of ddPCR mutation assay mixture to a maximum of 2 ng of ucfDNA. The ddPCR mutation assay consisted of wild-type and JAK2 V617F mutation probes labeled with HEX and FAM fluorochromes, respectively. A droplet was then generated from the reaction mixture using a QX200 droplet generator (Bio-Rad Laboratories). The droplet was then cycled as follows: denaturation at 95°C for 10 min, followed by 40 reaction cycles (94°C for 30 s and 55°C for 1 min), and a final cycle at 98°C for 10 min. The plate was incubated overnight at 12°C to stabilize the droplets (12). Droplet fluorescence was assessed using a QX200 droplet reader. DNA isolated from HEL cells (purchased from the Japan Bioresearch Bank, Izumisano, Japan), which are known to possess the JAK2 V617F mutation, was used as a positive control. ddPCR data were analyzed using the QuantaSoft software program (version 2.0; Bio-Rad Laboratories). The JAK2 V617F mutation positivity rate was calculated as follows: JAK2 V617F mutation-positive droplets/(JAK2 V617F mutation-positive droplets+wild-type positive droplets) (13). As reported previously, we defined a positive JAK2 V617F mutation as a fractional abundance of >0.1% (14).
Estimation of the JAK2 V617F allele ratio of gDNA derived from blood
To estimate the JAK2 V617F allele ratio in the blood, we performed quantitative PCR (qPCR) using 100 ng of genomic DNA obtained from the peripheral blood cells of each JAK2 V617F mutation-positive patient. The allele ratio was calculated based on the melting curve as previously described (15). JAK2 V617F allele ratio results were not obtained from a melting curve assay for two patients for whom blood samples were not collected during the study period. Instead, the results obtained at the diagnosis were used to assess the ratio in one case, and a ddPCR analysis of DNA extracted from the blood was used to assess the ratio in the other case.
Statistical analyses
All data are reported as the mean±standard error of the mean (SEM), unless otherwise noted. An unpaired t-test was used to determine the significance of intergroup differences in ucfDNA concentrations. The correlation of JAK2 V617F mutation positivity between blood-derived DNA and ucfDNA was evaluated using Pearson's correlation coefficient. p values of <0.05 were considered statistically significant. All statistical analyses were performed using the Excel (Microsoft, Redmond, USA) or GraphPad Prism 9 software programs (GraphPad Software, San Diego, USA).
Results
Patient characteristics
Twenty patients positive for JAK2 V617F mutations were included in this study, and 10 were diagnosed with PV, while 10 were diagnosed with ET (Table). Three (15%) patients developed secondary myelofibrosis. Twelve patients received hydroxyurea, and five received the JAK1/2 inhibitor ruxolitinib. The mean white blood cell count at urine collection was 8,880±828 /μL. The mean serum creatinine level and estimated glomerular filtration rate (eGFR) were 0.96±0.08 mg/dL and 53.7±3.4 mL/min/1.73 m2, respectively. Nine patients (45%) had an eGFR of <50 mL/min/1.73 m2. StreckⓇ Urine Preserve solution was used to collect urine samples from 10 patients (50%).
Table.
Clinical Characteristics of JAK2 V617F Mutation-positive Patients Whose Urine Cell-free DNA was Tested.
| Characteristics | Total (n=20) |
|---|---|
| Median age (range) | 72.5 (49-85) |
| Male, n (%) | 6 (30) |
| Disease | |
| Polycythemia vera, n (%) | 10 (50) |
| Essential thrombocythemia, n (%) | 10 (50) |
| Progression to secondary myelofibrosis, n (%) | 3 (15) |
| Therapy, n (%) | |
| Hydroxyurea | 12 (60) |
| Ruxolitinib | 5 (25) |
| Anagrelide1 | 2 (10) |
| No therapy | 2 (10) |
| Laboratory data at urine sample collection | |
| White blood cell count (/µL) | 8,880±828 |
| Red blood cell count (×104/μL) | 359±24 |
| Hematocrit (%) | 36.1±2.1 |
| Platelet (×104/μL) | 44.4±6.7 |
| Serum creatinine level (mg/dL) | 0.96±0.08 |
| Estimated glomerular filtration rate (mL/min/1.73 m2) | 53.7±3.4 |
| Median urinary pH (range) | 6 (6-6.5) |
| Urine specific gravity | 1.013±0.001 |
Laboratory data are shown as the mean±SEM (except urinary pH).
1One patient received a combination of hydroxyurea and anagrelide.
Four patients negative for the JAK2 V617F mutation were also analyzed. Two of these patients were diagnosed with PV, and two were diagnosed with myelofibrosis that developed from CALR mutation-positive ET.
ucfDNA concentration
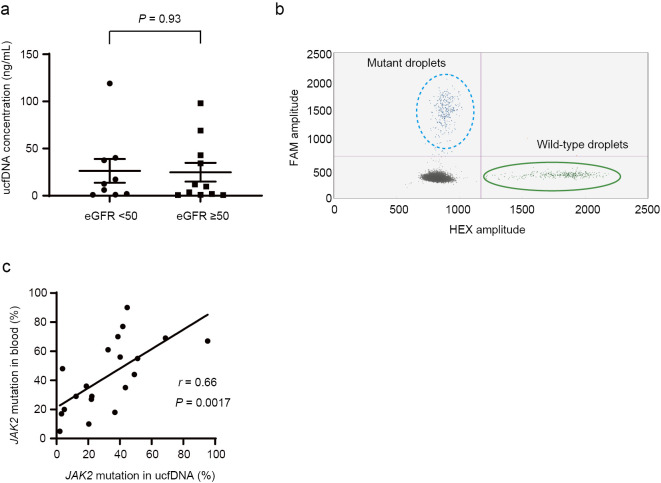
The mean ucfDNA concentration in all PV or ET patients was 25.5±7.7 (range: 0.7-119) ng/mL, whereas the mean ucfDNA concentration in the healthy volunteers was 2.0±0.8 (range: 0.6-4.2) ng/mL (Supplementary material). The mean ucfDNA concentrations were 28.3±10.4 and 22.7±11.7 ng/mL in the patients with PV and ET, respectively (p=0.72) ng/mL. There was no significant difference in the ucfDNA concentration between the patients with an eGFR of <50 and those with an eGFR of ≥50 (eGFR <50 group: 26.3±12.6 vs. eGFR ≥50 group: 24.9±9.9; p=0.93, Fig. 1a). There was also no significant difference in the ucfDNA concentration between the samples that were and were not collected using StreckⓇ Urine Preserve solution (p=0.44).
Figure 1.
(a) Urine cell-free DNA (ucfDNA) concentrations in JAK2 V617F mutation-positive patients with an estimated glomerular filtration rate (eGFR) of <50 or ≥50 mL/min/1.73 m2. (b) Representative scatterplots of JAK2 V617F mutation detection in ucfDNA using digital droplet polymerase chain reaction (ddPCR). In this sample, the ucfDNA concentration was 0.9 ng/mL. Although the ucfDNA concentration was <1 ng/mL, the JAK2 V617F mutation was detectable. Blue droplets: JAK2 V617F mutation-positive; green droplets: JAK2 wild-type. (c) Correlation of JAK2 V617F mutation (%) positivity between blood-derived DNA and ucfDNA among all JAK2 V617F mutation-positive patients.
JAK2 V617F detection in ucfDNA
ddPCR detected no JAK2 V617F mutation-positive droplets in any of the 5 healthy subjects. In contrast, JAK2 V617F mutations were detectable using ucfDNA in all 20 patients with the JAK2 V617F mutation. Although four patients had ucfDNA concentrations of <1 ng/mL, ddPCR still detected the JAK2 V617F mutation in these patients (Fig. 1b). No mutation-positive droplets were detected in any of the four JAK2 V617F mutation-negative patients (Supplementary material).
A comparison of the allele ratios of blood-derived DNA and positivity of ucfDNA for the JAK2 V617F mutation
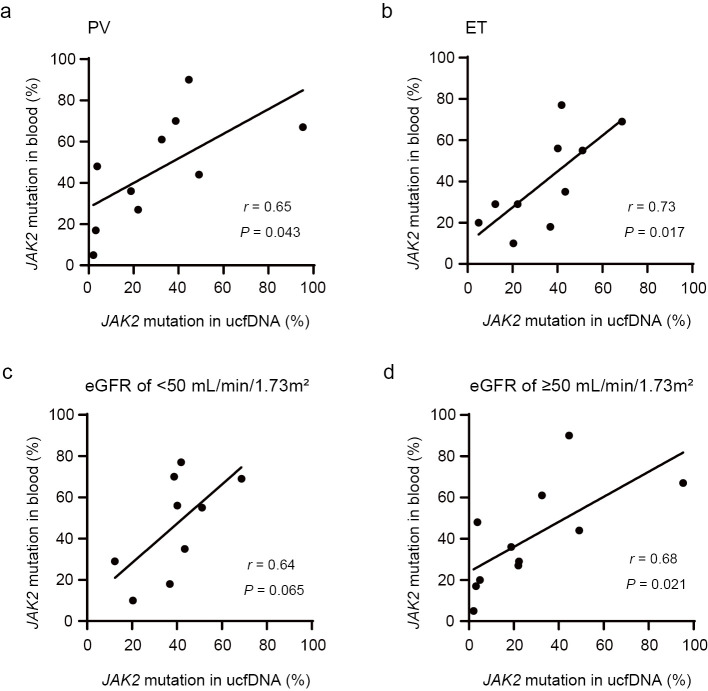
The correlation of the JAK2 V617F mutation positivity rates between DNA derived from blood and ucfDNA was assessed (Fig. 1c). The JAK2 V617F mutation positivity rate of ucfDNA correlated with the JAK2 V617F allele ratio in blood-derived DNA (r=0.66). The coefficient of correlation between the JAK2 V617F positivity rate of ucfDNA and the JAK2 V617F allele burden of blood-derived DNA was 0.65 and 0.73 in patients with PV and ET, respectively (Fig. 2a, b). There was a positive correlation between the JAK2 V617F allele ratio in blood and the JAK2 V617F positivity rate of ucfDNA in both eGFR groups (Fig. 2c, d).
Figure 2.
Correlation of JAK2 V617F mutation (%) positivity between blood-derived DNA and ucfDNA among JAK2 V617F mutation-positive patients with polycythemia vera (PV) (a), essential thrombocythemia (ET) (b), an eGFR of <50 mL/min/1.73 m2 (c), or an eGFR of ≥50 mL/min/1.73 m2 (d).
Discussion
This study showed that a hematopoietic tumor-specific genetic mutation, JAK2 V617F, could be detected in urine samples using ddPCR. In addition, the JAK2 V617F mutation positivity rate of ucfDNA was correlated with the JAK2 V617F allele frequency in the blood. The JAK2 V617F positivity rate of ucfDNA and the JAK2 V617F allele ratio of blood were correlated in both patients with eGFR values <50 and ≥50 mL/min/1.73 m2.
The detection of tumor-specific genetic mutations in the urine has been reported in solid tumors (7-10). Hematological tumor cells are found in the peripheral blood and may proliferate near blood or lymphatic vessels. ucfDNA is filtered into urine from the blood by the renal glomerular apparatus (16). Therefore, the amount of cell-free DNA derived from hematological tumors excreted in urine would be greater than the amount of cell-free DNA derived from solid tumors excreted in urine, suggesting that ucfDNA may be useful for detecting hematological tumor-specific mutations. However, there is little published information on the detection of genetic mutations in urine samples from patients with hematological tumors (11).
The detection rates of mutations in ucfDNA derived from solid tumors, such as lung, pancreatic, and colorectal cancers, have been reported. A previous study on patients with epidermal growth factor receptor (EGFR) mutation-positive non-small-cell lung cancer showed that the sensitivity of such EGFR mutations for detecting non-small-cell lung cancer was 67-72% (7). Another study reported KRAS mutations in ucfDNA in 48% of patients with pancreatic ductal adenocarcinoma (9). In patients with colorectal cancer, the sensitivity of detecting KRAS mutations in ucfDNA for diagnosing colorectal cancer was reported to be 33.3% (17). To our knowledge, only one study has detected genetic mutations in ucfDNA obtained from patients with hematological malignancies (18). That study detected the myeloid differentiation primary response gene 88 (MYD88) L265P mutation in primary central nervous system lymphoma (18). MYD88 mutations were detected in the ucfDNA of 75% of patients. Our study showed that the JAK2 V617F mutation was detected in the ucfDNA of all PV and ET patients that possessed this mutation. These results suggest that the detection rate of tumor-specific gene mutations using ucfDNA may be higher in hematological malignancies than in solid tumors.
The JAK2 V617F variant allele frequency (VAF) is associated with outcomes, including thrombosis and myelofibrotic progression in PV (4). In addition, a reduction in the JAK2 V617F VAF was found to be associated with hematological response in PV patients treated with interferon (19). Thus, information about not only the presence or absence of the JAK2 V617F mutation but also the mutation positivity rate is required during the treatment of PV. However, no previous studies of ucfDNA have reported correlations between VAF in plasma and the ucfDNA mutation positivity rate. We observed a positive correlation between the JAK2 V617F mutation positivity rate for blood-derived DNA and the JAK2 V617F mutation positivity rate for ucfDNA. Thus, our study suggests that urine can be used to monitor hematological malignancies in which tumor cells are typically found in bone marrow and blood cells.
ddPCR allows the detection of small amounts of mutated ucfDNA. As shown in Fig. 1c, the JAK2 V617F mutation was detectable in ucfDNA from PV and ET patients whose ucfDNA concentrations were <1 ng/mL. Our results suggest that ddPCR can detect disease-specific mutant genes in the urine of patients with hematological malignancies, even if the ucfDNA concentration is low.
Most ucfDNA originates from cfDNA in the plasma, which is subjected to glomerular filtration by the kidneys. Therefore, the glomerular filtration rate may affect the detection sensitivity of mutated genes in ucfDNA. Indeed, previous studies on solid tumors reported that the detection sensitivity of mutated genes in ucfDNA in patients with a creatine clearance rate (CCr) ≥70 mL/min was significantly lower than that in patients with a CCr <70 mL/min (9,17). In our study, the JAK2 V617F mutation was detectable in all patients using ucfDNA, regardless of the renal function. In addition, the JAK2 V617F positivity rates of blood and ucfDNA were correlated, even among patients with an eGFR ≥50 mL/min/1.73 m2. Thus, the effect of the renal function on mutation detection may be less important when detecting tumor-specific mutations in ucfDNA in hematological malignancies than solid tumors. Further investigations are needed to determine whether or not tumor-specific genetic mutations are more easily detected in ucfDNA in hematological malignancies than in solid tumors, regardless of the renal function, owing to the large number of tumor cells in the blood.
Urinary tests are noninvasive and easily repeated. To obtain plasma-derived cfDNA, blood must be collected in an expensive cfDNA collection tube. In contrast, urine-derived cfDNA samples can be collected simply by patients urinating at home. Therefore, this approach has merit in clinical practice. However, the use of such tests for analyzing genetic mutations appears to be limited, as ucfDNA is more quickly degraded than that found in the serum (20). Our study showed that urine samples could be used to detect the JAK2 V617F mutation for two hours after collection, without any preservation methods being required. A previous study showed that ucfDNA yields were unchanged by delaying sample processing for up to 45 min after urine collection in healthy volunteers (21). Furthermore, that study also demonstrated that the fragment size distribution of ucfDNA was unaffected for up to four hours before the addition of preservatives (21). Our present study suggests that cfDNA derived from urine can be utilized in routine clinical practice to detect mutations using ddPCR in patients with hematological malignancies. In addition, StreckⓇ Urine Preserve solution allows urine samples to remain unprocessed for several days. In our study, urine was collected during outpatient visits, the times of which varied. The ucfDNA fragment size distribution of the first void is reported to be similar to that of subsequent voids (21). In all of our patients, the JAK2 V617F mutation was detected in ucfDNA, suggesting that this gene mutation may be detectable in ucfDNA extracted from urine collected from patients with hematological malignancies, regardless of the time at which the urine is collected. The extraction of ucfDNA may be simpler than that of plasma-derived cfDNA.
In addition to the non-invasiveness of ucfDNA analyses, such analyses may be less markedly affected by foreign substances, such as cells and proteins, than blood-derived cfDNA analyses (16,22). This may reduce the detection of clonal hematopoiesis-related mutations and facilitate the detection of mutated genes in tumor-associated cfDNA (11,23). These advantages may facilitate the analysis of ucfDNA in addition to blood-derived cfDNA, even in patients with hematological malignancies.
Several limitations associated with the present study warrant mention. First, this was a single-center study with a small sample size. Second, the specificity of the ucfDNA mutation detection test could not be determined, as mutations were detected in all cases. Third, mutations in ucfDNA were only evaluated using ddPCR. Direct sequencing was not performed because the amount of cfDNA was low.
In conclusion, our study confirmed that JAK2 V617F could be detected in urine using ddPCR. Our results indicate that ucfDNA is a valuable tool for diagnosing and monitoring patients with MPN. These findings suggest that other disease-specific gene mutations in hematological malignancies can be detected using ucfDNA. A comparison of genomic mutations found in the blood and urine might be useful for understanding residual clones after therapeutic intervention in patients with hematological malignancies.
The authors state that they have no Conflict of Interest (COI).
Financial Support
This study was supported by a grant (KAKENHI 21K16248) from the Japan Society for the Promotion of Science (JSPS) and a grant from the Takeda Science Foundation to HH. This study was also supported by a grant (KAKENHI 22K20799) to YH, a grant (KAKENHI 19K08821) to TS from JSPS, and a grant from the Takeda Science Foundation to NH.
Acknowledgement
We thank the patients and clinical and laboratory staff at Wakayama Medical University Hospital for participating in this study.
References
- 1.Spivak JL. Myeloproliferative neoplasms. N Engl J Med 376: 2168-2181, 2017. [DOI] [PubMed] [Google Scholar]
- 2.How J, Garcia JS, Mullally A. Biology and therapeutic targeting of molecular mechanisms in MPNs. Blood 141: 1922-1933, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tefferi A, Barbui T. Polycythemia vera and essential thrombocythemia: 2021 update on diagnosis, risk-stratification and management. Am J Hematol 95: 1599-1613, 2020. [DOI] [PubMed] [Google Scholar]
- 4.Moliterno AR, Kaizer H, Reeves BN. JAK2V617F allele burden in polycythemia vera: burden of proof. Blood 141: 1934-1942, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley JC, Diehn M. Detection and diagnostic utilization of cellular and cell-free tumor DNA. Annu Rev Pathol 16: 199-222, 2021. [DOI] [PubMed] [Google Scholar]
- 6.Dang DK, Park BH. Circulating tumor DNA: current challenges for clinical utility. J Clin Invest 132: e154941, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 11: 1690-1700, 2016. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi Y, Fujita K, Matsuzaki K, et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci 110: 1771-1779, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terasawa H, Kinugasa H, Ako S, et al. Utility of liquid biopsy using urine in patients with pancreatic ductal adenocarcinoma. Cancer Biol Ther 20: 1348-1353, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellini B, Pejovic N, Feng W, et al. ctDNA MRD detection and personalized oncogenomic analysis in oligometastatic colorectal cancer from plasma and urine. JCO Precis Oncol 5, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tivey A, Church M, Rothwell D, Dive C, Cook N. Circulating tumour DNA - looking beyond the blood. Nat Rev Clin Oncol 19: 600-612, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowlands V, Rutkowski AJ, Meuser E, Carr TH, Harrington EA, Barrett JC. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep 9: 12620, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterhouse M, Follo M, Pfeifer D, et al. Sensitive and accurate quantification of JAK2 V617F mutation in chronic myeloproliferative neoplasms by droplet digital PCR. Ann Hematol 95: 739-744, 2016. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe J, Natsumeda M, Okada M, et al. High detection rate of MYD88 mutations in cerebrospinal fluid from patients with CNS lymphomas. JCO Precis Oncol 3: 1-13, 2019. [DOI] [PubMed] [Google Scholar]
- 15.Lay M, Mariappan R, Gotlib J, et al. Detection of the JAK2 V617F mutation by LightCycler PCR and probe dissociation analysis. J Mol Diagn 8: 330-334, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salfer B, Li F, Wong DTW, Zhang L. Urinary cell-free DNA in liquid biopsy and cancer management. Clin Chem 68: 1493-1501, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta R, Yamada T, Sonoda H, et al. Detection of KRAS mutations in circulating tumour DNA from plasma and urine of patients with colorectal cancer. Eur J Surg Oncol 47: 3151-3156, 2021. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe J, Natsumeda M, Kanemaru Y, et al. Comparison of circulating tumor DNA between body fluids in patients with primary central nervous system lymphoma. Leuk Lymphoma 60: 3587-3589, 2019. [DOI] [PubMed] [Google Scholar]
- 19.Gisslinger H, Klade C, Georgiev P, et al. Ropeginterferon alfa-2b versus standard therapy for polycythaemia vera (PROUD-PV and CONTINUATION-PV): a randomised, non-inferiority, phase 3 trial and its extension study. Lancet Haematol 7: e196-e208, 2020. [DOI] [PubMed] [Google Scholar]
- 20.Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene 590: 142-148, 2016. [DOI] [PubMed] [Google Scholar]
- 21.Markus H, Zhao J, Contente-Cuomo T, et al. Analysis of recurrently protected genomic regions in cell-free DNA found in urine. Sci Transl Med 13: eaaz3088, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su YH, Wang M, Brenner DE, et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn 6: 101-107, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R, Zang J, Xie F, et al. Urinary molecular pathology for patients with newly diagnosed urothelial bladder cancer. J Urol 206: 873-884, 2021. [DOI] [PubMed] [Google Scholar]