Abstract
Transcriptional heterogeneity of tumor-associated macrophages (TAMs) has been investigated in individual cancers, but the extent to which these states transcend tumor types and represent a general feature of cancer remains unclear. We performed pan-cancer single-cell RNA sequencing analysis across nine cancer types and identified distinct monocyte/TAM composition patterns. Using spatial analysis from clinical study tissues, we assessed TAM functions in shaping the tumor microenvironment (TME) and influencing immunotherapy. Two specific TAM clusters (pro-inflammatory and pro-tumor) and four TME subtypes showed distinct immunological features, genomic profiles, immunotherapy responses, and cancer prognosis. Pro-inflammatory TAMs resided in immune-enriched niches with exhausted CD8+ T cells, while pro-tumor TAMs were restricted to niches associated with a T-cell-excluded phenotype and hypoxia. We developed a machine learning model to predict immune checkpoint blockade response by integrating TAMs and clinical data. Our study comprehensively characterizes the common features of TAMs and highlights their interaction with the TME.
Subject terms: Cancer microenvironment, Tumour biomarkers
Introduction
The tumor microenvironment (TME) plays an important role in clinical outcomes and response to therapy1. It is now recognized that the TME contains numerous immune cells, of which tumor-associated macrophages (TAMs) tend to be the most abundant2. We previously reported that TAMs are associated with multiple potent functions, including but not limited to tumor cell invasion, cancer stem cells, therapy resistance, and crosstalk with other cancer cells3–5. Connections between the molecular and functional diversities of TAMs are emerging. It is crucial to elucidate the heterogeneity of TAMs by identifying their molecular signatures to ensure their precise targeting.
Data illustrating the cellular heterogeneity in cancer have increased in recent years owing to the widespread application of single-cell RNA-sequencing (scRNA-seq) technology. Recent pan-cancer studies have characterized the molecular diversity of TAMs6–8; however, the considerable heterogeneity observed among these cells prompts the need for more in-depth analysis. It is also crucial to examine the biological relevance and clinical implications of TAM subsets in larger cohorts of patients, particularly concerning their role in immunotherapy. Additionally, how TAMs shape the TME by interacting with other TME components remains to be determined. Recent advances in the spatial transcriptome have allowed the simultaneous identification of diverse cells in the TME; therefore, combining single-cell transcriptome data with spatial transcriptome data will contribute to an improved understanding of the inter-cellular communication within the TME.
Recent studies have identified an association between increased TAM infiltration, along with the regulation of T-cell activation, and its effect on immunotherapy response in tumors9,10. Furthermore, the clinical association between TAMs and their relationship to immunotherapy remains to be established, and there remains a need to understand the biological properties and signaling pathways activated by TAMs in the TME, which, in turn, may influence the response to immunotherapy.
Herein, we used scRNA-seq data and classification of The Cancer Genome Atlas (TCGA) database to identify specific transcriptional, immunological and genomic features for monocyte/TAM clusters across cancers, with a particular focus on immunotherapy-related TAMs. Additionally, we used a spatial profiling assay to assess the multifaceted functions of TAMs in molding the TME. Finally, by fully integrating TAMs, a machine learning algorithm was developed to accurately predict the probability of immunotherapy response in patients.
Result
Identification and molecular characterization of monocyte/TAM clusters
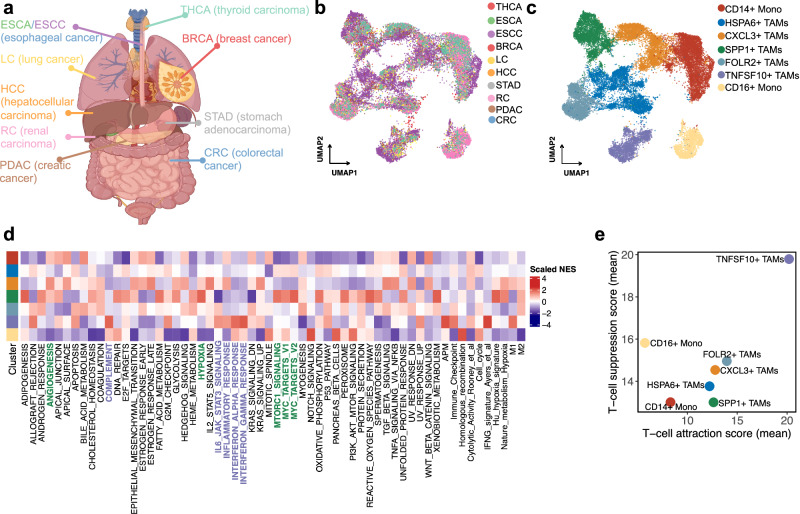
We analyzed the scRNA-seq data of monocytes/macrophages across nine cancer types through the combination of eight scRNA-seq datasets (Fig. 1a). Filtering out adjacent normal tissue, peripheral blood, and nonsignificant clusters, the integrated data contained 24,983 monocytes/macrophages from primary treatment-naïve tumor tissues. High-resolution clustering of the integrated data identified seven clusters, comprising two groups of Monos, and five groups of TAMs (Fig. 1b, c). Significantly, each subtype contained cells from a different tumor type, indicating that the clusters and expression states were largely consistent and did not represent patient-specific subpopulations or batch effects, although they do vary in their proportions across nine cancer types (Supplementary Fig. 1a). Next, we conducted differentially expressed gene (DEG) analysis (Supplementary Fig. 1b) and gene set enrichment analysis (GSEA) (Fig. 1d) between Mono/TAM clusters. CD14+ Mono, indicative of classical monocytes, were characterized by high expression levels of CD14 and S100A8/9, typically recruited during inflammation. These cells exhibited expression of monocyte trafficking factors such as SELL, which is involved in endothelial cell adhesion. CD16+ Mono, representing non-classical monocytes, were identified by their low expression of CD14 coupled with high expression of FCGR3A and CDKN1C, making them less abundant. HSPA6+ TAMs display a complex expression profile that includes components of the major histocompatibility complex class II (HLA-DPB1, HLA-DRA, HLA-DPA1, and HLA-DRB1) and molecular chaperones (HSPA1A and HSPA6), underscoring their role in antigen presentation and stress response. CXCL3+ TAMs are characterized by the expression of G0S2, indicative of their involvement in lipid metabolism and apoptosis regulation. FOLR2+ TAMs are marked by the expression of FOLR2 and a diverse array of genes such as SLC40A1, C1QC, C1QA, and SEPP1, indicating their involvement in iron homeostasis, activation of the complement system, and selenium metabolism, respectively. SPP1+ TAMs showed preferential expression of genes involved in hypoxia, angiogenesis, epithelial-to-mesenchymal transition (EMT), and the mammalian target of rapamycin signal pathway, which promote multiple aspects of tumor progression (Fig. 1d and Supplementary Fig. 1C). Using SCENIC, we found enhanced expression of the transcription factors TP53, PBX3, and MITF in SPP1+ TAMs (Supplementary Fig. 1d). Moreover, TNFSF10+ TAMs expressed genes associated with interferon (IFN)-regulated, complement, IL-6/JAK/STAT3, APM, and canonical M1 signatures, highlighting TNFSF10+ TAMs as important sources of inflammatory response and immunoregulatory molecules (Fig. 1d and Supplementary Fig. 1e). High expression of immune checkpoint molecules, such as CD274, as well as LGALS9, PDCD2, and HAVCR2 in TNFSF10+ TAMs can be regulated by STAT1, indicating that they contribute to T-cell suppression (Supplementary Fig. 1d, f).
Fig. 1. Comprehensive analysis of pan-cancer single-cell transcriptome and functional characteristics of monocytes/TAMs.
a Schematic representation of pan-cancer single-cell transcriptome of monocytes/TAMs. Figures were created using BioRender (biorender.com) with authorized permission. b, c The UMAP plot of all monocyte/TAM clusters. d Cancer cell pathway and immune-related pathways enrichment analysis in seven monocyte/TAM clusters. e Scatterplot of the mean T cell-attraction score versus the mean T cell-suppression score for monocyte/TAM clusters.
We next sought to define gene signatures with the aim of inferring the abundance and functions of SPP1+ and TNFSF10+ TAMs within other scRNA datasets and bulk tumor transcriptomes. To this end, we conducted a single-cell DEG analysis, before filtering the DEGs list to identify the most significant DEGs, screening each of the DEGs based on their ability to selectively discriminate specific TAM clusters (Supplementary Table 1 and the “Methods” section). The top expressed genes in the TNFSF10+ TAM signature included the guanylate-binding family proteins GBP1, GBP4, and GBP5, which are induced in IFN-γ-activated macrophages and promote inflammatory signaling within the innate immune system via inflammasome assembly11. Genes associated with IFN responsiveness (MX1, IFIT3, RSAD2, PARP14, CXCL9-10, and STAT1), innate immune responses (SAMD9L), and antigen presentation (PSMB9 and TAP1) were also identified in the TNFSF10+ TAM signature. We next quantified the T-cell-attractive and T-cell-suppressive properties of all monocyte/TAM clusters, the results of which showed that TNFSF10+ TAMs have a strong potential to recruit T cells and subsequently suppress T-cell function (Fig. 1e). The core signatures of SPP1+ TAMs can be represented by the expression of ECM deposition and remodeling factors (SPP1, FN1, CTSB, and CTSD), suggesting a role in promoting metastasis. Transcriptional signatures specific to SPP1+ TAMs were also identified, including lipid-related genes (APOC1 and APOE)12, TREM213, and SDC214, which may actively suppress antitumor immune responses. We refined these signatures by probing the expression of these transcripts across multiple monocytes/macrophages across nine cancer types, only retaining genes specific for TNFSF10+ and SPP1+ TAM clusters (Supplementary Fig. 2a). We then validated these features in independent gastric and large pan-cancer cohorts (GSE183904 and PMID: 35931863). As a result, we identified two distinct clusters of macrophages, which were identified as SPP1+ and TNFSF10+ TAMs based on scoring cells for our previously derived gene signatures (Supplementary Fig. 2b, c).
Based on their signature genes, enriched pathways, and predicated function, we named these TNFSF10+ and SPP1+ TAMs pro-inflammatory TAMs and pro-tumor progression TAMs (pro-inflam TAMs and pro-tumor TAMs), respectively. Through a cross-sectional comparison of three recently published monocyte/macrophage scRNA-seq datasets, cluster similarity analysis showed that our monocyte/TAM clusters shared similarities but were distinct in specific cancer types (Supplementary Fig. 2d). Thus, our analysis expanded our understanding of common pan-cancer monocyte/macrophage characteristics.
Four distinct subtypes based on pro-inflamed and pro-tumor TAMs across pan-cancer levels
Next, we conducted a pan-cancer analysis in the GTEx and TCGA consortia to compare pro-inflam and pro-tumor TAMs between non-disease healthy tissue and tumor tissue. We discovered that pro-inflam and pro-tumor TAMs were consistently low across normal tissues, but upregulated in a variety of tumors (Supplementary Fig. 3a, b). Compared with pro-inflam TAMs, pro-tumor TAMs were superior in discriminating patient survival within multiple cancer types (Supplementary Fig. 3c). We implemented the CIBERSORT algorithm to infer the differential abundance of immune cells associated with pro-inflam and pro-tumor TAM signatures from 9164 TCGA tumor specimens. Consistent with findings that macrophage-derived CXCL9 and CXCL10 are required for T-cell infiltration15, pro-inflam TAMs were associated with a high level of M1 macrophages and T cells (Supplementary Fig. 3d). Pro-tumor TAMs were related to TME lacking effector T cells and with a mixture of M0, M1, and M2 macrophages (Supplementary Fig. 3d).
To stratify various types of cancer based on pro-inflam and pro-tumor TAMs, we used RNA-seq data from a cohort of 9164 TCGA tumors across 32 cancer types. We first applied consensus clustering to identify two groups of robustly co-expressed pro-inflam and pro-tumor TAM genes to be used for subtyping (Supplementary Fig. 4a). Then, we confirmed that these markers obtained from scRNA-seq could be used in bulk RNA-seq data by measuring gene co-expression (Supplementary Fig. 4b). Furthermore, our analyses showed a positive correlation between the expression of pro-inflam and pro-tumor TAM markers in bulk tumors (Supplementary Fig. 4c). Significantly, we identified no correlation between pro-inflam and pro-tumor TAM marker scores and tumor purity, indicating that tumor purity is not a pertinent confounder for these transcriptomic signatures (Supplementary Fig. 4d). To further clarify the potentially relevant immune cells for pro-inflam and pro-tumor TAM markers, we identified the immune cell predictors for these TAM markers by random forest (RF) analysis. M1 macrophages were found to be the important variable for predicting pro-inflam TAMs, whereas M2 macrophages predicted pro-tumor TAMs (Supplementary Fig. 4e, f). In summary, we showed that the expression of our pro-inflam and pro-tumor TAMs marker gene sets derived from scRNA-seq data are highly specific for macrophages and sufficient to further sample stratification by bulk RNA-seq.
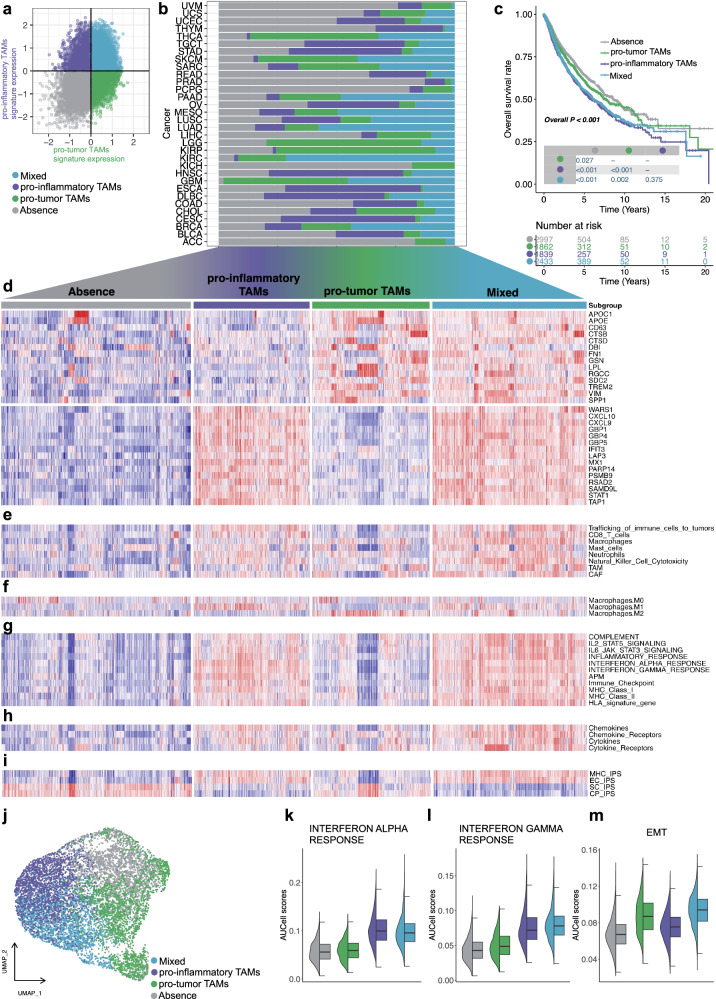
Next, we calculated the median expression levels of co-expressed pro-inflam and pro-tumor TAM genes for each sample and used them to assign one of the following four distinct subtypes associated with these two TAMs: absence, pro-inflam TAMs, pro-tumor TAMs, and mixed (Fig. 2a, d and Supplementary Fig. 2e). These TAM subtypes existed in 28 cancer types at varying abundances (Fig. 2b). The estimated 5-year overall survival (OS) rate were 64.4% (absence, 95% confidence interval (CI): 61.9–66.9%), 60.8% (pro-tumor TAMs, 95% CI: 57.7–64.1%), 53.9% (pro-inflam TAMs, 95% CI: 50.7–57.3%), and 55.4% (mixed, 95% CI: 52.6–58.2%), respectively. Pro-inflam TAM-enriched cases were associated with the shortest OS (Fig. 2c), consistent with the role of the inflammatory TME in tumor progression16. Interestingly, a survival benefit was observed in the absence of pro-inflam and pro-tumor TAM expression (Fig. 2c). The enrichment scores for immune infiltration (Fig. 2e), M1 macrophages infiltration (Fig. 2f), inflammatory potential (Fig. 2g), antigen presentation (Fig. 2g, i), cytokines, and chemokines (Fig. 2h) were greater in the pro-inflam TAM-enriched subtypes, and were lower in the pro-tumor TAMs and absence subtypes. Additionally, the pro-tumor TAM subtype was devoid of T-cell infiltration (Fig. 2e). In summary, the pro-inflam TAM-enriched subtypes were enriched in immunologically ‘hot’ tumors, and pro-tumor TAMs are an important determinant of a T-cell-excluded tumor phenotype across different cancer types within TCGA.
Fig. 2. Expression levels, subtype distribution, survival analysis, and immune-related signatures of pro-inflammatory and pro-tumor TAMs across pan-TCGA samples.
a Scatter plot showing median expression levels of co-expressed pro-inflammatory and pro-tumor TAMs genes in each pan-TCGA sample. b Stacked bar plot of four subgroups distribution across 32 different cancer types. c Survival analysis of four subtypes in the pan-TCGA dataset. The statistical significance was determined by a log-rank test. d Heatmap depicting expression levels of co-expressed pro-inflam and pro-tumor TAMs genes across four subgroups. e Heatmap depicting expression levels of immune cell scores calculated by ssGSEA across four subgroups. f Heatmap depicting expression levels of M0/1/2 macrophage scores calculated by ssGSEA across four subgroups. g Heatmap depicting expression levels of inflammatory and antigen-presentation-related signatures calculated by ssGSEA across four subgroups. h Heatmap depicting expression levels of chemokines, cytokines, and receptor scores calculated by ssGSEA across four subgroups. i Heatmap depicting expression levels of IPS across four subgroups. j UMAP display color-coded by four subtypes using Louvain clustering to cluster patients in pan-TCGA patients. Each dot represents a single patient. k–m Violin plots depicting AUCell scores for IFN-alpha response, IFN-alpha response, and EMT across four subgroups.
Next, we performed principal component analysis (PCA) through co-expressed pro-inflam and pro-tumor TAM genes to further characterize the overall function of pro-inflam and pro-tumor TAMs across pan-cancer. The four subtypes of the 9164 TCGA tumors mentioned above were separated (Supplementary Fig. 5a). We projected these markers to the PCA space to validate the relationships between co-expressed pro-inflam and pro-tumor TAM genes and found that pro-inflam and pro-tumor TAM markers that were away from the origin were grouped (Supplementary Fig. 5b). Using uniform manifold approximation and projection (UMAP) clustering based on the principal components as input, the four subtypes identified above could be well separated and did not indicate cancer-specific subgroups (Fig. 2j and Supplementary Fig. 5c). We confirmed that pro-inflam and pro-tumor TAM-derived signatures were largely restricted to pro-inflam and pro-tumor TAM-enriched subtypes, respectively (Supplementary Fig. 5d). Consistent with the single-cell results, the pro-inflam TAM-enriched subtype (pro-inflam TAMs and mixed) scored highest for IFN response signatures (Fig. 2k, l), inflammatory response, and complement activation (Supplementary Fig. 5e, f), while the pro-tumor TAM-enriched subtype (pro-tumor TAMs and mixed) scored highest for EMT (Fig. 2m) and angiogenesis (Supplementary Fig. 5g), showing a pro-metastatic pattern. Additionally, the high expression of TGF-β observed in pro-tumor TAMs (Supplementary Fig. 5h) aligns with previous research linking TGFβ with immune cell exclusion in tumors17. A series of stepwise events named the “cancer-immunity cycle” involved in the anti-cancer immune response can effectively kill cancer cells18. Notably, in the subtype enriched in pro-inflam TAMs (pro-inflam TAMs and mixed), activities of the majority of the cancer-immunity cycle were identified as upregulated, including the antigen release and presentation (Supplementary Fig. 5i–l), trafficking (Supplementary Fig. 5m), and infiltration (Supplementary Fig. 5n) of immune cells to tumors. GSEA on the MsigDB Hallmarks gene sets revealed that tumor progression and inflammatory response were closely associated with the subtypes of pro-inflam TAMs and pro-tumor TAMs, respectively (Supplementary Fig. 6a). Moreover, TEAD119, TWIST120, RUNX221, and TCF1222, which are preferentially activated in TAMs with tumor growth properties, were elevated in the pro-tumor TAM subtype (Supplementary Fig. 6b, c). Additionally, STAT123, a transcription factor that stimulates macrophage-mediated inflammatory responses and induces macrophage differentiation towards the pro-inflammatory direction, was highly expressed in the pro-inflam TAM subtype consistent with decoupleR analyses (Supplementary Fig. 6b, c).
Tumor genomic profiles of the four distinct subtypes based on pro-inflam and pro-tumor TAMs
To gain further insights into the genomic landscape within the tumor, which has been shown to affect antitumor immunity and TME activity, we investigated the links between genomic and TAM classification. As a result, we observed that the pro-inflam TAM subtype exhibited a higher tumor mutation burden (TMB) compared to other subtypes (Supplementary Fig. 7a, b) and markedly more mutations in APOBEC-related signatures (Supplementary Fig. 7a). Among the frequently mutated genes (>5%), the pro-inflam TAM subtype had more mutations of TP5324, TTN25, and KMT2D26, which have been previously reported to be associated with the effectiveness of immunotherapy, while the absence subtype was abundant in mutations in APC. Furthermore, no clearly mutated genes were found in the pro-tumor TAM subtype (Supplementary Fig. 7a). Pro-inflam TAMs were negatively associated with tumor purity (Supplementary Fig. 7d), which correlates with previously observed “cancer-immunity cycle” properties. Pro-inflam TAMs were also enriched in ploidy (Supplementary Fig. 7c), homologous recombination deficiency (HRD) (Supplementary Fig. 7e), loss of heterozygosity (LOH) (Supplementary Fig. 7f), neoantigen (Supplementary Fig. 7g), and aneuploidy (Supplementary Fig. 7h). Copy number alterations were highlighted for comparison, and the pro-inflam TAM subtype harbored genomic amplification in the IFN-γ pathway, including JAK1, JAK2, PIK3CA, CASP8, CASP9, and STAT1, as well as copy number loss of CDK5, indicating a response to immune checkpoint blockade (ICB) (Supplementary Fig. 7i)27,28. The additional T-cell recognition observed in the pro-inflam TAM subtype may be linked to the amplification of genes associated with antigen-presentation machinery (Supplementary Fig. 7j). Subsequently, a multinomial logistic regression analysis was performed to evaluate the association between single-base mutational signatures and TAM subtypes. The pro-inflam TAM subtype had significantly more mutations in the context of APOBEC-related signatures (SBS2 and SBS13) compared to other subtypes, which were previously associated with ICB response and tumor aggressiveness29 (Supplementary Fig. 7k). Moreover, the pro-tumor TAM subtype had a relatively high frequency of mutations in SBS25 and SBS11 (Supplementary Fig. 7l). Concordantly, a high contribution of APOBEC-related signatures and SBS25 were associated with the mixed subtype (Supplementary Fig. 7m). Ultimately, the integration of transcriptomic- and genomic alteration-based TAM classification reveals the potential value of TAMs subtypes in terms of tumor progression and response to immunotherapy.
Identifying pro-inflam and pro-tumor TAMs related to immunotherapy
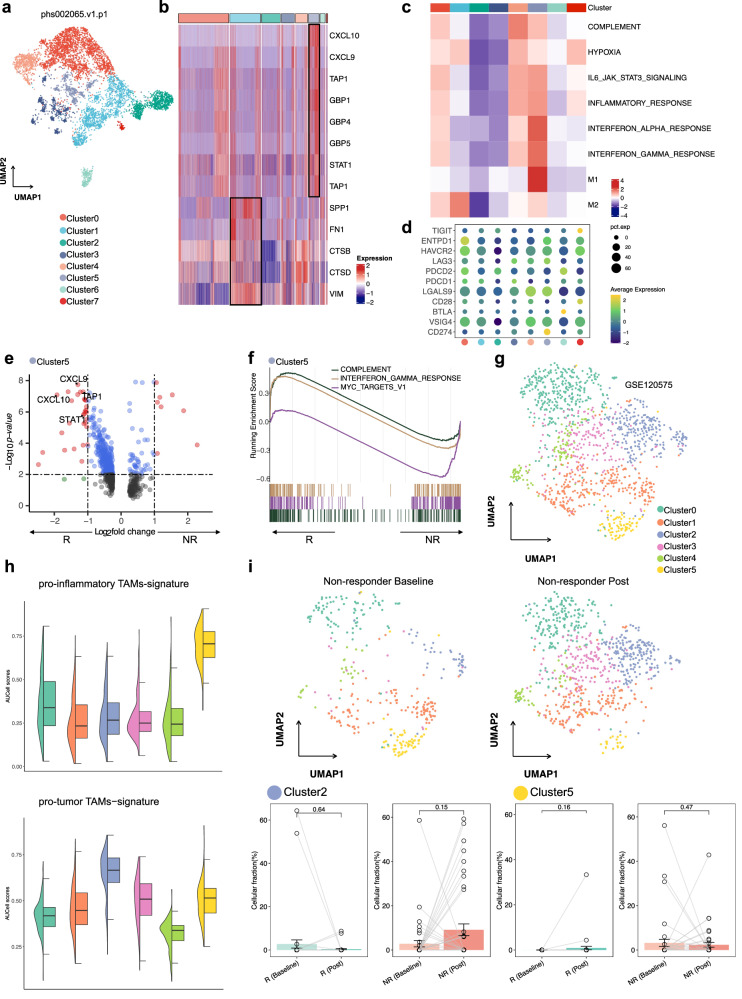
ICB has yielded encouraging results in numerous types of cancers. Our aforementioned analyses suggested that pro-inflam and pro-tumor TAMs play a central role in ICB therapy. To better understand the mechanism underlying the ICB response, we performed pro-inflam and pro-tumor TAM-derived signatures on two scRNA-seq datasets (phs002065.v1 and GSE120575) to identify the TAMs related to ICB response. In the scRNA-seq data analysis (phs002065.v1), these monocytes/macrophages were redivided into eight clusters (Fig. 3a), in which 1324 macrophages of Cluster1 were identified as pro-tumor TAMs, and 451 macrophages of Cluster5 were identified as pro-inflam TAMs (Fig. 3b). Meanwhile, pro-inflam and pro-tumor TAM-derived signatures were highly restricted to Cluster5 and Cluster1, respectively (Supplementary Fig. 8a, b). Functional enrichment analysis confirmed that inflammatory-related pathways, such as hallmark IFN-γ response, IFN-α response, and IL-6/JAK2/STAT3, were activated in Cluster5, while the hypoxia pathway was activated in Cluster1, with M2-like features that could contribute to ICB resistance (Fig. 3c). To better understand the roles of Cluster5 and Cluster1 in the course of ICB, we examined their expression of immune checkpoint genes and immune-related transcriptional programs. The results revealed that the co-inhibitory receptors CD274, LGALS9 (encoding Galectin 9), and LAG3 were detected in a larger proportion of Cluster5 (Fig. 3d). Moreover, genes previously identified as a pro-inflam TAM signature (CXCL9, CXCL10, TAP1, and STAT1), which mark immune cell infiltration, were pronounced in Cluster5 from cells from responders compared with those from non-responders (Fig. 3e). Furthermore, Cluster5 from responders upregulated gene sets associated with IFN-γ response and complement, indicating a response to pro-inflam TAMs in responders (Fig. 3f).
Fig. 3. Analysis of monocyte/TAMs clusters and pathway activities, and immunotherapy response in the phs002065 and GSE120575 datasets.
a UMAP visualization of all re-clustered monocytes/macrophages in the phs002065 dataset. b Heatmaps for monocytes/macrophages clusters in the phs002065 dataset for select pro-inflam and pro-tumor TAMs markers highlighted. c Heatmap shows the difference in pathway activities scored by GSVA per cell between different monocytes/macrophage clusters in the phs002065 dataset. d The expression of co-inhibitory molecules of monocytes/macrophages clusters in the phs002065 dataset. e Volcano plot showing upregulation of markers specific for pro-inflam TAMs in the Cluster5 (ICB responders) in comparison with the Cluster5 (ICB non-responders). R: responders including ICB PR patients; NR: non-responders including ICB SD/PD patients. f GSEA shows enriched pathways in the Cluster5 (ICB responders). R responders including ICB PR patients, NR non-responders including ICB SD/PD patients. g UMAP visualization of all re-clustered monocytes/macrophages in the GSE120575 dataset. h Violin plots depicting AUCell scores for gene signatures derived for pro-inflam and pro-tumor TAMs across all re-clustered monocytes/macrophages in the GSE120575 dataset. i UMAP plots showing the distribution of pro-inflammatory and pro-tumor TAMs in non-responders after immunotherapy (top); fraction of cells belonging to each immunotherapy phase for in Cluster2 and Cluster5; each point represents one sample; R responders including ICB CR/PR patients, NR non-responders including ICB SD/PD patients. Percentage corrected according to the total number of cells per sample; error bars indicate the 95% CI for the calculated relative frequencies; p value using the default Wilcoxon rank sum test (bottom).
Next, we reclassified monocytes/macrophages in the GSE120575 dataset into six clusters (Fig. 3g) and observed systematic qualification of pro-inflam and pro-tumor TAM-derived signatures in Cluster5 and Cluster2, respectively (Fig. 3h and Supplementary Fig. 8c). Similar results were obtained in that Cluster5 highly expressed immune-checkpoint genes (HAVCR2, LGALS9, and CD274; Supplementary Fig. 8d). GSEA revealed upregulation of IFN responses, IL-6/JAK2/STAT3, and ‘M1-like’ phenotype in Cluster5 (Supplementary Fig. 8e). Comparing ICB responders with non-responders, Cluster2 upregulated the hypoxia pathway and M2-like signature in non-responders (Supplementary Fig. 8f). To interpret Cluster5 and Cluster2 subset dynamics following immunotherapy and their relationship with immunotherapy response, we examined the regulatory effects of immunotherapy on these subsets. As a result, the non-responders showed decreased levels of Cluster5, which was accompanied by elevated levels of Cluster2 following ICB treatment (Fig. 3i). To further understand the specific monocyte/TAM clusters associated with ICB responses, we performed Scissor on our integrated scRNA-seq dataset containing only gastric cancer (GC) cells with GC bulk patients with known immunotherapy response information (PRJEB25780) (Supplementary Fig. 8g). We determined 77.1% of Cluster5 as being Scissor+ cells, which was associated with a favorable ICB response, and 83.1% of Cluster3 as Scissor− cells, which are related to unfavorable ICB responses (Supplementary Fig. 8h). Collectively, such observations imply a close correlation of pro-inflam and pro-tumor TAMs with immunotherapy.
Spatially mapping pro-inflam and pro-tumor TAMs
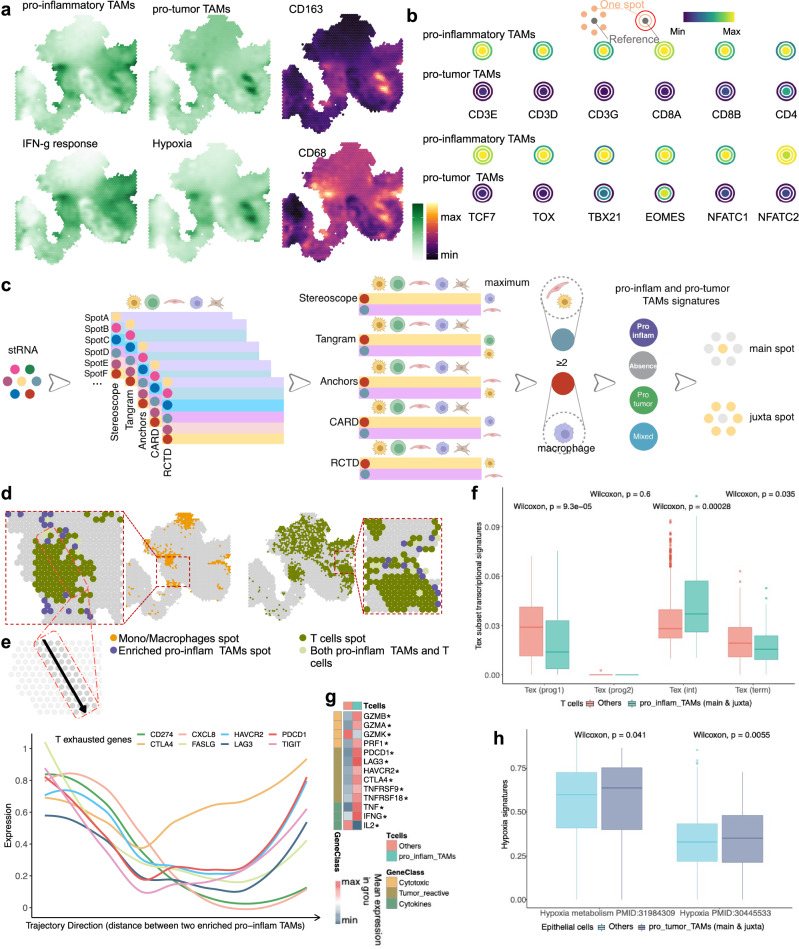
To identify the spatial distribution of the pro-inflam and pro-tumor TAMs illustrated above and their co-localization to the defined TME, we next performed spatially resolved transcriptomic RNA-sequencing (stRNAseq) on formalin-fixed, paraffin-embedded (FFPE) tissue. Treatment-naive tissue samples were obtained from four patients with primary GC from our clinical study of neoadjuvant therapy (ClinicalTrials.gov: NCT04341857), and the datasets contained a total of 4435 sequenced spots. After spatially mapping the expression profiles based on pro-inflam and pro-tumor TAM signatures, the results confirmed the co-localization of classical macrophage markers and the aforementioned functional features (Fig. 4a and Supplementary 9a). To gain further insights into the spatial distribution of pro-inflam and pro-tumor TAMs, we examined the pro-inflam and pro-tumor TAM signatures in spatially resolved transcriptomics of other cancer samples with pathological annotation. The results showed that both signatures tended to be enriched in tumors compared with nontumor sections at the level of spatial transcriptomics clusters in primary liver cancer (Supplementary Fig. 9c). Among the three kidney tumor-normal interface samples, both TAMs were enriched in the tumor core rather than the normal and interface regions (Supplementary Fig. 10a). As expected, the spatial distribution of pro-inflam and pro-tumor TAMs dispersed across invasive cancer, stroma, and lymphocyte regions in breast cancer (Supplementary Fig. 9b). Neighborhood analysis found that spots enriched for the pro-inflam TAM signature were enriched for CD8+ T, with key markers of exhaustion (e.g., TCF7 and NFATC2) (Fig. 4b). In contrast, spots enriched for the pro-tumor TAM signature exhibited no T-cell gene enrichment. We next integrated our spatial transcriptomics data with re-annotated scRNA-seq data for 10 major cell types and sought to map spots to specific cell types (Fig. 4c). Generally, deconvolution methods yield high correlations (Supplementary Fig. 9d). The spatial proximity of enriched pro-inflam TAMs coupled with T cells is confirmed in Fig. 4d. Compared with pro-tumor TAMs, both the main and juxta spots of enriched pro-inflam TAMs were enriched with immune cells, including T cells identified using Tangram (Supplementary Fig. 10b-c). To further demonstrate the spatial dependence of T-cell exhaustion (Tex) on enriched pro-inflam TAMs, we quantified the putative distance between the two cell types. Spots containing T cells that were highly expressed for Tex markers were adjacent to enriched pro-inflam TAMs, suggesting that pro-inflam TAMs are associated with Tex (Fig. 4e). Spots containing T cells located in the main and juxta of enriched pro-inflam TAMs exhibited a high TexInt score compared with others, implying that T cells surrounding these TAMs possessed the potential to regain cytotoxicity after immunotherapy (Fig. 4f). Elevated expression levels of cytotoxic enzymes, including GZMB and GZMA, hallmarks of tumor reactivity, and cytotoxic profiles, including IFNG and TNF, were observed in spots containing T cells located in the main and juxta of enriched pro-inflam TAMs (Fig. 4g). Moreover, a high hypoxia score was expressed in epithelial cells located in the main and juxta spots of enriched pro-tumor TAMs compared with others (Fig. 4h). These findings support our original observations in that pro-inflam TAMs resided in spatially immune-enriched niches alongside exhausted CD8+ T cells, forming pro-inflam TAM–Tex niches, while pro-tumor TAMs were restricted to niches associated with hypoxia. Despite their exhausted phenotype, T cells in pro-inflam TAM–Tex niches may still exert cytolytic effector functions and retain antitumor activity.
Fig. 4. Spatial distribution and topological analysis of immune and hypoxia markers, TAMs, and Tex.
a Surface plot of INF-γ, hypoxia, pro-inflam, and pro-tumor TAMs signatures, and classical markers in the S1 tumor sample. b Schematic illustrating topological analysis on the Visium spot hexagonal grid: reference spot depicted in gray shows the average gene expression, with adjacent spots one hexagon away in orange and those two hexagons away in red, demonstrating average expression of the same gene. Average expression of Tex markers and various T-cell markers surrounding spots enriched for pro-inflam and pro-tumor TAMs signatures. c Graphical summary of deconvolution and integration of spatial transcriptomics spots; main spot, TAMs-localized spot; juxta spot, TAMs-neighboring spot (spots of distance less than or equal to 100 µm). d Mapping of monocytes/macrophages, enriched pro-inflam TAMs, and T cells in S1 tumor sample. e Line plots illustrate the distance between two spots that have been randomly selected for their enrichment in pro-inflam TAMs. The x-axis represents the trajectory direction, with the distance units (µm) away from the starting enriched pro-inflam TAM spot. The two ends of the x-axis represent the positions of the two selected enriched pro-inflam TAM spots. y-Axis represents the expression of Tex markers. f Boxplots showing the score of Tex processes signatures in the spots containing T cells. Tex Prog1 T cell exhaustion progenitors, Tex Prog2 T cell exhaustion progenitors 2, TexInt T cell exhaustion intermediate, Tex term T cell exhaustion terminally. g Heatmap showing the expression of select markers of cytotoxic potential, tumor activity, and cytokine secretion for spots containing T cells. The Wilcoxon Rank Sum test was utilized to identify DEGs between groups of spots, with an asterisk (*) indicating a significance level of p < 0.05. h Boxplots showing the enrichment score of hypoxia signaling pathways in the spots containing epithelial cells.
Based on cell type deconvolution and integration, the potential mediators of pro-inflam TAM–CD8+ T-cell interactions could then be modeled. Spatial analysis of the interaction expression of ligand-receptor pairs that positively correlated (r > 0.4) with the pro-inflam TAM signature was mapped (Supplementary Fig. 11a). Interestingly, multicellular pair interactions revealed that pro-inflam TAMs provided the ligands, while the T cells in pro-inflam TAM–Tex niches expressed the receptors: pro-inflam TAM expression of HLA-C, CXCL9, CXCL11, TNFSF10 (TRAIL), and CD274 (PD1) as ligands for LILRA1/B1/B2, CXCR3, TNFRSF10A (TRAILR1), and PDCD1 on T cells (Supplementary Fig. 12a), with the finding that pro-inflam TAMs were the main producer of the above ligands in the integrated scRNA-Seq data (Fig. 1f and Supplementary Fig. 11b). The spatial proximity of the pro-inflam TAM ligands coupled to the neighboring T-cell receptors was confirmed (Supplementary Fig. 11c). The interaction expression of ligand-receptor pairs was high in TRG0 patients, implying that these pairs are strongly associated with immunotherapy benefit (Supplementary Fig. 12b). Additionally, we discovered genes that were highly expressed in spots with pro-inflam TAM–Tex niche-enriched pathways that contribute to the recognition and regulation of T cells, and were enriched in antigen binding, receptor ligand activity, and CXCR chemokine receptor binding (Supplementary Fig. 12c). Given that the expression of ligand-receptor pairs in different cell types partially overlap, and given that each spot contains other cell types, we further used mIHC to visualize those proteins and observed contacts between CD68 + TNFSF10 + TAMs (pro-inflam TAMs) and CD8 + PD1 + T cells (Supplementary Fig. 12d). We further confirmed higher infiltration of CD68 + TNFSF10+ TAMs and CD8 + PDCD1 T cells in TRG0 patients than in TRG1-3 patients (Supplementary Fig. 12e). These results suggest that these pro-inflam TAM–Tex niches have the potential to restore antitumor activity through immunotherapy, which may be critical for treatment efficacy (Supplementary Fig. 12f).
Pro-inflam and pro-tumor TAMs correlate with immunotherapy efficacy
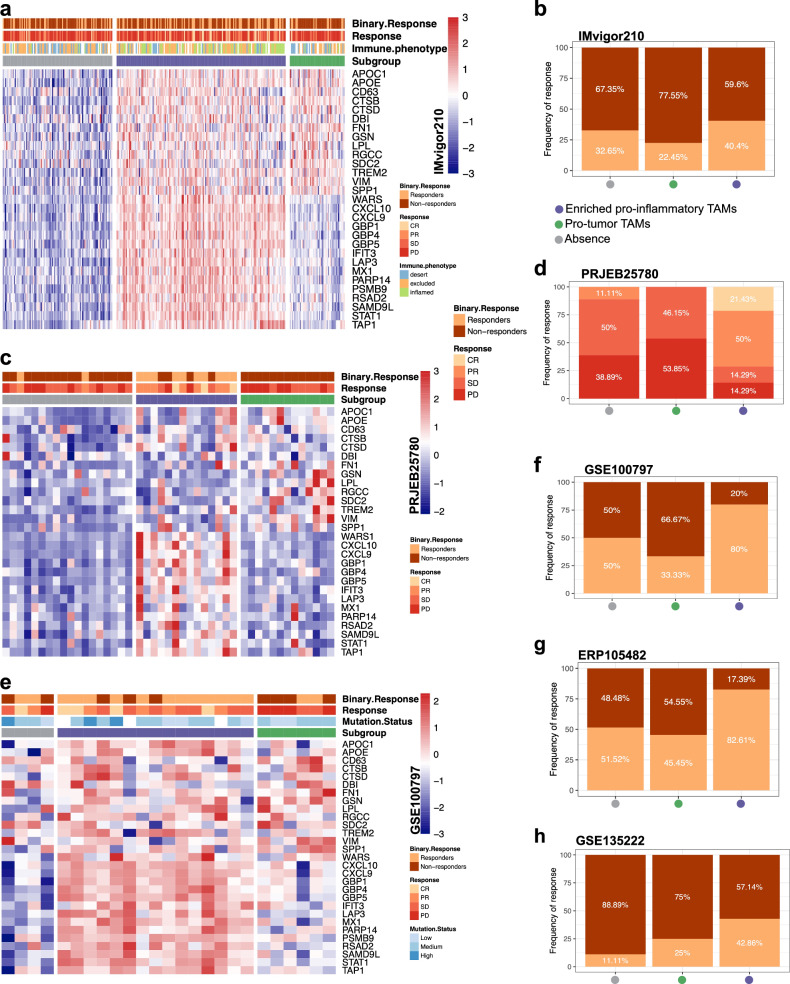
To identify whether TAM classification can be applied as a tool to predict the response to ICB, the responses to ICB were undertaken across the three TAM subtypes, in which we combined pro-inflam and mixed TAMs into the pro-inflam TAM-enriched subtype. In an independent metastatic melanoma cohort (n = 73) treated with anti-PD-1 alone or combined anti-PD-1 and anti-CTLA-4 immunotherapy, patients were classified into three TAM subtypes (Fig. 5a), with the pro-inflam TAM subtype found to be significantly associated with response (complete response/partial response + long-term survival with stable disease) to ICB (Fig. 5b). Similar findings were observed with independent cohorts of patients with GC (n = 45; Fig. 5c, d), melanoma (n = 115; Fig. 5e–g and Supplementary Fig. 13a), and lung cancer (n = 27; Supplementary Fig. 13b and Fig. 5h). The high mutational burden, the synergistic effects of combined anti-PD-1/CTLA-4 therapies, and the immunogenicity of melanoma currently make it more sensitive to combinatorial ICB treatment compared to other tumors30,31. The patients of these cohorts received anti-PD(L)-1, anti-CTLA-4, adoptive T-cell therapy, or anti-PD(L)-1 plus anti-CTLA-4, suggesting that the TAM classification system can be applied to diverse immune-based therapies as a potential biomarker of response. When examining whether pro-inflam and pro-tumor TAMs are relevant to ICB, subtypes enriched in pro-inflam TAM were found to exhibit the longest OS in patients receiving immunotherapy (Supplementary Fig. 13c–e). Moreover, patients with pro-tumor TAM subtypes treated with immunotherapy had a poorer prognosis than those with other subtypes (Supplementary Fig. 13c–e). Notably, in lung cancer, the PFS following anti-PD(L)-1 was the longest in the pro-inflam TAM subtype (Supplementary Fig. 13f). In contrast, in melanoma and bladder cancer, patients with the pro-inflam TAM-enriched subtype who were not treated with immunotherapy displayed an inferior prognosis (Supplementary Fig. 13g and Supplementary Fig. 14a), highlighting the importance of immunotherapy in patients with the pro-inflam TAM-enriched subtype.
Fig. 5. Clinical response to immunotherapy for different TAM subtypes across multiple datasets.
a Heatmap of pro-inflam and pro-tumor TAMs co-expressed genes for three different TAMs subtypes classification in the IMvigor210 dataset. b Rate of clinical response to immunotherapy in three different TAM subtypes in the IMvigor210 cohort. c Heatmap of pro-inflam and pro-tumor TAMs co-expressed genes for three different TAMs subtypes classification in the PRJEB25780 dataset. d Rate of clinical response to immunotherapy in three different TAM subtypes in the PRJEB25780 cohort. e Heatmap of pro-inflam and protumor TAMs co-expressed genes for three different TAMs subtypes classification in the GSE100797 dataset. f Rate of clinical response to immunotherapy in three different TAM subtypes in the GSE100797 cohort. g Rate of clinical response to immunotherapy in three different TAM subtypes in the ERP105482 cohort. h Rate of clinical response to immunotherapy in three different TAM subtypes in the GSE135222 cohort.
As expected, the pro-inflam TAM-enriched subtype was recognized as an immune-inflamed phenotype (Supplementary Fig. 14b) and higher neoantigen burden (Supplementary Fig. 14c). Furthermore, the immune-favorable TME type non-fibrotic (IE) was higher in the pro-inflam TAM-enriched subtype that saw the greatest benefit from immunotherapy (Supplementary Fig. 14d–g). In contrast, the immunosuppressive subtype F, which is characterized by low T-cell infiltration and lower response rates to immunotherapy, was found to be more prevalent in the pro-tumor TAM-enriched subtype (Supplementary Fig. 14d–g). We then assessed whether pro-inflam and pro-tumor TAMs were altered in patients with distinct responses during immunotherapy in the ERP105482 cohort. Tumors that attained a response tended to be enriched for pro-inflam TAMs (Supplementary Fig. 14j), which increased in a stepwise manner with the introduction of immunotherapy (Supplementary Fig. 14k). In contrast, in non-responders, tumors lacked pro-inflam and pro-tumor TAMs (Supplementary Fig. 14l), which increased by a small margin with the introduction of immunotherapy. Additionally, the increase in pro-tumor TAMs was greater than that in pro-inflam TAMs (Supplementary Fig. 14m), further supporting the notion that these cell populations are critical in the response to ICB.
Performance of TAM predictors of response to immunotherapy
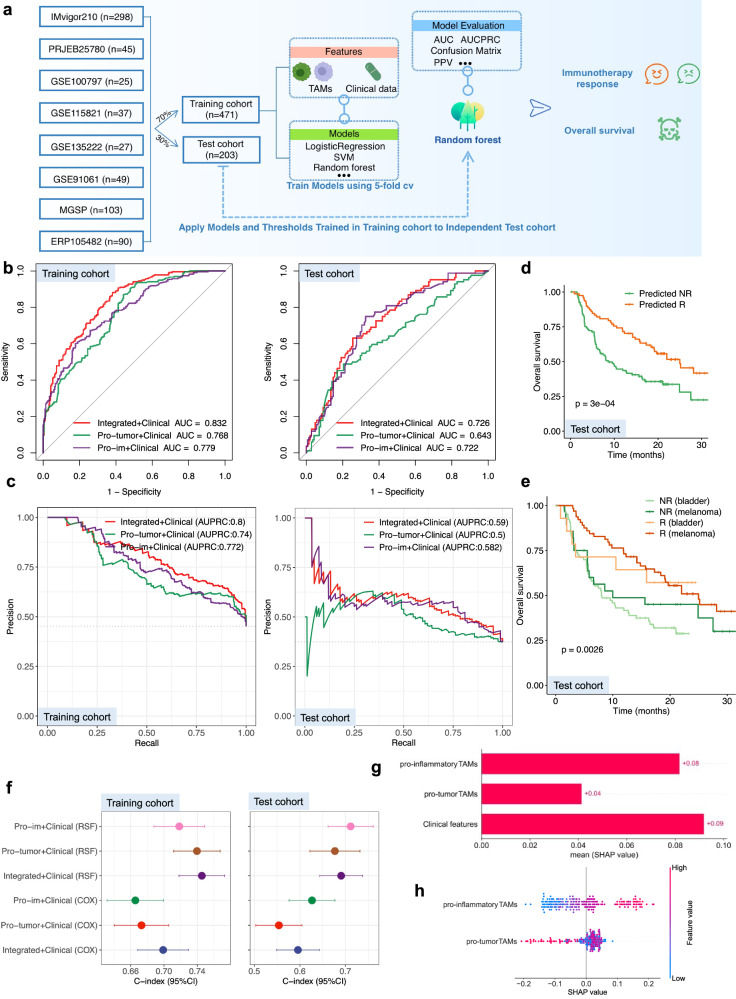
To investigate the predictive value of pro-inflam and pro-tumor TAMs as an improved gene expression-based predictor, we collected bulk RNA-Seq data and clinical information from eight ICB cohorts grouped into two datasets, the training set (n = 471) and testing set (n = 203). Approximately 45.1% of the patients had melanoma, 44.2% had bladder cancer, and the remaining 10.69% had non-small-cell lung cancer and GC (Supplementary Fig. 15a). These patients were treated with tumor-infiltrating lymphocytes, PD-1/PD-L1 inhibitors, CTLA-4 blockade, or a combination of both PD-1/PD-L1 and CTLA-4 agents (Supplementary Fig. 15a). We integrated the above clinical information (cancer type and drug class) and pro-tumor and pro-inflam TAMs, and derived a series of three immunotherapy response prediction models, including different feature combinations, using the following: (1) clinical information and pro-tumor and pro-inflam TAMs; (2) clinical information and pro-inflam TAMs; and (3) clinical information and pro-tumor TAMs. We trained the model with five machine-learning algorithms and applied fivefold cross-validation (CV) for the parameter optimization of each model. The RF model achieved the highest accuracy and was selected as the successor model. The flowchart of the analysis process is shown in Fig. 6a. We attempted to assess the performance of the integrated clinical-TAMs RF model in terms of multiple metrics. The integrated model achieved superior performance, as indicated by the area under the curve (AUC) in discriminating responders and non-responders compared with pro-tumor or pro-inflam TAMs alone in both the training (0.83, 0.77, and 0.78, respectively) and test cohorts (0.73, 0.64, and 0.72, respectively) (Fig. 6b). We further verified the higher area under the precision-recall curve (AUPRC) achieved by the integrated RF model than pro-tumor or pro-inflam TAMs alone in both the training and test cohorts (Fig. 6c). We also performed subgroup analysis in the test cohort. In addition to the GSE135222 cohort, which presented with the lowest AUC of 0.63, in other cohorts, the integrated RF models all showed robust, generalizable ability to predict response, with AUCs of >0.65 (Supplementary Fig. 15b–h). These results prompted us to use a machine learning framework to incorporate pro-tumor and pro-inflam TAMs into a predictive model of response. The TMB has been approved by the FDA as a biomarker to predict ICB efficacy in solid tumors, and we also compared the performance of the integrated model with TMB. The integrated model showed better performance than the TMB in both the training and test cohorts (Supplementary Fig. 15i, j). We further compared the performance of the integrated model with previous well-established predictive gene signatures, including our own established GC stem-like cell-related score (GCScore)3. The majority of these pan-cancer signatures exhibited desirable performance in only one or two cohorts. For instance, the AUC of the CXCL9:SPP1 ratios attained 0.92 in GSE135222 (Supplementary Fig. 16g), and 0.75 in GSE100797 (Supplementary Fig. 16i) but declined to 0.58 in GSE91061 (Supplementary Fig. 16f). Nevertheless, our TAM signature reached adequate performance in all cohorts (Supplementary Fig. 16), across four cancer types, which further highlights its potential as a pan-cancer ICB response prediction model.
Fig. 6. Machine learning framework for predicting immunotherapy response and survival outcomes based on TAM subtypes.
a Schematic of the machine learning framework. b ROC curves and corresponding AUC values for three feature types in the training and test cohorts. c The corresponding AUPRC values of three feature types in the training and test cohorts. d Kaplan–Meier curves comparing OS between responders and non-responders in the testing cohort. e OS stratified between responders and non-responders in patients in the testing cohort with different cancer types. f Comparison of C-index of multiple prediction models in training and testing cohorts. g, h Feature importance of pro-inflam and pro-tumor TAMs in the integrated TAMs RF model in the testing cohort.
Furthermore, a prolonged OS was consistently observed for patients predicted as ICB responders using our RF-based ML in the test cohort with OS data available (Fig. 6d), regardless of cancer type (Fig. 6e) and treatment regimens (Supplementary Fig. 17a). To test whether pro-tumor and pro-inflam TAM signatures could also predict OS, we evaluated the performance of the classical Cox proportional hazards (Cox) model and the random survival forest (RSF) model using the concordance index (C-index) and integrated brier score. We found that the C-index of the integrated TAM RSF model (0.75, 95% CI: 0.72–0.77) was significantly higher than others in the training cohort and that the C-index of the pro-inflam TAM RSF model (0.71, 95% CI: 0.66–0.76) was higher than others in the test cohort (Fig. 6f). Compared with the CXCL9:SPP1 ratio, our integrated model exhibited enhanced prognostic performance (Supplementary Fig. 17b). The integrated TAM RSF model showed good calibration with low Brier scores (Supplementary Fig. 17c). Additionally, the time-dependent AUC suggested that the integrated TAM RSF model had considerable value in predicting OS (Supplementary Fig. 17d).
Importance and interaction analysis of pro-inflam and pro-tumor TAMs
We next explored the importance of, and interactions between pro-inflam and pro-tumor TAMs in the integrated clinical-TAMs RF model using Shapley additive explanations (SHAP) and PDPbox. The effect of clinical information, pro-inflam, and pro-tumor TAMs on the output of the model was investigated using the SHAP value. We found that pro-tumor TAMs were effective for response prediction only at the highest values, in contrast to pro-inflam TAMs, which displayed significant discriminative power across all values (Fig. 6g, h). Based on PDPbox, a significantly increased response was observed when pro-inflam TAMs were higher than approximately 0 (Supplementary Fig. 18a). Similar to the amount of pro-tumor TAMs that can affect the response to immunotherapy (Fig. 6h), the negative effects on immunotherapy occur when the pro-tumor TAMs are >1.5 (Supplementary Fig. 18a). We also identified interactions between pro-inflam and pro-tumor TAMs, in that they were significantly positively correlated (Supplementary Fig. 18b). Consistent with PDPbox (Supplementary Fig. 18c), a high pro-tumor TAM combined with highly pro-inflam TAM was more predictive of responses, supporting the differentiation state from pro-tumor TAMs toward pro-inflam TAMs for patient responses (Supplementary Fig. 18b). These results were in a good agreement with the above-mentioned result concerning the immune profile of pro-inflam and pro-tumor TAMs.
Discussion
As one of the most abundant immune cell types in tumors, macrophages have multiple potent functions highlighting their heterogeneous nature32. With the wide application of single-cell technologies, a few studies have delved into the heterogeneity of monocyte/macrophage clusters through pan-cancer analysis6,7,33. Although these studies have revealed significant transcriptomic diversity in monocyte/macrophages, a unifying annotation of their molecular functional characteristics remains lacking. Considering that monocytes and macrophages exhibit a high degree of complexity and tissue specificity, we excluded those from blood, normal tissue, and lymph nodes when integrating the scRNA-Seq database, focusing only on tumor-enriched monocytes and macrophages. In the present analysis, all monocyte/TAM clusters were observed in each cancer type, although certain preferences were evident. An in-depth cross-sectional comparison of three monocyte/macrophage scRNA-seq datasets through CellTypist has significantly contributed to our understanding of TAM heterogeneity in cancer. We identified pro-tumor TAMs sharing characteristic features of TAMs, including TREM2 and C1Q, which promote tumor growth34,35 and general immunosuppression13. Our approach to generating pro-tumor TAMs is broader than previously reported, encompassing wide tumorigenic potential and further inducing immunosuppressive and tumor growth-promoting functions. Our “pro-inflam TAMs” exhibited more elevated activities than those traditionally ascribed to inflammatory macrophages. These TAMs act not only as potent inflammatory signals but also act as specialists of immune regulation, similar to specialized subsets such as CXCL10-Hi TAMs and IL4I1 TAMs7. Such insights not only break paradigms but are at the core of the designing of TAM-targeted therapies and at improving our ability to predict patient responses to immunotherapy. Our study diverged from the pan-cancer approaches of Cheng et al.6 and Combes et al.8 by introducing a novel computational framework that integrates scRNA data with spatial transcriptomics. This approach sheds light on the spatial distribution of TAMs and reveals new interactions between pro-inflam TAMs and T cells, underlining their pivotal role in modulating responses to immunotherapy. While Mulder et al.7 delineated five TAM subsets in various cancers, our work went a step further by detailing the interactions between these subsets and other immune cells, thereby offering novel predictions for immunotherapy outcomes by combining TAM signatures with clinical data. Ma et al.32 emphasized the importance of unified nomenclature for TAM diversity and highlighted the advancements in single-cell omics technologies that have enriched our understanding of TAM molecular diversity. The absence of consensus on TAM subset terminology has hindered the full exploitation of data wealth from various studies. Our study aims to bridge this gap by providing a detailed analysis of two TAM subsets, pro-tumor and pro-inflammatory, characterized by their distinct gene expression profiles and potential spatial interactions with other immune cells, notably T cells. These interactions hold significant implications for tumor progression and response to immunotherapy, thus offering a fresh perspective on TAM diversity that enriches the existing literature. Contrary to the broad categorizations offered by Chen et al.6 and Mulder et al.7, our study proposed a nuanced perspective on TAM roles in immunotherapy. In doing so, we aligned with Ma et al.‘s call for a more refined understanding of TAM diversity, considering both molecular and spatial dimensions.
A recent review concluded that IFN-primed TAMs act as immunosuppressive macrophages highly expressing IFN-regulated genes, such as CXCL10, PDL1, and M1-like markers across many tumor types32. Similar to these previous findings, we found that pro-inflam TAMs were characterized by high expression of IFN-regulated genes and immune checkpoint molecules. However, we further observed that pro-inflam TAMs exhibited inflammatory phenotypes that actively recruit and regulate immune cells. Pro-inflam TAMs constitute an inflamed immune environment, which mediates Tex through potential ligand-receptor axes including TRAIL-TRAILR1, PD1-PDCD1, and CXCL9/CXCL11-CXCR3. These axes have been implicated in the recruitment and development of Tex36. Indeed, the possibility of modulating these axes to decrease the exhausted states of T cells has been investigated in early-stage clinical trials37. We show that enrichment of pro-inflam TAMs is associated with different clinical outcomes, including poor prognosis in patients who were not treated with ICB and better prognosis in patients receiving immunotherapy. This may be partly explained by the identification of an association between an exhaustion-like T-cell phenotype and an inflamed immune environment with altered cytotoxic and proliferation potential, consistent with previous results38,39. Hypoxia and EMT induce a series of biological alterations that promote angiogenesis and are associated with resistance to immunotherapy40,41. The SPP1 gene encodes osteopontin, which is associated with EMT, hypoxia, and tumor progression42,43. Consistent with previous studies linking SPP1 + TAMs to angiogenesis enriched in hypoxic regions in the TME6,44, pro-tumor TAMs marked by the expression of SPP1, which were associated with angiogenesis, EMT, and hypoxia, predicted a poor prognosis. Significantly, we investigated the generalized characteristics of T-cell-excluded phenotype in the pro-tumor TAM-enriched TME, which explains the poor effect of immunotherapy. Our investigation markedly enhances the existing paradigm concerning TAMs by meticulously addressing three critical dimensions: infiltration/number, transcriptional diversity, and spatial localization. We advance beyond traditional quantitative assessments of TAM densities, as conducted by Mantovani45 and Nixon et al.10, by stratifying these cells into functionally distinct pro-inflammatory and pro-tumor phenotypes, thus elucidating their differential impacts on oncogenesis and immune regulation. Using single-cell RNA sequencing, we map out the transcriptional heterogeneity inherent to TAM populations, revealing specific gene expression signatures that indicate their functional states within the TME. Furthermore, by applying spatial transcriptomics and mIHC, we precisely chart the locational dynamics of TAMs, showing how their spatial arrangements relative to other tumor and immune cells critically influence their biological roles. This integrative analysis not only deepens our understanding of TAM functionalities but also highlights their potential as versatile targets for therapeutic strategies, capable of influencing tumor progression through nuanced manipulation of their spatial and transcriptional landscapes.
Our main conclusions hold upon the integration of scRNA-Seq ICB cohorts, TCGA pan-cancer transcriptomic analysis, and genomic landscapes. In the scRNA-Seq ICB cohorts, pro-inflam TAMs were associated with a favorable ICB response, while pro-tumor TAMs were related to unfavorable ICB responses. TCGA pan-cancer transcriptomic analysis showed increased infiltration of immune cells, especially CD8+ T cells, in tumors with the pro-inflam TAM subtype across different cancer types. Genomic features suggest that the pro-inflam TAM subtype was correlated with TP53 mutations, TMB, HRD, APOBEC mutational signatures, and chromosomal instability. These ICB biomarkers have been previously reported24,46–48 and should prompt a search for potential mechanisms associated with pro-inflam TAMs.
Our findings indicate that pro-tumor and pro-inflam TAMs can more effectively predict ICB response and accurately distinguish patients with survival benefits. Predictive modeling of tumor immunotherapy using Scikit-learn and associated machine-learning algorithms has garnered widespread validation in the scientific community. Studies such as Kong et al.‘s network-based approach49, Wiesweg et al.‘s PD-L1–independent response predictions50, and Polano et al.‘s pan-cancer analysis51 demonstrate the robust application of these tools in predicting outcomes of cancer immunotherapies. These models utilize various machine learning algorithms implemented in Scikit-learn, yielding significant insights into treatment responsiveness and enhancing predictive accuracies across different types of cancers. We utilized a series of machine learning algorithms to analyze the role of TAMs in immunotherapy response. These algorithms were selected based on their proven efficacy in previous studies for handling complex datasets and delivering reliable predictive performance. The validation of these tools is well-documented in oncological research, ensuring that our methodology is thoroughly rigorous and reproducible. The observed overlap and correlation between pro-inflam and pro-tumor TAM signatures do not diminish their value for patient stratification; instead, they underscore the significance of acknowledging the spectrum of TAM phenotypes within tumor biology. While individual studies have underscored the prognostic value of markers like CXCL9 and SPP152–54, our integrated model, which encompasses a broader range of TAM-associated biomarkers, showed improved predictive capability over a variety of datasets and cancer types. Moreover, our analysis revealed that our TAM signature demonstrates greater consistency and reliability across all evaluated cohorts when compared to the CXCL9:SPP1 ratio55. This suggests that while individual biomarkers are insightful for understanding the tumor immune environment, an aggregated signature that captures various facets of TAM activity may be a more robust tool for predicting patient responses to immunotherapy. In addition to being satisfied with the effectiveness of the model, we have given more thought to the reasons for the model’s effectiveness. Furthermore, the importance of pro-inflam and pro-tumor TAMs in the model and how each feature affects the final prediction results were analyzed. Subject to further validation, we aim to pursue clinical implementation of our approach in the near future. Our identification of TAM subsets not only aligns with the diversity highlighted by Mulder et al.7 but also extends beyond the classifications proposed by Chen et al.6 by elucidating their functional implications across various cancer types. The comprehensive analysis underscores the call by Ma et al.32 for a more nuanced understanding of macrophage diversity, unveiling unique TAM functions that could guide therapeutic strategies. Our research emphasizes the critical role of TAM spatial distribution, as explored by Qu et al.56 and Qi et al.54, in influencing immune responses, thereby underlining the necessity of incorporating TAM localization into therapeutic design considerations.
Our study does have some limitations that warrant discussion. First, while we have characterized the common characteristics of two TAM clusters, experimental validation remains necessary to determine whether these functional TAMs play a similar role and to explore the potential mechanism across different cancer types. Second, due to the limited number of patients meeting the inclusion criteria (treatment-naïve sample, gastroscopy sample resectability, and postoperative pathological regression data), only four clinical trial patients were recruited for our ST cohort. Additionally, the ST technique has not yet achieved single-cell resolution. Due to inherent limitations in the method, it is not always possible to differentiate between variations in gene expression and differences in cell type composition within spot transcriptomes. Our study design and the insights drawn from the analysis of these four samples are in line with accepted practices in the field of spatial transcriptomics, where meaningful biological insights can still be derived from a limited number of samples, particularly when studying the spatial architecture and cellular interactions within the TME57,58. For instance, a pro-inflam TAM–Tex niche interaction may occur either because T cells have upregulated these receptors or because T cells with these receptors have migrated to this location. However, in either scenario, a pathogenic mechanism is identified, offering therapeutic value. Future studies will aim to expand the dataset and utilize more robust statistical methods to confirm the observed trends and refine our understanding of pro-inflam TAM–Tex niche interactions in tumor tissues responding to immunotherapy. Thirdly, the mechanism by which immunotherapy induces functional changes in the two TAMs remains unclear. Additional experimental validation is necessary to confirm whether this state shift in TAMs is a result of altered differentiation or population changes.
In summary, we present a comprehensive atlas of two TAM clusters that are commonly observed across cancer types, including their phenotypic and biofunctional categorization, as well as their association with immunotherapy and prognosis. Additionally, we identify exhaustion-associated TME characteristics, which may facilitate the discovery of new therapeutic targets and highlight the value of TAMs as a novel biomarker for response stratification and prediction.
Methods
scRNA‑Seq data analysis
The outputs of the cell range from eight publicly available scRNA‑Seq datasets were processed using the Read10X and CreateSeuratObject functions within the Seurat package (V 4.1.0) to construct Seurat objects. In this study, we selected eight single-cell RNA sequencing datasets based on strict criteria for robustness and relevance across different cancer types. Inclusion and exclusion of studies were set according to data sets up to a cut-off of April 2022, chosen to represent the latest technological advances in single-cell technologies before our analysis, and to represent the latest findings relevant to the current state of the art in cancer research. According to our pan-cancer study design, we selected representative databases for diverse cancer types, balanced by cell number, to minimize overrepresentation bias and to mitigate the possibility of one single cancer type affecting the conclusions of the study. Each dataset underwent several quality checks, including the removal of duplicate sequences, strict cell detection, exclusion of lowly expressed genes, and overall quality controls such as UMI count, the number of detected genes, and the proportion of mitochondrial genes. These procedures are important to maintain a high level of data integrity and to make it suitable for advanced analytical procedures. In this study, we applied the most stringent of these criteria to include only those datasets that ensure the most reliable insights into cellular dynamics within the TME, providing a solid foundation for our understanding of TAM heterogeneity and its implications across different cancers. The adjacent normal tissue, lymph node, and peripheral blood were filtered out, and only the primary treatment-naïve tumor tissue was kept. Then, cells with <1000 unique molecular identifiers, >6000 expressed genes, or a mitochondrial gene count >10% were filtered. We extracted monocytes/macrophages based on the cell type annotations provided by some of the authors and conducted a re-clustering analysis of all cells based on monocyte/macrophage markers (CD14, CD16, CD163, and CD68). Following the removal of low-quality cells, high-quality cells were normalized and scaled using the sctransform wrapper in Seurat, before being batch-corrected for the individual dataset using Harmony (V 1.0.0). We applied principal component analyses to reduce the dimensionality of the data using the top 3000 most variable genes in the dataset. The FindClusters function was performed to generate different clustering results with resolutions ranging from 0.1 to 0.8. An appropriate resolution was determined based on cluster stability with clustree (V 0.5.0)59. The non-significant clusters were filtered based on both statistical and biological considerations to ensure the relevance and specificity of our findings. This process involved two key criteria: (1) removing clusters containing fewer than 200 cells to exclude statistically underpowered clusters, and (2) identifying and removing potential doublets with DoubletFinder (V 2.0.4) to reduce technical noise. Seven clusters, including 24,983 cells, were retained for the subsequent analysis. The FindAllMarkers function provided by Seurat was used to perform differential gene expression analysis with a Wilcoxon rank-sum test to define marker genes for each monocyte/TAM cluster. Cancer hallmark gene sets from the Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org/) were used in the GSVA analysis. Genelists for the M1 and M2 signatures were adopted from Azizi et al.60. Signatures related to hypoxia and immunity, among others, were collected from the IOBR package (V 0.99.9) and compiled from other studies61. GSVA was performed on these DEGs with the GSVA package (V 1.46.0). We assembled T-cell–attractive and T-cell–suppressive properties of monocyte/TAM clusters using gene signatures assembled from the literature38. The activity of individual cells for each gene set was estimated using the AUCell package (V 1.20.2)62.
We selected the phs002065.v1 and GSE120575 datasets for their well-documented immunotherapy response information. Initial exploratory data analysis involved assessing the quality of the datasets as described above, normalizing gene expression data, and identifying highly variable genes as potential markers for cell clustering. To classify cells into meaningful clusters, we employed the Leiden algorithm, chosen for its effectiveness in revealing the intricate cellular landscape based on transcriptional profiles. We adjusted the Leiden algorithm’s resolution parameter, a critical step in defining the granularity of our clustering, allowing us to capture the cellular diversity within the TME accurately. Upon integrating our pro-inflam and pro-tumor TAM-derived signatures with the clustering results, we identified eight clusters within the phs002065.v1 dataset and six within GSE120575.
SCENIC analysis
After annotating each cell type according to the characteristics of cell type marker genes, we used the pySCENIC package (V 0.12.0) to identify the enriched transcriptome factors (TF) of monocyte/TAM clusters62. SCENIC assesses the activity of transcription factors and their downstream target genes within individual cells. The key aspect of SCENIC is its ability to move beyond the expression level of individual transcription factors; it determines their regulons (the group of genes they regulate) by integrating co-expression analysis with motif enrichment analysis. The raw count matrix as the input was the normalized expression matrix output from Seurat. The Benjamini–Hochberg procedure was used to correct multiple hypotheses.
Single-cell pseudo-time analysis
A recently developed algorithm, PAGA, was performed on monocyte and macrophage clusters to define imputed pseudotime trajectories in monocytes/TAMs63. The computations were conducted using default parameters. Monocle3 (V 1.3.1) was also applied to construct the differentiation trajectory of monocytes/TAMs, and trajectory construction was performed with default parameters. Additionally, to evaluate the most likely trajectories of cell progression among monocytes/TAMs, we applied the algorithm slingshot (V 2.6.0)64. Each cell was given a pseudo-time value according to its predicted position along the predicted trajectories with default parameters.
Comparative analysis of cross-sectional studies using CellTypist
To perform comparisons of cross-sectional studies, we trained logistic regression models with pan‑cancer scRNA‑Seq datasets and cell annotations as the training data using the CellTypist (V 1.3.0)65. We used LR models optimized using the stochastic gradient descent algorithm to predict the identities of cells in three previously published datasets7,39,66 and compared the similarities of the monocyte/TAM clusters to the previously reported annotations.
Pro-inflam and pro-tumor TAM-specific signatures
The DEGs for pro-inflam and pro-tumor TAM clusters were identified as genes with an adjusted p value < 0.05 and logFC > 2 comparing all other clusters in the dataset. Next, we screened each DEG based on its ability to discriminate between individual clusters and selected only those with an AUC > 0.65.
SCISSOR analysis
We used SCISSOR (V 2.0.0) to integrate phenotypic data from bulk RNA-seq experiments with known immunotherapy response information (PRJEB25780) into our integrated single-cell data67. SCISSOR was run on the primary tumor cells of each patient individually according to the SCISSOR tutorial using mutation data (logistic regression) and OS (Cox regression) as dependent variables.
Pan-TCGA: immune microenvironment characterization
Comparisons of pro-inflam and pro-tumor TAM signatures between tumors from TCGA and their host tissues from GTEx were downloaded from the GTEx groups and screened for tumor types with significant differences between normal and tumor tissues68. The infiltration levels of pro-inflam and pro-tumor TAMs were quantified using the single sample gene set enrichment analysis (ssGSEA) implemented in the GSVA package (V 1.46.0) based on pro-inflam and pro-tumor TAM-specific signatures.
For immune infiltrating cell subpopulation analysis, we used the CIBERSORT algorithm to quantify the relative abundance of 22 types of immune cells in the Pan-Cancer Atlas of TCGA69. The TAM-related immune profiles were established by determining the Pearson correlation coefficients between pro-inflam and pro-tumor TAM signatures and CIBERSORT results. Additionally, we applied RF analysis to evaluate the importance of 22 immune cells for the interpretation of pro-inflam and pro-tumor TAMs using the rfPermute package (V 2.5.1). To estimate the importance of these diverse immune cells, we used the mean decrease in the Gini coefficient of variables, in which the higher the value of the mean decrease in the Gini score implies more important variables.
The immunophenoscore was determined as previously described70. Briefly, to depict the activation of immune microenvironment-related gene signatures in each tumor sample, we used ssGSEA to determine pathway activity using signatures that were summarized and compiled by the IOBR package (V 0.99.9)61. The signatures from previously published studies were retrieved using the signature_collection_citation function within the IOBR package.
Pan-TCGA: survival analysis
Kaplan–Meier survival curves with the cumulative event table and the cumulative number of censored subjects table were generated using the survminer package (V 0.4.9). The Cox proportional hazards regression model was used to calculate the hazard ratios between the good- and poor-outcome prediction groups within the survival package (V 3.5.0).
Pan-TCGA: consensus clustering
Consensus clustering analysis was performed on the R package ConsensusClusterPlus (V 1.62.0) using the following settings: maxK = 6, reps = 1000, pItem = 0.95, pFeature = 1, clusterAlg = “hc,” and distance = “Pearson.” Consensus clustering analysis used the K-means method to identify the optimum number of clusters in 9164 pan-TCGA patients based on the expression of pro-inflam and pro-tumor TAM-specific signatures. We used consensus clustering to determine two groups of robustly co-expressed genes for subtype classification to select genes that are co-regulated within each TAM, thus enhancing the specificity and discriminative power of our subtype definitions. The hard cutoffs refer to the classification of subtypes based on whether the median values of co-expressed pro-inflam and pro-tumor TAM are greater than or less than 0. Four subtypes were determined for each sample based on the scaled mean of co-expressed pro-inflam and pro-tumor TAM genes, namely absence (co-expressed pro-inflammatory and pro-tumor TAM genes ≤ 0), mixed (co-expressed pro-inflammatory and pro-tumor TAM genes > 0), pro-inflammatory TAMs (co-expressed pro-inflammatory TAM genes > 0 and co-expressed pro-tumor TAMs < 0), and pro-tumor TAMs (co-expressed pro-inflammatory TAM genes < 0 and co-expressed pro-tumor TAMs > 0) subgroups.
Pan-TCGA: dimension reduction
PCA was performed with the factoextra R package (V 1.0.7) using co-expressed pro-inflam and pro-tumor TAM genes. Of note, the top five components were used for non-linear dimension reduction to generate the UMAP for visualization.
Pan-TCGA: differential expression
We stratified patients into binary phenotypes based on pro-inflam and pro-tumor TAMs. Differential expression was performed on raw gene count data using edgeR (V 3.40.0) to identify groups of genes that are highly or lowly expressed in different binary phenotypes71. After generating a ranked list of DEGs for any comparisons of interest, gene set enrichment was performed using the camera statistical method in edgeR. The annotated gene sets provided within the MSigDB were used as the input to this GSEA method.
Pan-TCGA: transcription factor activity
We used DoRothEA (V 1.10.0) and decoupleR (V 2.9.1) to calculate the TF activity for the four subgroups in TCGA’s Pan-Cancer Atlas72,73. TF activity was assessed using analytic rank-based enrichment analysis, as part of DoRothEA. The normalized enrichment score for each TF regulator was derived from the DoRothEA tutorial. To assess TF enrichment scores within our dataset, we employed the univariate linear model approach as part of decoupleR. This methodology entails fitting a linear model for each sample in our dataset (denoted as ‘mat’) against each TF in our network (denoted as ‘net’), where the model predicts observed gene expression levels based solely on the interaction weights between the TF and its target genes. The t-value associated with the slope of each fitted model serves as the enrichment score for the corresponding TF.
Pan-TCGA: copy-number driver analysis, APOBEC enrichment estimation, and mutational signatures
To process the TCGA pan-cancer somatic data, we employed the maftools package (V 2.14.0). We utilized GISTIC 2.0 to identify significant events of genomic deletion or amplification, applying a refined computational approach that assesses somatic copy number alterations. For each tumor, an APOBEC mutagenesis enrichment score was estimated using trinucleotideMatrix function within maftools (V 2.14.0), and the samples were divided into APOBEC enriched and non-APOBEC enriched74. Signature decomposition was conducted on the bulk exome sequencing mutation data using deconstructSigs (V 1.9.0). The Wellcome Trust Sanger Institute Mutational Signature Framework was used as a reference to reconstruct the mutational signature of each tumor. Associations between these mutational signatures and TAM subtypes were determined using multinomial logistic regression models.
Spatial transcriptomics: patients and tissue acquisition
All samples were obtained from the Affiliated Cancer Hospital of Zhengzhou University. Four primary tissues were obtained from the gastroscopic sampling of clinical trial (ClinicalTrials.gov: NCT04341857) patients who had a pathological diagnosis of GC before receiving neoadjuvant immunotherapy75. The clinical stages of these patients were all cT3N + M0. Pathological regression in the primary tumor after surgery was graded according to the Ryan criteria for TRG as follows76: TRG0 for two patients, TRG1 for one patient, and TRG3 for one patient.
This study was approved by the Affiliated Cancer Hospital of Zhengzhou University Ethics Committee and written informed consent was obtained from all patients (IRB: 2023-KY-0018).
Spatial transcriptomics: library construction and sequencing
RNA quality was assessed by an Agilent 2100 Bioanalyzer (G2943CA). Four FFPE samples were used to prepare the spatial transcriptome for construction and sequencing. The Visium Spatial Gene Expression for FFPE (10× Genomics) protocol was used to perform the deparaffinization, hematoxylin, and eosin (H&E) staining, and decrosslinking. GEX libraries were generated for each section and spatial transcriptomic sequencing was conducted on a NovaSeq PE150 platform according to the manufacturer’s instructions (Illumina). FASTQ files together with the corresponding H&E-stained images in tiff format were processed by Space Ranger (V 1.3, 10× Genomics) with default parameters to generate the gene expression matrices.
Spatial transcriptomics: spatial gene signatures and neighborhood expression profiles
Visium data were analyzed with Seurat, and the data were normalized with SCTransform77. Given that each spot may contain approximately 8–20 cells, we developed a signature-based strategy to score the enrichments of the two TAMs in each spot. Specifically, we defined the average log-transformed normalization expression values of the genes in the signature as the corresponding TAM scores.
The neighborhood of a spot was defined as spots of a distance of ≤100 µm, resulting in sets of ≤6 neighbors per spot. Neighborhood expression profiles including the spot itself were generated by taking the top 5th percentile of spots for a given signature as the reference spots and then averaging the expression of neighborhood spots around the reference spot. Figure 4B shows the average of these profiles across all of the reference spots considered and standardized across the signatures.
Spatial transcriptomics: deconvolution and integration of spatial transcriptomics spots
To facilitate cell type deconvolution and the integration of spatial transcriptomics spots, spatial transcriptomics data was integrated with the largest reference scRNA-seq data for human GC to date using established methods78–80. To verify the robustness of the applied methods, integration methods were conducted that were conceived to integrate single-cell and spatial transcriptomics data. Specifically, CARD (V 1.0)81, RCTD (V 2.2.0)82, Tangram (V 1.0.3)83, Stereoscope (V 0.3)84, and anchors, as implemented in Seurat77, were performed. Tangram is an integrated deep-learning framework constructed for the projection of sc/snRNA-seq data onto spatially localized transcriptomic data. This allows alignment of profiles obtained from the sc/snRNA-seq method against spatially collected data from the same tissue; high-resolution, full-transcriptomic spatial mapping at single-cell resolution becomes feasible. To identify the overall concordance between the distinct integration methods tested, the results of the scores for each cell type were intercompared with each other in a correlation matrix for all spots using Pearson’s correlation. The score for each spot of each method was extracted from the resulting object, and the cell corresponding to the maximum value of each spot score was considered the cell type identified using the method. Considering that spatial transcriptomics spots are not at a single-cell resolution, at least ≥2 of the cell types identified by the method are considered specific cells (Fig. 4C). The spots containing macrophages were resolved into four types (absence, mixed, pro-inflam TAMs, and pro-tumor TAMs) based on pro-inflam and pro-tumor TAM-specific signatures, as described above. We combined the spots containing pro-inflam and mixed TAMs into enriched pro-inflam TAMs spots. Spots containing pro-tumor and mixed TAMs were taken as enriched pro-tumor TAM spots.
Tex process and hypoxia signatures were collected from previous studies85–87. Spots containing T cells and epithelial cells were scored for the expression of the Tex processes and hypoxia signatures, respectively, using the AUCell package (V 1.20.2)62.
Spatial transcriptomics: deciphering pro-inflam TAMs and CD8+ T-cell interactions
Spatial co-expression analysis of matching ligand-receptor pairs was based on information from CellTalkDB88. The ligand-pair analysis used in our study is adapted from established frameworks89. Initially, the expression of each ligand and receptor outlined in CellTalkDB was identified for each spot in the dataset. The expression of matched ligands and receptors was evaluated for each spot and its neighborhood spots. Hence, for each ligand-receptor pair, we identified whether, for a specific spot expressing partner A, partner B was expressed in the same spot or in one of the neighborhood spots (maximum six). Matched pairs co-expressed in the same spot or directly in neighborhood spots were deemed to have a high probability of interaction. The expression of the interacting pairs of ligands and receptors was inferred, and defined as the average between the mean expression of partner A and partner B on a given spot transcriptome and its neighborhood spots. Visualization of the interaction pairs was performed using the R package SPATA2 (V 0.1.0)90.
To link niche-specific TAM-to-cell communications networks to these interactions, the Pearson correlation between the interaction expression value for each ligand-receptor pair and enrichment of pro-inflam and pro-tumor TAM signatures was determined. We retrieved the interacting pairs of ligands and receptors satisfying the Pearson correlation coefficient ≥ 0.4. To account for the spatial location of niche-specific pro-inflam TAM-to-cell communications, we further classified spots including T cells based on their location concerning pro-inflam TAMs (juxta spot, pro-inflam TAM-neighboring spot). Given that each spot contains other cell types, spots without T cells and mono/macrophages were determined based on the cell type that appears most frequently according to the aforementioned deconvolution and integration methods. Next, the Wilcoxon rank-sum test was performed between the TRG0 and TRG1-3 groups to determine the interactions that were significantly altered between different pathological regressions (TRG0 and TRG1-3). GO enrichment analysis was performed using the clusterProfiler R package (V 4.6.0), and the cnetplot function was used to visualize the enrichment results of the GO biological process91.
Spatial transcriptomics: spatial trajectory
Dynamic gene expression changes along trajectories were evaluated using the plotTrajectoryGenes function within SPATA290. The SPATA2 package offers an innovative methodology for conducting spatial trajectory analysis in spatial transcriptomics, facilitating the visualization and examination of gene expression alterations along designated trajectories within tissue specimens. This technique permits investigators to interactively draw trajectories, emphasize them, designate them for later reference, and preserve them in the SPATA object for subsequent analysis. Utilizing the ‘plotTrajectoryGenes‘ function, we can visualize and analyze the variation in gene expression along spatially defined trajectories. This strategy yields critical insights into how expression levels evolve across various regions of interest and pinpoints genes exhibiting analogous patterns. Such a method is exceptionally advantageous for detecting gene expression patterns associated with gradients of cell differentiation or migration within the tissue environment.
Multiplex immunofluorescence staining
To quantify and spatially visualize pro-inflammatory TAMs, fluorescent multiplex immunohistochemistry was performed on FFPE tissue samples from 16 patients with GC before receiving neoadjuvant immunotherapy. Among these, 12 patients were classified as having TRG 1–3, while four were categorized as TRG0. The multiplex immunofluorescence staining was conducted using a Bond RX Autostainer (Leica Biosystems). FFPE tissue slides were sequentially incubated with primary antibodies targeting CD8a (85336, CST, 1:600), PDCD1 (ab237728, Abcam, 1:500), TNFSF10 (3219, CST, 1:3000), CXCL9 (ab290643, Abcam, 1:6000), CD68 (76437, CST, 1:5000), and PanCK (ab7753, Abcam, 1:200), followed by incubation with secondary antibodies and reactive Opal fluorophores (Opal480, Opal520, Opal620, Opal650, Opal690, Opal780, and Opal570). The multiplex IHC staining was performed sequentially using the following order of antibodies: CD68, PD1, TNFSF10, CD8, panCK, and CXCL9. After completing the sequential reactions, the slides were counterstained with DAPI. Once staining was completed, the slides were scanned using a fluorescence slide scanner (Leica, VERSA8). Digital whole slide multispectral images in. qptiff file format and hotspot deconvoluted images in. tiff format were uploaded to the Indica Lab HALO platform (V 3.0) for cell quantification. The HALO Highplex FL analysis module (Indica Labs; Albuquerque, NM, USA) was used with the Halo software for subsequent analysis. During the analysis process, the parameters for each analysis module were adjusted according to the actual conditions of the images. Typically, three representative sections of intact tissue from each patient were evaluated to provide a complete picture of the TME. However, the exact number of sections and area analyzed per section will depend on the sample size and tissue heterogeneity. The number of macrophages and T cells assessed will depend on the density of these cells in the selected tissue sections. For quantitative purposes, immune cell-rich sections are usually analyzed to ensure that the data accurately reflect the cellular composition and interactions in the TME. After determining the optimal parameters, the same analysis template was used for all similar images to avoid the influence of subjective factors. Each annotation of different regions represented an analysis layer, the names of which could be customized to identify the different staining signals of cells within the analysis field of view. Upon completion of the single-layer quantitative analysis for each image, the overall information of all results was statistically summarized. For each defined cell type, the number of positive cells in the sample and the percentage of that cell type among the total cells in the sample were calculated.
ICI response prediction model training
We implemented a machine learning framework using the scikit-learn package (V 1.1.0) built on Python (V 3.9.1)92. To generate the training (70%) and test (30%) datasets, we partitioned the dataset using the train_test_split function, which randomly divides the dataset into training and test subsets using the test_size = 0.3 parameter. The model features were used as discrete values (cancer type and drug class) or continuous values (median expression levels of pro-inflam and pro-tumor TAM signatures). In one-hot encoding, we used the get_dummies function to create dummy variables of the discrete values. Patients were categorized into responders, defined as CR, PR, or SD, with OS > 12 months, and non-responders, defined as PD or SD, with OS ≤ 12 months. We trained the ICI response classification model with model features using five common machine learning (ML) algorithms, namely the support vector machine, RF, logistic regressions, k-nearest neighbors, and decision trees. Fivefold CV was adopted for hyperparameter tuning using GridSearchCV to optimize the performance of the model derived from the training dataset, and the RF classification model (hyperparameters: max_depth = 5, n_estimators = 200, and min_samples_split = 2) with the best performance was chosen as the final model.
Evaluating machine learning classifiers
We used the precrec package (V 0.14.1) to generate the AUC and area under the precision-recall curve (AUPRC) values for RF models with three feature types, namely integrated pro-inflam and pro-tumor TAMs and clinical data; pro-inflam TAMs and clinical data; and pro-tumor TAMs and clinical data. The optimal thresholds for the response probability calculated with the RF model were determined using Youden’s index method with the coords function from the pROC package (V 0.18.0).
Comparing TAM signatures with other published signatures
To further evaluate the predictive value of TAM signatures, we compared the TAM signatures with previously reported ICI response signatures, including IFN-γ93, GCScore3, EMT94, and TME-based markers (Treg95, T.cell.inflamed93, cancer-associated fibroblast96, and M1 TAM60).
Machine learning explainability
To enhance the interpretability of our model, we used the SHAP (V 0.41.0) and PDPbox (V 0.2.1) packages built on Python, which offer insights into the importance of each variable in the interpretation of data. Shapley values were computed to measure the contribution of each parameter to the performance of the prediction model.
ICI response survival analysis model
Next, we constructed our machine learning-based survival models, including the Cox proportional hazards and RSF. Model discrimination was assessed using the concordance index (C-index), and calibration was evaluated using Brier scores calculated using the pec package (V 2.5.4). The Kaplan–Meier plot and log-rank p values were generated using the survminer package.
Pan‑cancer scRNA‑Seq datasets
ScRNA-seq datasets containing monocytes/macrophages from BRCA60, ESCC97, PDAC98, ESCA6, THCA6, RC6, and CRC99 were downloaded from the gene expression omnibus (GEO; GSE114725, GSE145370, GSE154778, GSE154763, and GSE146771,). The dataset describing STAD100 was downloaded from https://dna-discovery.stanford.edu/research/datasets/, while that describing LC101 was downloaded under the BioProject accession number PRJNA591860. The accession number for HCC102 is CNP0000650 (https://db.cngb.org/ search/project/CNP0000650).
Two other scRNA-seq cohorts, STAD78, and pan-cancer103, were used to verify the macrophage-conserved nature of TAM signatures. The data of these two cohorts were accessed through the GEO accession numbers GSE183904 and GSE203612.
scRNA‑Seq and Bulk RNA-seq ICI cohorts
Data of independent scRNA-seq ICI cohorts were accessed through the accession numbers phs002065.v1.p139 and GSE120575104 with comprehensive immunotherapy response information.
We retrospectively analyzed the gene expression profiles from eight ICI datasets. Transcriptomic datasets were obtained under accession numbers IMvigor21017, PRJEB25780105, GSE100797106, GSE115821107, GSE135222108, GSE91061109, MGSP110, and ERP105482111.
Pan‑cancer TCGA dataset
Transcriptomic data of the TCGA pan-cancer cohort were obtained from UCSC Xena (https://xena.ucsc.edu/) to investigate pro-inflam and pro-tumor TAM subtypes across 32 cancer types. Clinical, copy number variation data, and mutation data (mc3.v0.2.8.PUBLIC.maf) were downloaded from the GDC TCGA data portal (https://portal.gdc.cancer.gov/). The TMB, neoantigen load, HRD, tumor purity/ploidy, aneuploidy scores, and LOH, were obtained from the respective supplemental materials112.
Spatial transcriptomics datasets
Spatially resolved transcriptomics data from Alexander et al.113, Thomas et al.114, and Chen et al.115 can be downloaded from the Zenodo data repositories 10.5281/zenodo.4739739, 10.17632/g67bkbnhhg.1 and http://lifeome.net/supp/livercancer-st/data.htm, respectively. All datasets used in the paper are stored in Supplementary Table 2.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82373271), the National Natural Science Fund Youth Fund of China (82003041); Joint Funds of the National Natural Science Foundation of China (U2004132); the Key Scientific and Technological Projects in Henan Province (222102310324); the Project of Tackling Problems in Medical Science and Technology of Henan Province (LHGJ20190631).
Author contributions
N.L. and C.W. planned and designed the study. C.W. wrote the manuscript and carried out the data analysis. J.Y.M., Y.M.W., Y.S.W., and Y.W.Y. conducted the sample preparation. X.Q.X. performed pathology assessments. L.S.D., Y.W.D., Y.L.B., C.Z., and W.S. recruited patients, gathered the clinical data for the study, and made conceptual contributions to the study. N.L. and X.S.L. supervised the project. All authors read and approved the final manuscript.
Data availability
The processed count matrices derived from the raw ST data generated in this study have been deposited in the GSA-Human (Genome Sequence Archive for Human in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, https://ngdc.cncb.ac.cn/gsa-human/) under the accession code PRJCA024136. The data are available under controlled access. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Code availability
All code generated for analysis is available from the authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chen Wei, Yijie Ma, Mengyu Wang, Siyi Wang.
Contributor Information
Qingxin Xia, Email: 13838173710@139.com.
Suxia Luo, Email: zlyyluosuxia0361@zzu.edu.cn.
Ning Li, Email: lining97@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00660-4.
References
- 1.Whiteside, T. L. The tumor microenvironment and its role in promoting tumor growth. Oncogene27, 5904–12 (2008). 10.1038/onc.2008.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassetta, L. & Pollard, J. W. A timeline of tumour-associated macrophage biology. Nat. Rev. Cancer23, 238–57 (2023). 10.1038/s41568-022-00547-1 [DOI] [PubMed] [Google Scholar]
- 3.Wei, C. et al. Characterization of gastric cancer stem-like molecular features, immune and pharmacogenomic landscapes. Brief Bioinform.23, bbab386 (2022). 10.1093/bib/bbab386 [DOI] [PubMed] [Google Scholar]
- 4.Wei, C. et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer18, 64 (2019). 10.1186/s12943-019-0976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang, C. et al. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. J. Am. Soc. Gene Ther.29, 2088–107 (2021). 10.1016/j.ymthe.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell184, 792–809.e23 (2021). 10.1016/j.cell.2021.01.010 [DOI] [PubMed] [Google Scholar]
- 7.Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity54, 1883–900.e5 (2021). 10.1016/j.immuni.2021.07.007 [DOI] [PubMed] [Google Scholar]
- 8.Combes, A. J. et al. Discovering dominant tumor immune archetypes in a pan-cancer census. Cell185, 184–203.e19 (2022). 10.1016/j.cell.2021.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol.14, 399–416 (2017). 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nixon, B. G. et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity55, 2044–58.e5 (2022). 10.1016/j.immuni.2022.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara, Y. et al. Guanylate-binding protein 5 is a marker of interferon-γ-induced classically activated macrophages. Clin. Transl. Immunol.5, e111 (2016). 10.1038/cti.2016.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill, L. A., Kishton, R. J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol.16, 553–65 (2016). 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong, D., Wang, Y. & You, M. A gene expression signature of TREM2(hi) macrophages and γδ T cells predicts immunotherapy response. Nat. Commun.11, 5084 (2020). 10.1038/s41467-020-18546-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loftus, P. G. et al. Targeting stromal cell Syndecan-2 reduces breast tumour growth, metastasis and limits immune evasion. Int. J. Cancer148, 1245–59 (2021). 10.1002/ijc.33383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.House, I. G. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res.26, 487–504 (2020). 10.1158/1078-0432.CCR-19-1868 [DOI] [PubMed] [Google Scholar]
- 16.Yang, L. & Lin, P. C. Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. Semin. Cancer Biol.47, 185–95 (2017). 10.1016/j.semcancer.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature554, 544–8 (2018). 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity39, 1–10 (2013). 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Q. et al. Interrogation of the microenvironmental landscape in spinal ependymomas reveals dual functions of tumor-associated macrophages. Nat. Commun.12, 6867 (2021). 10.1038/s41467-021-27018-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, Y. et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene36, 1157–66 (2017). 10.1038/onc.2016.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell179, 829–45.e20 (2019). 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 22.Lu, Y. et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat. Commun.13, 4594 (2022). 10.1038/s41467-022-32283-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Investig.122, 787–95 (2012). 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assoun, S. et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer132, 65–71 (2019). 10.1016/j.lungcan.2019.04.005 [DOI] [PubMed] [Google Scholar]
- 25.Jia Q, Wang J, He N, He J, & Zhu B. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight4, e127901 (2019). [DOI] [PMC free article] [PubMed]
- 26.Zhang, P. & Huang, Y. Genomic alterations in KMT2 family predict outcome of immune checkpoint therapy in multiple cancers. J. Hematol. Oncol.14, 39 (2021). 10.1186/s13045-021-01050-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorand, R. D. et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science353, 399–403 (2016). 10.1126/science.aae0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao, J. et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell167, 397–404.e9 (2016). 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedegaard, J. et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell30, 27–42 (2016). 10.1016/j.ccell.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 30.Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell165, 35–44 (2016). 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hargadon, K. M. The role of interferons in melanoma resistance to immune checkpoint blockade: mechanisms of escape and therapeutic implications. Br. J. Dermatol185, 1095–104 (2021). 10.1111/bjd.20608 [DOI] [PubMed] [Google Scholar]
- 32.Ma, R. Y., Black, A. & Qian, B. Z. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol.43, 546–63 (2022). 10.1016/j.it.2022.04.008 [DOI] [PubMed] [Google Scholar]
- 33.Qian, J. et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res.30, 745–62 (2020). 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, X. et al. TREM2 promotes glioma progression and angiogenesis mediated by microglia/brain macrophages. Glia71, 2679–95 (2023). 10.1002/glia.24456 [DOI] [PubMed] [Google Scholar]
- 35.Roumenina, L. T. et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol. Res.7, 1091–105 (2019). 10.1158/2326-6066.CIR-18-0891 [DOI] [PubMed] [Google Scholar]
- 36.McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol.37, 457–95 (2019). 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- 37.Chow, A., Perica, K., Klebanoff, C. A. & Wolchok, J. D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol.19, 775–90 (2022). 10.1038/s41571-022-00689-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tietscher, S. et al. A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer. Nat. Commun.14, 98 (2023). 10.1038/s41467-022-35238-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi, K. et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell39, 649–61.e5 (2021). 10.1016/j.ccell.2021.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulle, A. & Lim, K. H. Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther.5, 249 (2020). 10.1038/s41392-020-00341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, W. R. & Hay, M. P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer11, 393–410 (2011). 10.1038/nrc3064 [DOI] [PubMed] [Google Scholar]
- 42.Denhardt, D. T., Noda, M., O’Regan, A. W., Pavlin, D. & Berman, J. S. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Investig.107, 1055–61 (2001). 10.1172/JCI12980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, J. et al. Characterizing intercellular communication of pan-cancer reveals SPP1+ tumor-associated macrophage expanded in hypoxia and promoting cancer malignancy through single-cell RNA-seq data. Front. Cell Dev. Biol.9, 749210 (2021). 10.3389/fcell.2021.749210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pombo Antunes, A. R. et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci.24, 595–610 (2021). 10.1038/s41593-020-00789-y [DOI] [PubMed] [Google Scholar]
- 45.Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov.21, 799–820 (2022). 10.1038/s41573-022-00520-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristescu, R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science362, eaar3593 (2018). 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiMarco, A. V. et al. APOBEC mutagenesis inhibits breast cancer growth through induction of T cell-mediated antitumor immune responses. Cancer Immunol. Res.10, 70–86 (2022). 10.1158/2326-6066.CIR-21-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aspeslagh, S., Chabanon, R. M., Champiat, S. & Postel-Vinay, S. Understanding genetic determinants of resistance to immune checkpoint blockers. Semin. Cancer Biol.65, 123–39 (2020). 10.1016/j.semcancer.2019.12.020 [DOI] [PubMed] [Google Scholar]
- 49.Kong, J. et al. Network-based machine learning approach to predict immunotherapy response in cancer patients. Nat. Commun.13, 3703 (2022). 10.1038/s41467-022-31535-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiesweg, M. et al. Machine learning reveals a PD-L1–independent prediction of response to immunotherapy of non-small cell lung cancer by gene expression context. Eur. J. Cancer140, 76–85 (2020). 10.1016/j.ejca.2020.09.015 [DOI] [PubMed] [Google Scholar]
- 51.Polano, M. et al. A pan-cancer approach to predict responsiveness to immune checkpoint inhibitors by machine learning. Cancers11, 1562 (2019). 10.3390/cancers11101562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seitz, S. et al. CXCL9 inhibits tumour growth and drives anti-PD-L1 therapy in ovarian cancer. Br. J. Cancer126, 1470–80 (2022). 10.1038/s41416-022-01763-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang, Y. K. et al. CXCL9 is a potential biomarker of immune infiltration associated with favorable prognosis in ER-negative breast cancer. Front Oncol.11, 710286 (2021). 10.3389/fonc.2021.710286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat. Commun.13, 1742 (2022). 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bill, R. et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science381, 515–24 (2023). 10.1126/science.ade2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qu, Y. et al. Baseline frequency of inflammatory Cxcl9-expressing tumor-associated macrophages predicts response to avelumab treatment. Cell Rep.32, 107873 (2020). 10.1016/j.celrep.2020.107873 [DOI] [PubMed] [Google Scholar]
- 57.Liu, Y. et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol.78, 770–82 (2023). 10.1016/j.jhep.2023.01.011 [DOI] [PubMed] [Google Scholar]
- 58.Ozato, Y. et al. Spatial and single-cell transcriptomics decipher the cellular environment containing HLA-G+ cancer cells and SPP1+ macrophages in colorectal cancer. Cell Rep.42, 111929 (2023). 10.1016/j.celrep.2022.111929 [DOI] [PubMed] [Google Scholar]
- 59.Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience7, giy083 (2018). 10.1093/gigascience/giy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell174, 1293–308.e36 (2018). 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng, D. et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol.12, 687975 (2021). 10.3389/fimmu.2021.687975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–6 (2017). 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol.20, 59 (2019). 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom.19, 477 (2018). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suo, C. et al. Mapping the developing human immune system across organs. Science376, eabo0510 (2022). 10.1126/science.abo0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med.27, 820–32 (2021). 10.1038/s41591-021-01323-8 [DOI] [PubMed] [Google Scholar]
- 67.Sun, D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol.40, 527–38 (2022). 10.1038/s41587-021-01091-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature550, 204–13 (2017). 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun.4, 1–11 (2013). 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charoentong, P. et al. Pan-cancer Immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep.18, 248–62 (2017). 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 71.McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res.40, 4288–97 (2012). 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia-Alonso, L., Holland, C. H., Ibrahim, M. M., Turei, D. & Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res.29, 1363–75 (2019). 10.1101/gr.240663.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Badia-i-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinforma. Adv.2, vbac016 (2022). 10.1093/bioadv/vbac016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayakonda, A., Lin, D. C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res.28, 1747–56 (2018). 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li, N. et al. Efficacy and safety of neoadjuvant sintilimab in combination with FLOT chemotherapy in patients with HER2-negative locally advanced gastric or gastroesophageal junction adenocarcinoma: an investigator-initiated, single-arm, open-label, phase II study. Int J. Surg.110, 2071–2084 (2024). 10.1097/JS9.0000000000001119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryan, R. et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology47, 141–6 (2005). 10.1111/j.1365-2559.2005.02176.x [DOI] [PubMed] [Google Scholar]
- 77.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–902.e21 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar, V. et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov.12, 670–91 (2022). 10.1158/2159-8290.CD-21-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet.22, 627–44 (2021). 10.1038/s41576-021-00370-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet.53, 1698–711 (2021). 10.1038/s41588-021-00972-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol.40, 1349–59 (2022). 10.1038/s41587-022-01273-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol.40, 517–26 (2022). 10.1038/s41587-021-00830-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods18, 1352–62 (2021). 10.1038/s41592-021-01264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol.3, 565 (2020). 10.1038/s42003-020-01247-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beltra, J. C. et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity52, 825–41.e8 (2020). 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu, X. J. et al. The genome landscape of tibetan sheep reveals adaptive introgression from argali and the history of early human settlements on the Qinghai–Tibetan plateau. Mol. Biol. Evol.36, 283–303 (2019). 10.1093/molbev/msy208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ye, Y. et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab.1, 431–44 (2019). 10.1038/s42255-019-0045-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shao, X. et al. CellTalkDB: a manually curated database of ligand–receptor interactions in humans and mice. Brief. Bioinforma.22, bbaa269 (2021). 10.1093/bib/bbaa269 [DOI] [PubMed] [Google Scholar]
- 89.Kaufmann, M. et al. Identification of early neurodegenerative pathways in progressive multiple sclerosis. Nat. Neurosci.25, 944–55 (2022). 10.1038/s41593-022-01097-3 [DOI] [PubMed] [Google Scholar]
- 90.Kueckelhaus, J et al. Inferring spatially transient gene expression pattern from spatial transcriptomic studies. Preprint at bioRxiv10.1101/2020.10.20.346544.
- 91.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol.16, 284–7 (2012). 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res.12, 2825–30 (2011). [Google Scholar]
- 93.Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig.127, 2930–40 (2017). 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan, T. Z. et al. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med.6, 1279–93 (2014). 10.15252/emmm.201404208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Givechian, K. B. et al. Identification of an immune gene expression signature associated with favorable clinical features in Treg-enriched patient tumor samples. NPJ Genom. Med.3, 14 (2018). 10.1038/s41525-018-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nurmik, M., Ullmann, P., Rodriguez, F., Haan, S. & Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer146, 895–905 (2020). 10.1002/ijc.32193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng, Y. et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat. Commun.11, 6268 (2020). 10.1038/s41467-020-20019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin, W. et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med.12, 80 (2020). 10.1186/s13073-020-00776-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell181, 442–59.e29 (2020). 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 100.Sathe, A. et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res.26, 2640–53 (2020). 10.1158/1078-0432.CCR-19-3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maynard, A. et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell182, 1232–51.e22 (2020). 10.1016/j.cell.2020.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun, Y. et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell184, 404–21.e16 (2021). 10.1016/j.cell.2020.11.041 [DOI] [PubMed] [Google Scholar]
- 103.Barkley, D. et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet.54, 1192–201 (2022). 10.1038/s41588-022-01141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell175, 998–1013.e20 (2018). 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim, S. T. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med.24, 1449–58 (2018). 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 106.Lauss, M. et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun.8, 1738 (2017). 10.1038/s41467-017-01460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Auslander, N. et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med.24, 1545–9 (2018). 10.1038/s41591-018-0157-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung, H. et al. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun.10, 4278 (2019). 10.1038/s41467-019-12159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Riaz, N. et al. Tumor and microenvironment evolution during Immunotherapy with Nivolumab. Cell171, 934–49.e16 (2017). 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med.25, 1916–27 (2019). 10.1038/s41591-019-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gide, T. N. et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer cell35, 238–55.e6 (2019). 10.1016/j.ccell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 112.Thorsson, V. et al. The immune landscape of cancer. Immunity48, 812–30.e14 (2018). 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet.53, 1334–47 (2021). 10.1038/s41588-021-00911-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li, R. et al. Mapping single-cell transcriptomes in the intra-tumoral and associated territories of kidney cancer. Cancer Cell40, 1583–99.e10 (2022). 10.1016/j.ccell.2022.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu, R. et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci. Adv.7, eabg3750 (2021). 10.1126/sciadv.abg3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed count matrices derived from the raw ST data generated in this study have been deposited in the GSA-Human (Genome Sequence Archive for Human in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, https://ngdc.cncb.ac.cn/gsa-human/) under the accession code PRJCA024136. The data are available under controlled access. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All code generated for analysis is available from the authors upon request.