Abstract
To understand the high variability of the asymptomatic interval between primary human immunodeficiency virus type 1 (HIV-1) infection and the development of AIDS, we studied the evolution of the C2-V5 region of the HIV-1 env gene and of T-cell subsets in nine men with a moderate or slow rate of disease progression. They were monitored from the time of seroconversion for a period of 6 to 12 years until the development of advanced disease in seven men. Based on the analysis of viral divergence from the founder strain, viral population diversity within sequential time points, and the outgrowth of viruses capable of utilizing the CXCR4 receptor (X4 viruses), the existence of three distinct phases within the asymptomatic interval is suggested: an early phase of variable duration during which linear increases (∼1% per year) in both divergence and diversity were observed; an intermediate phase lasting an average of 1.8 years, characterized by a continued increase in divergence but with stabilization or decline in diversity; and a late phase characterized by a slowdown or stabilization of divergence and continued stability or decline in diversity. X4 variants emerged around the time of the early- to intermediate-phase transition and then achieved peak representation and began a decline around the transition between the intermediate and late phases. The late-phase transition was also associated with failure of T-cell homeostasis (defined by a downward inflection in CD3+ T cells) and decline of CD4+ T cells to ≤200 cells/μl. The strength of these temporal associations between viral divergence and diversity, viral coreceptor specificity, and T-cell homeostasis and subset composition supports the concept that the phases described represent a consistent pattern of viral evolution during the course of HIV-1 infection in moderate progressors. Recognition of this pattern may help explain previous conflicting data on the relationship between viral evolution and disease progression and may provide a useful framework for evaluating immune damage and recovery in untreated and treated HIV-1 infections.
Human immunodeficiency virus type 1 (HIV-1) infection is characterized by an asymptomatic period of highly variable length between acute infection and AIDS (32, 38, 42, 53–55, 72). Among the viral factors that may affect the rate of HIV disease progression, HIV strains with a syncytium-inducing (SI) phenotype on MT-2 cells (now referred to as X4 or R5/X4 for viruses with dualtropism that includes CXCR4 [3]) have long been associated with faster disease progression, although the mechanism of this effect is unclear (7, 13, 22, 36, 64, 70, 78). Also unclear is the role of viral evolutionary change in affecting the duration of the asymptomatic period. While several studies have found an inverse relationship between the rate of viral diversification and disease progression (15, 16, 23, 45, 46, 73, 82, 83), others have not (26, 49, 52); one study found instances of both inverse and direct relationships (51).
To understand more clearly the relationship of viral evolutionary changes, the emergence of X4 viral phenotypes, and the rate of progression over the course of HIV-1 disease, we studied the evolution of the C2-V5 region of the HIV-1 env gene in nine homosexual men enrolled in the Multicenter AIDS Cohort Study (MACS) (34). These nine men constituted a subset of individuals previously selected for a study of changes in plasma viremia, T-cell subsets, and cytotoxic memory T-cell responses in MACS participants who had a downward inflection in CD3+ T-cell numbers (65). A high prevalence (75%) of such inflections has been found in MACS participants who have developed AIDS, occurring a median of 1.7 years prior to the onset of AIDS (24). The C2-V5 region of env was chosen for the analysis of viral changes because it encodes an important target for immune responses, determines coreceptor specificity, and exhibits a high degree of phylogenetically informative variability (41, 42). An average of 12 time points per person were studied, covering 6 to 12 years of infection.
As the present study is one of the most comprehensive longitudinal studies of HIV-1 evolution in vivo to date, it provides unique insights into the patterns of viral change over time, as well as the emergence and representation of X4 viruses and changes in T-cell subsets associated with disease progression. The observed patterns and associations may explain inconsistencies among previous reports relating HIV-1 sequence evolution to disease progression and enhance our understanding of the factors that lead to progression.
MATERIALS AND METHODS
Study participants and clinical specimens.
The MACS is an ongoing prospective cohort study, with over 5,000 homosexual men participating, whose study design has been described in detail (34). At semiannual visits, participants provide responses to structured, interviewer-administered questionnaires (including detailed information about therapies taken since the last visit) and provide blood specimens that are stored at −70°C (serum and plasma) or −135°C (peripheral blood mononuclear cells [PBMCs]) at a central repository. T-cell subsets were determined by flow cytometry by previously described methods (25, 68), with antibodies obtained from Becton Dickinson Immunocytometry Systems (San Jose, Calif.) and an ELITE flow cytometer (Coulter Electronics, Hialeah, Fla.).
The selection of participants for the present study has been described elsewhere (65). Briefly, 306 seroconverters, enrolled between 1983 and 1984 with less than 8 months between their last seronegative and first seropositive visits, were ranked for their decline in longitudinal CD3+ cell counts with data gathered through 1994. Individuals were required to develop and maintain low CD3+ cell levels for at least two visits and to have frozen plasma and PBMCs available at most or all semiannual visits. Six of the nine men (all except participants 8, 9, and 11) selected for this study were among those with the most well-defined inflections in CD3+ lymphocyte counts. One man (participant 8) had a less well-defined inflection of CD3+ lymphocytes but progressed to a CD4+ lymphocyte count of <200 cells/μl. Two men (participants 9 and 11) were initially selected to be part of a comparison group with stable lymphocyte counts but exhibited signs of progression with subsequent follow-up. Clinical and/or immunologic AIDS (11) was observed in seven men, and five men had died by the end of 1998. Seven men received antiretroviral treatment (primarily zidovudine) (see Fig. 2d) during the course of study, and participant 8 reported the use of potent antiretroviral therapy at the last study time point. Subject 11, the slowest progressor studied, did not take any antiretroviral therapy or prophylactic antibiotics during the time period analyzed in this study.
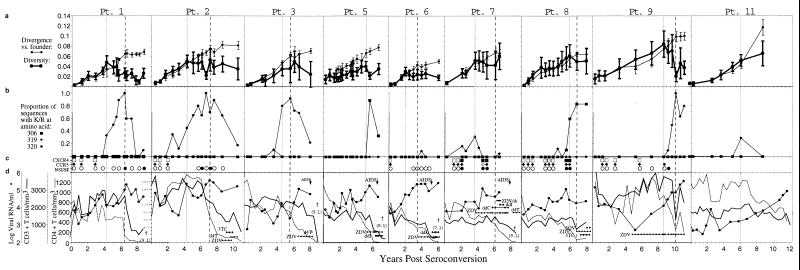
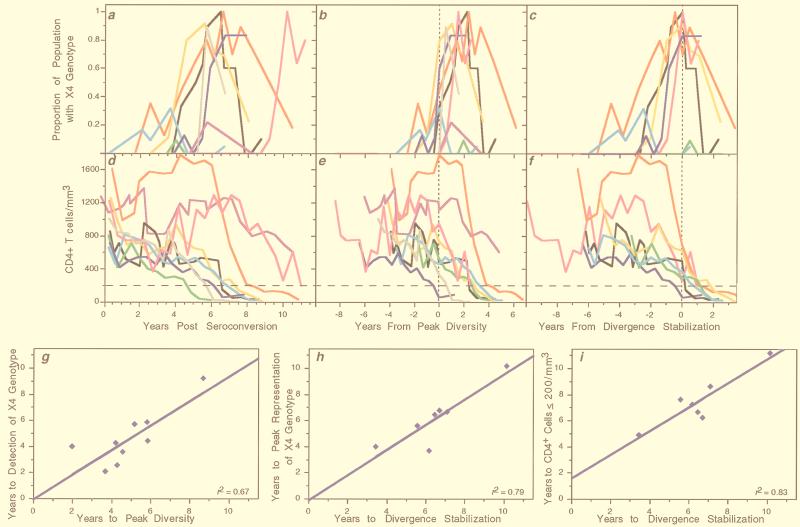
FIG. 2.
Summary of sequences, viral phenotypes, and T-cell measures. Participant identifiers above each panel are from a previous study (65); participants 4 and 10 were excluded from the current study due to a lack of available specimens. The horizontal axis of each panel indicates the time relative to seroconversion. (a) Genetic distances of the combined population of viral DNA and RNA sequences relative to the founder strain (divergence, thin lines) and within individual time points (diversity, thick lines). The left (dotted) vertical line for each participant (and the single dotted line for participants 5 and 11) indicates the time of peak viral diversity, at which a significant stabilization or decrease in the slope of population diversity growth was observed as defined in the text. The final time point from participant 7 had an increase in overall viral diversity due to the appearance of unique divergent virus populations in the plasma versus the those in the PBMCs, although these populations taken individually had much lower levels of diversity (see appendix at reference 71a). The right (dashed) vertical line for each participant (except participants 5 and 11) indicates the time at which the divergence from the founder strain began to slow down or stabilize (divergence stabilization). (b) Viral genotype analysis. Each panel shows the proportions of deduced amino acid sequences containing mutations predictive of the X4 phenotype, based on encoding a basic amino acid (lysine or arginine, indicated by K/R in the figure) at residues 306, 319, or 320 within the envelope glycoprotein. For participant 7, mutations at both position 319 and position 320 (open symbol) were found at one time point. (c) Viral phenotype and coreceptor usage analysis. Data taken from Rinaldo et al. (65). The squares indicate CXCR4 coreceptor usage, and the diamonds indicate CCR5 coreceptor usage by virus isolates derived from the indicated time points. Open symbols indicate no growth, and filled symbols indicate growth on cells expressing the specified coreceptor plus CD4 (see Materials and Methods). The filled circles indicate syncytium formation (SI phenotype), and the open circles indicate a lack of syncytium formation (NSI phenotype) when the virus isolates were added to MT-2 cells. (d) Clinical progression. Data was taken from Rinaldo et al. (65). CD4+ T-cell levels are shown with a dotted line. Patient 2 had exceptionally high CD4+ T-cell levels early in infection which are plotted with a different scale as indicated in the panel. CD3+ T-cell numbers are shown with the thick line, whereas plasma RNA levels are shown with the filled circles connected by a thin line. Antiretroviral treatment, AIDS diagnosis by development of opportunistic infections (AIDS), and survival time are also indicated. Participants’ visits, at which time the antiretroviral drugs were prescribed, are indicated (ZDV, zidovudine; d4T, stavudine; 3TC, lamivudine; ddI, didanosine; SQV, saquinavir). Five participants died after the period of analysis shown, and this time (in years) is indicated in parentheses below the dagger (†).
PCR amplification, cloning, and sequencing.
Ficoll-Hypaque-purified PBMCs were lysed by the addition of quick lysis buffer (10 mM Tris [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20) and digested with 100 μg of proteinase K per ml at 56°C for 1 h. Lysates from PBMCs were treated with GeneReleaser (BioVentures, Murfreesboro, Tenn.) according to the manufacturer’s protocol and used for PCR. Nested PCR with hot start (19) was used on serial dilutions of the lysate, with amplification of the entire gp160 in the first round followed by amplification of the C2-V5 region in the second round. Template quantitation was done by measuring the frequency of positive and negative reactions at different dilutions (66). Primer sequences for ED3, ED12, ED31, DR7, and DR8 have been described previously (14, 17, 45); primer Nef3 is 9028-5′-TAAGTCATTGGTCTTAAAGGTACC, where the number corresponds to the position in the NL4-3 genome (GenBank accession no. M19921) of the 5′ nucleotide. First-round primers, ED3 and Nef3, were used with the following cycling conditions: 94°C for 2 min; 3 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min; 36 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min; and a final incubation at 72°C for 10 min. The second-round PCR included a 1:20 dilution of the first-round products in fresh reaction buffer plus primers DR7 and DR8 with the following cycling conditions: 94°C for 2 min; 36 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final incubation at 72°C for 10 min. Plasma samples were processed for RNA extraction and cDNA synthesis as previously described (45). cDNA was amplified first with primers ED12 and ED31, followed by a second round as for the PBMC samples.
All PCR amplifications were done with procedural safeguards, including the aliquoting of all reagents and the physical separation of sample processing and post-PCR handling steps. Replicate control amplifications with no template were included in every PCR experiment to test for carryover contamination. Amplifications with 10 genome equivalents of 8E5 cell (21) DNA were also performed as an internal control to monitor the efficiency of the PCR. To avoid template resampling (44), we pooled first-round PCR products from an average of 50 viral templates and reamplified the pool in a second round for derivation and sequence analysis of an average of 13 clones per time point. Sequences were also derived from participant 7 by selecting a single clone per PCR for sequencing. The PCR products were cloned into a TA vector (Invitrogen, Carlsbad, Calif.) and sequenced on an ABI 370 or 377 sequencer (Foster City, Calif.) by dye-primer and dye-terminator protocols.
Sequence analysis.
Sequences bearing open reading frames (∼90% of all sequences determined) were first aligned with the Pileup program in the GCG suite (Genetics Computer Group, Madison, Wis.) and then manually edited. Pairwise evolutionary nucleotide distances (excluding gaps in the pairwise alignment) were estimated with the Kimura two-parameter model of evolution (35) with a transition/transversion ratio of 2 as implemented in MEGA (40) or PHYLIP, version 3.5 (20). DNA distances were also estimated with a general time-reversible model with site-to-site variation in substitution rates (discrete approximation of a γ distribution with a shape parameter, α = 0.5 with four bins [77]). Identical trends were seen with either method, although as expected, the Kimura two-parameter method tended to underestimate DNA distances at higher divergence levels. This comparison is available online (71a).
Analysis of viral divergence and diversity.
Both viral divergence from the founder strain and viral diversity were estimated at each time point. To estimate viral diversity at a given time point, we determined the mean and standard deviation for pairwise nucleic acid distances between all sequences obtained at that time point. To estimate viral divergence at a given time point, a founder sequence was approximated as the sequences found at the initial virus-positive time point. Nearly identical values for viral divergence were observed when these distances were estimated with a single consensus sequence from the first time point. The mean and standard deviation for all pairwise comparisons between sequences from the first positive time point and subsequent time points were then calculated. The rate of substitution was measured by estimating the expected number of substitutions that accumulated along the branch from an ancestral sequence (77) as described below.
At time t = t0, we estimated the rate of substitution by estimating the expected number of substitutions that accumulate along the branch from an ancestral sequence to a sequence sampled at time t = t1 and dividing by the elapsed time, i.e.,
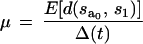 |
1 |
where μ is the rate of substitution per site per unit time, Δ(t) = t1−t0, and E[d(sa0,s1)] is the expected number of substitutions per site between a sequence at t1, s1 and the ancestral sequence at t0, sa0. However, with a small sample of sequences taken from a much larger population, it is unlikely that we have sampled the ancestral sequence sa0. Therefore, to reexpress equation 1 so that the expectation containing sa0 is removed, we can use the fact that, under an additive model of sequence evolution, d(sa0, s1) = d(s0,s1) − d(sa0, s0), where s0 is a sequence sampled at t0. Therefore, we can rewrite the numerator of equation 1 as E[d(s0,s1)]−E[d(sa0,s0)]. Under a neutral model of evolution, sa0 is one randomly selected sequence, and E[d(sa0,s1)] = E[d(s0x,s0y)] where the right-hand term is the expected distance between any pair of sequences sampled randomly from the population at t0. If selection is operating, then E[d(sa0,s0)] = E[d(s0x, s0y)] + ɛ. Using this, we can show that
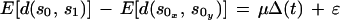 |
2 |
Since the viral population at the first time point is effectively homogeneous, let E[d(s0x,s0y)] ≈ 0. E[d(s0,s1)] can then be estimated by the average pairwise distances between all sequences from t1 and t0, and equation 2 allows us to solve for μ by linear regression.
Mantel’s generalized regression permutation procedure (47, 62, 74, 80) was used to test the hypothesis that the average pairwise differences between all possible pairs of sequences within a time point were significantly smaller than average pairwise differences between all possible pairs of sequences between time points. Two matrices were constructed. The first was a matrix, E, of pairwise evolutionary distances constructed using the Kimura two-parameter model (35). The second matrix, M, with elements M(i,j) = |x(i) − y(j)| if x ≠ y and M(i,j) = 0 otherwise, corresponds to the elements of E representing the pairwise distance between sequence i and j, where x(i) and y(j) are the ordinal values associated with each time point from which sequences i and j were obtained. This form of the M matrix imposes a temporal structure on the genetic diversity of env sequences, such that sequences separated by a longer sampling interval have greater evolutionary distances. The statistic of interest is r2, the square of the Pearson correlation coefficient, calculated between all pairs of elements of E and M excluding the diagonal. The null distribution of r2 was obtained by randomly permuting the row and column labels of M 1,000 times and calculating the value of r2 between E and the permuted M. For all individuals, the estimated value of r2 was never exceeded by any value generated by the permutation procedure (P < 0.001; data not shown). These results imply that sequences from each time point are most similar to those from the same time point. Furthermore, the imposition of a temporally ordered null matrix suggests that sequences from each time point are most similar to those from closer versus more distant time points.
In evaluating the evolutionary patterns over time, it was apparent that after a period of linear increase there was a time point at which the mean genetic diversity plateaued or decreased, and another time point at which the mean genetic divergence appeared to slow down or to stabilize in some individuals. It was of interest to evaluate the association of these evolutionary time points with time points of disease markers (i.e., estimated time of CD3+ T-cell inflection and the time at which CD4+ T-cell counts decreased to 200/μl) and the times of emergence and peak representation of X4 viruses. To accomplish this, times of peak diversity and divergence stabilization were estimated relative to the time of seroconversion (seroconversion being defined by the midpoint between the last seronegative and the first seropositive visits) by manual review of the data. Peak diversity and divergence stabilization were taken as the point at which the mean distance between sequences from a given time point (diversity) or from the founder sequence (divergence) either was at its maximum or slowed its rate of increase. The changes in diversity trajectory were substantiated by statistically significant (P < 0.01) quadratic trends over time. The same point of divergence stabilization or slowdown was evident when comparing sample divergences from time points other than the founder population (data not shown). However, the stabilization of divergence could not be statistically substantiated by quadratic regression analysis, and in two cases (participants 7 and 8) was suggested by only a single data point. The times of CD3+ inflection points were initially chosen by manual review, and were within 1 year of those predicted by a subsequently developed automated algorithm (24). The association of the timing of different events was evaluated by linear regression. The time between events was evaluated by analyzing the paired (within-individual) differences of event times with t tests and Wilcoxon’s signed-rank tests.
Coreceptor usage and SI phenotype prediction and determination.
The occurrence of viruses utilizing the CCR5 and CXCR4 coreceptors was determined by one or more of three different assays. (i) Virus isolates derived from fresh or frozen viable PBMCs were tested for syncytium induction on MT-2 cells (31). (ii) The phenotypic use of coreceptors was determined by the growth of viral isolates cultured with cells expressing CD4 and coreceptors (cells graciously provided by D. Littman, Skirball Institute of Biomolecular Medicine, New York, N.Y.) (65). (iii) The X4 phenotype was predicted from viral sequence data, based on deduced basic amino acid substitutions at positions 306, 319, and 320 of the mature gp120 protein (3, 4, 7, 13, 22, 36, 64, 70, 78). None of these methods distinguished between X4 viruses and dualtropic R5/X4 viruses. For simplicity, viruses predicted (via sequence analysis) or observed (via phenotypic culture) to use the CXCR4 coreceptor are referred to as X4.
Nucleotide sequence accession numbers.
All sequences described were deposited in the GenBank database under accession no. AF137629 to AF138163, AF138166 to AF138263, and AF138305 to AF138703.
RESULTS
Consistent trends in viral evolution.
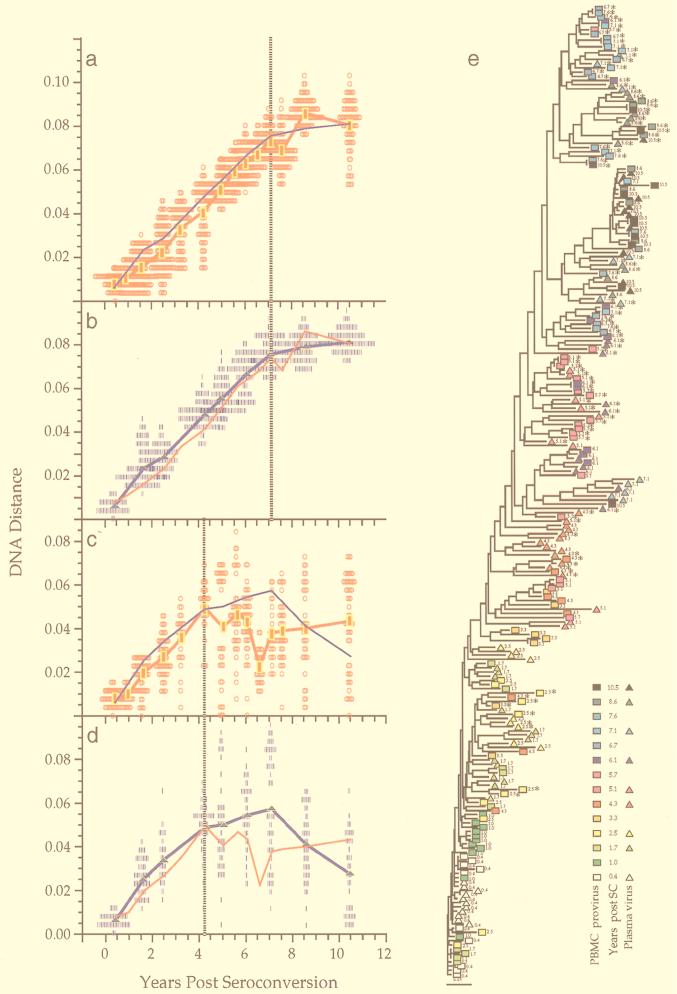
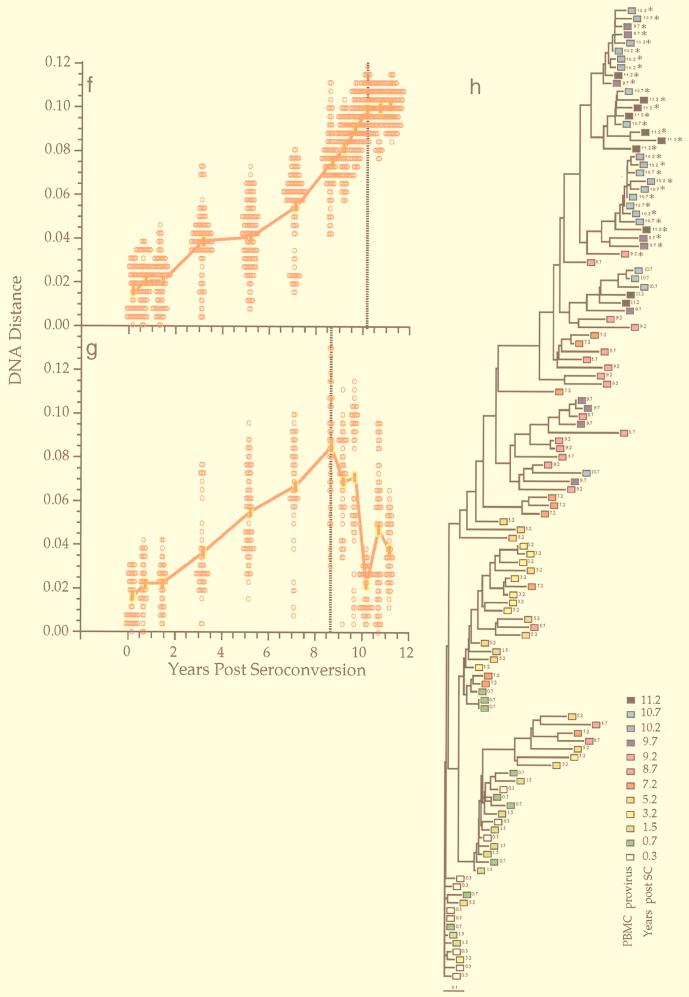
A total of 1,300 sequences bearing an open reading frame were obtained from the nine men at a total of 106 time points, for an average of 12.3 sequences from each time point. Paired PBMC and plasma samples were examined at 25 time points. Figure 1 shows sequence comparisons for two participants, illustrating three features of env gene evolution that were observed among all participants (for complete data sets, see reference 71a). First, the changes in viral divergence (from an approximated founder strain; see Materials and Methods) were essentially the same in both cell-associated viral DNA (Fig. 1a) and plasma RNA (Fig. 1b). The patterns of increase in viral diversity (the breadth of population at a given time point) were also similar in the viral DNA and RNA populations (Fig. 1c and d), although at later times they appeared to diverge in participant 2, perhaps due to the unavailability of RNA sequences from the two populations at all time points. Thus, divergence and diversity measurements were not affected by the different half-lives of cells versus virions, in agreement with another recent report (2). For simplicity, therefore, we combined DNA and RNA data for the analyses shown in subsequent figures. Second, divergence increased linearly for several years after seroconversion but then appeared to slow or stabilize late in infection, again at approximately the same times in DNA and RNA (Fig. 1a, b, and f). Third, the breadth of viral population diversity at a given time point increased in parallel with divergence for a few years after seroconversion before reaching a peak and then leveling off or decreasing prior to the point of divergence stabilization. Figures 1a to d, f, and g also illustrate the variability in pairwise comparisons of divergence and diversity around mean values. The data were distributed approximately symmetrically around the means with some evidence of a multimodal distribution in diversity measures at later time points. The evolutionary relationships between all of these sequences showed a significantly time-ordered, progressive evolutionary trend away from the founder strain (P < 0.001) (Fig. 1e and h).
FIG. 1.
Viral population analyses for participants 2 and 9. Analyses of sequences from participant 2 (a to e) and participant 9 (f to h) are shown. Genetic distances between all possible pairs of nucleotide sequences for viral DNA in PBMC (a, c, f, and g) and viral RNA in plasma populations (b and d) are shown (means at each time point indicated by symbols connected by thick lines) as a function of time since seroconversion. Parallel estimates with sequences sampled from plasma are shown with a thin blue line (a and c) and those from PBMC as a thin red line (b and d). Divergence from the founder strain (i.e., the first virus-positive time point; see Materials and Methods) is shown (a, b, and f). Viral population diversity (i.e., distances between all pairs of sequences within each time point; see Materials and Methods) is shown (c, d, and g). Dotted vertical lines correspond to a peak or a slowdown in the rate of change of each measure. Neighbor-joining phylograms (e and h) are shown for participants 2 and 9, respectively, derived from maximum-likelihood distances between all the sequences in each patient with PHYLIP (20). Sequences are represented by a square for PBMC sequences or a triangle for plasma sequences, with an arbitrary color gradient corresponding to the time of sampling following seroconversion, as indicated in the key. An asterisk (∗) indicates sequences with a basic amino acid substitution in the V3 loop specifying the X4 genotype.
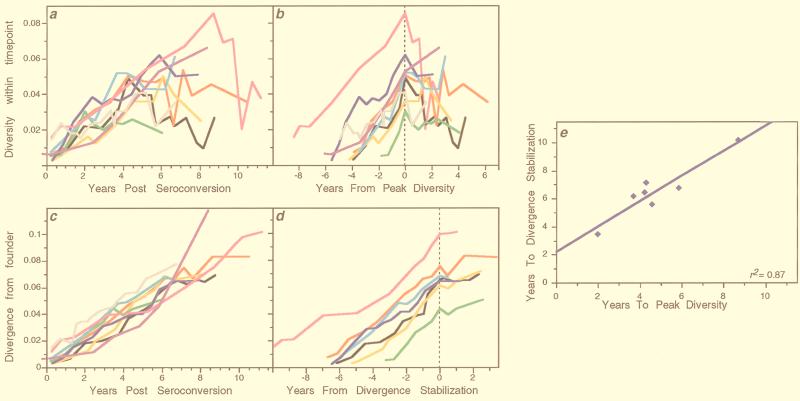
Figure 2a shows corresponding data from all participants, summarized with points and vertical bars reflecting the mean and standard deviation at each point. Two highly consistent patterns were evident in comparing trends across individuals (Fig. 3). First, viral population diversity increased and then stabilized or declined in all participants (Fig. 2a and 3a and b). Before reaching their peak, the increases in virus population diversity for each individual were highly linear (mean slope = 0.92% ± 0.27% per year) and consistent (mean r2 = 0.62). After the peak in diversity, the patterns declined on average, with a slope of −0.35% ± 0.69% per year, and exhibited more variability (mean r2 = 0.09; range, −0.01 to 0.26).
FIG. 3.
Trends of viral diversification across individuals. Data from the nine participants shown in Fig. 2 are replotted to show the similarities of viral population intra-time-point diversity (a and b) and divergence from founder strains (c and d) across individuals. Participant 1 is shown with a black line, 2 with red, 3 with orange, 5 with gray, 6 with green, 7 with cyan, 8 with blue, 9 with magenta, and 11 with purple. (a and b) Viral population intra-time-point diversity plotted relative to the time of seroconversion (a) or the time of peak or stabilization of diversity (vertical dotted lines) (b). (c) For each participant, all of the sequences from the first virus-positive visit (the founder strain) were compared to all sequences sampled at each subsequent time point. Similar slopes were also generated when mean values at each time point were used to calculate a combined slope (data not shown). (d) Viral population divergence from the founder strain as shown in panel c but plotted relative to the estimated time of divergence stabilization (vertical dotted line). Participants 5 and 11, who had no evident stabilization, are omitted. (e) Relationship between the times from seroconversion to peak viral diversity and to divergence stabilization.
Second, the pattern of population divergence from the founder strain was remarkably linear and consistent across subjects (Fig. 2a). The increase (mean ± standard deviation) in divergence was 0.91% ± 0.14% per year when all data were used (Fig. 3a). However, in most subjects divergence appeared to slow or stabilize late in infection (in participants 7 and 8, stabilization was suggested by only a single data point, and it was not detected in participants 5 and 11, the latter a possible late progressor). The mean increase in divergence was 1.02% ± 0.14% per year before stabilization—almost the same as the increase in diversity—and 0.15% ± 0.25% after stabilization (Fig. 3d). The linear correlation was also much greater for data before stabilization (mean r2 = 0.82; range, 0.75 to 0.92) than afterwards (mean r2 = 0.08; range, 0.00 to 0.30).
Although the phenomenon of divergence stabilization was not as well defined as peak diversity, there was a close temporal relationship between the times of peak diversity and divergence stabilization (for those participants in whom the latter could be estimated) (Fig. 3e). Linear regression of these two times yielded a high r2 (0.87) and a slope (0.90 ± 0.16) that was statistically indistinguishable from unity (P = 0.54), supporting the interpretation that these events were strongly linked. The mean time between peak diversity and divergence stabilization was 1.76 years (95% confidence interval [CI], 1.06 to 2.46).
Association of viral evolutionary change and X4 viruses.
Mutations predictive of the X4 viral phenotype (basic amino acid substitutions at positions 306, 319, and 320 of the mature gp120 protein) were detected at one or more time points in all nine individuals (Fig. 2b). Individual participant data, plotted in Fig. 2b, demonstrated a pattern in which X4 viruses were initially absent, rose to constitute >80% of the viral population in six cases, and then declined. To determine if these patterns of X4 virus prevalence were related to times of seroconversion, peak diversity, and divergence stabilization, data from all individuals were plotted relative to those events. Relative to seroconversion (Fig. 4a), the times of X4 emergence were spread out over the course of infection. However, X4 emergence appeared to be closely associated with, and usually just prior to, the time of peak diversity (Fig. 4b) (mean = −0.32 years; 95% CI, −1.26 to 0.63). Moreover, the peak prevalence of X4 viruses appeared to be closely associated with and just prior to the time of divergence stabilization (Fig. 4c) (mean = −0.35 years; 95% CI, −1.26 to 0.56). The strength of these associations is quantified by the correlations between the time of emergence of X4 viruses and the time to peak diversity (Fig. 4g) (r2 = 0.67) and the time of peak X4 with the time to divergence stabilization (Fig. 4h) (r2 = 0.79).
FIG. 4.
Trends of X4 genotype representation and CD4+ T-cell numbers across individuals. Data from all nine participants shown in Fig. 2 are replotted with the proportion of the virus population with an X4 genotype (a to c) or levels of circulating CD4+ T cells (d to f), relative to the time of seroconversion (a and d), time of peak viral population diversity (b and e), and time of divergence stabilization (c and f). Participant color-coding is the same as described in the legend to Fig. 3. The dotted vertical lines indicate the times of peak diversity (b and e) or divergence stabilization (c and f). The horizontal line across the lower panels indicates a CD4+ lymphocyte level of 200 cells/μl. (g to i) Relationships between the times from seroconversion to the times of the indicated viral population events. Slope = 0.94 ± 0.25 (g), 0.96 ± 0.22 (h), and 0.91 ± 0.19 (i).
In general, the predictions based on mutation analysis agreed with the determined phenotype (Fig. 2c and Table 1). Of a total of 38 virus isolates whose phenotype was determined on MT-2 cells, 27 were concordant. Seven others were non-syncytium-inducing (NSI) following virus isolation despite the abundant detection of a basic substitution at position 320 in the starting PBMC population (participants 1 and 2), and one isolate was NSI when the position 319 mutation was detected in a single clone (participant 6). Of 38 CCR5 and CXCR4 coreceptor tropism determinations, 31 were concordant with expectations (Table 1), including 4 of the 5 CXCR4+ isolates that were dually positive for CCR5 and CXCR4 (2 in participant 7 [V9 and V15] and 2 in participant 8 [V19 and V20]). One isolate from participant 2 was unexplainably dually negative on CCR5 and CXCR4 (as well as CCR3 and CCR2b) (data not shown) when the genotype of the PBMC viral DNA suggested an absence of CXCR4 usage. Participant 7 had two virus isolates that were both SI and CXCR4 positive (one was also CCR5 negative) following a period of detection of a low level of viruses with the position 320 substitution.
TABLE 1.
Virus phenotypes predicted from changes in V3 loop sequences compared to phenotypes observed in virus culturea
| Predicted virus phenotype | No. of isolates with phenotype
|
|||||||
|---|---|---|---|---|---|---|---|---|
| NSI | SI | CCR5− | CCR5+ | CXCR4− | CXCR4+ | |||
| R5 | 20 | 3 | 2 | 15 | 14 | 3 | ||
| X4 | 8 | 7 | 0 | 2 | 0 | 2 | ||
| P value for association | 0.030 | 1.00 | 0.058 | |||||
For each HIV isolate tested, the predicted phenotype based on the absence (R5) or presence (X4) of basic amino acid substitutions at positions 306, 319, or 320 of gp120 is compared to the results of tissue culture evaluations of viral growth on MT-2 cells (columns 1 and 2), CD4 and CCR5 cells (columns 3 and 4), and CD4 and CXCR4 cells (columns 5 and 6). The P values (from Fisher’s exact test) measure the association of viral phenotypes predicted from changes in V3 loop sequences and phenotypes observed in virus cultures. The table shows that the V3 loop phenotype was associated with the SI culture phenotype (P = 0.030) and marginally associated with the CXCR4 culture phenotype (P = 0.058) but not associated with the CCR5 culture phenotype (P = 1.00).
Relationship of disease progression with viral evolutionary changes and X4 viruses.
Across all participants, consistent relationships were noted between times of the viral events described earlier (i.e., peak diversity, divergence stabilization, and emergence and peak representation of X4 viruses) and changes in T-cell populations. The participants experienced different rates of decline of CD4+ T cells over time (Fig. 4d). However, seven of nine subjects (all except participants 9 and 11) transitioned to ≤200 CD4+ T cells/μl within 2 years following the point of divergence stabilization, with these two events highly associated in time (r2 = 0.83; mean interval = 0.97 years; 95% CI, 0.18 to 1.76) (Fig. 4i). The CD4+ T cells in subject 9 dropped to 202 cells/μl 1 year following the point of divergence stabilization (included in Fig. 4i), at which time he began highly active antiretroviral therapy (HAART). Subject 11 experienced the most protracted decline in CD4+ T cells and did not reach a level of 200 CD4+ T cells or a point of divergence stabilization as of the last time point studied.
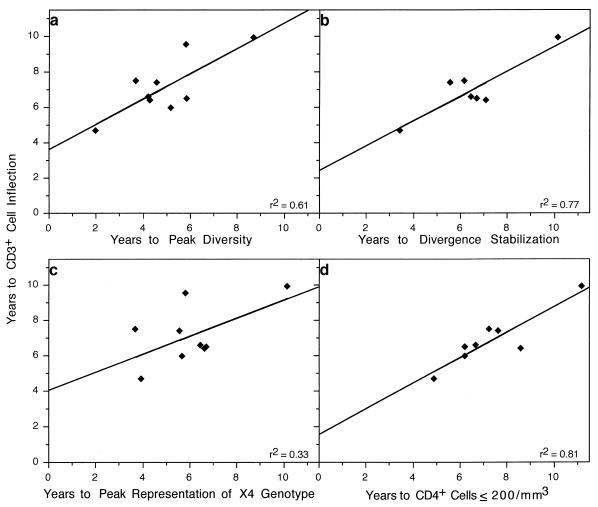
Consistent relationships were also noted among subjects between each of the above measures (times of peak viral diversity, divergence stabilization, transition to ≤200 CD4+ T cells per μl, peak representation of X4 viruses, and the failure of T-cell homeostasis [represented by an inflection in CD3+ cells] [65]), as shown in Fig. 5. Relative to failure of T-cell homeostasis, peak viral diversity occurred a mean of 2.2 years earlier (95% CI, 1.32 to 3.13 years earlier; P = 0.0005) (Fig. 5a), while peak X4 representation occurred a mean of 1.08 years earlier (95% CI, −0.20 to 2.36 years earlier; P = 0.087) (Fig. 5c) and divergence stabilization occurred a mean of 0.46 years earlier (95% CI, −0.44 to 1.36; P = 0.25) (Fig. 5b). CD4+ T cells transitioned to the ≤200 cells/μl threshold a mean of 0.48 years after the CD3+ T-cell inflection (95% CI, −0.23 to 1.19; P = 0.16) (Fig. 5d).
FIG. 5.
Relationships between times from seroconversion to CD3+ T-cell inflection and times to viral evolutionary changes and CD4+ T-cell numbers. The time of the CD3+ cell inflection point is plotted relative to the time of peak viral diversity, slope = 0.71 ± 0.22 (a); divergence stabilization, slope = 0.70 ± 0.17 (b); peak representation of X4 viral genotypes, slope = 0.51 ± 0.28 (C); and transition of the CD4+ T-cell levels to ≤200/μl, slope = 0.72 ± 0.14 (d).
Evolutionary patterns in late progressors.
Two participants initially selected as nonprogressors experienced evidence of disease progression with further follow-up. One of those (participant 9) was studied in more detail after this progression occurred. After a period of 8 years of stable CD3+ and CD4+ T-cell counts and a 4-year period of increasing viral load, he experienced a rapid decline in CD4+ T cells to 202 cells/μl at the last time point available before receiving HAART. Preceding the decrease in CD4+ T cells, a peak in viral diversity and an outgrowth of X4 viruses were observed. Participant 11 had stable T-cell counts and low viral loads for 7 years and remained asymptomatic through 1998 without drug therapy. However, attenuation in viral diversity and a minor outgrowth of X4 viruses were noted at 6 years after infection. These were followed by a decline in CD4+ and CD3+ T cells, indicating that he was experiencing disease progression.
DISCUSSION
Our results suggest that in the individuals studied, the time between primary HIV-1 infection and AIDS could be subdivided into three phases based on patterns of viral evolution over the C2-V5 region of env. The phases were defined by changes in viral divergence from the founder strain and intra-time point diversity; these definitions in turn were reinforced by observed changes in the prevalence of X4 viruses (probably including dualtropic X4/R5 viruses). In the first phase, both viral population divergence and diversity increased linearly, with diversification essentially keeping pace with divergence at a rate of about 1% per year. The beginning of the second phase was signaled by a leveling off, or decrease, in diversity. In this phase, the viral population continued to diverge from the founder strain at the same rate, while diversity continued to plateau or constrict. Finally, the increase in divergence also slowed or stabilized, marking the beginning of the third phase, the latter being characterized by a decline in diversity.
The peaking of diversity was strongly supported by statistical analysis. Although similar support for the point of divergence stabilization was not found, the validity of this concept, as well as the general framework put forward here, is strongly supported by the consistency of the patterns described among participants and by the close linkages observed between the three defined phases and the patterns of X4 virus prevalence and T-cell subset landmarks. Thus, the second phase was strongly associated with X4 viruses, which in most participants first appeared very close to the beginning of this phase and peaked in prevalence very close to the end of it. The third phase in turn was closely associated in time with the decline of CD4+ T cells to <200/μl and with the failure of T-cell homeostasis and a loss of phenotypically naive (CD45RA+RO−) CD4+ T cells (48a). The proposed framework is also generally consistent with previous sporadic reports of constriction of HIV diversity (15, 51, 58) and similar rates of divergence from the founder strain (39, 43, 45, 46, 82–84). Here, we have extended these previous reports by systematically documenting the consistency of these patterns and rates.
Other arguments support the idea that the proposed framework, though derived from a small number of participants, may have broad applicability. (i) Participants were studied in substantial depth from seroconversion to advanced disease in those developing disease progression. (ii) The findings were quite consistent. (iii) Participants were representative of most HIV-1-infected people in terms of the rate of disease progression (median survival time = 9.1 years after seroconversion compared to 10.2 years among the MACS participants as a whole [56]) and occurrence of T-cell homeostasis failure before AIDS (seen in ∼75% of MACS participants who develop AIDS [24, 34, 48]). (iv) The patterns were seen even in two participants who were initially selected for lack of progression of HIV disease but exhibited evidence of progression as follow-up advanced.
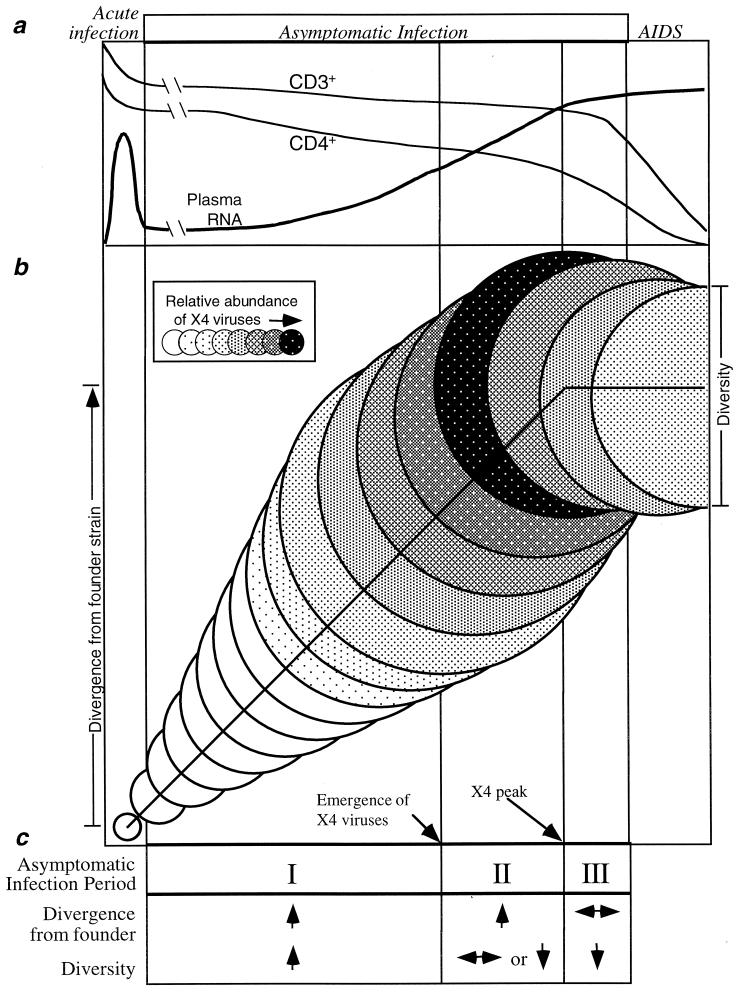
Taken together, a summary sequence of events can be inferred (Fig. 6) that describes the virological and T-cell changes that occur in the asymptomatic period of HIV infection. It should be stressed that these phases constitute general trends found in the six individuals we studied who developed AIDS during the period of analysis, although all nine experienced the initial events we described. The sequence begins with the detection of X4 viruses and the attainment of peak viral diversity at a variable time after seroconversion (4 to 9 years in these participants). If the time of X4 virus appearance is taken as time zero, then peak diversity occurs an average of 0.3 years later, followed by X4 peak representation at 1.5 years, divergence stabilization at 2.2 years, T-cell homeostasis failure at 2.5 years, and CD4+ cell transition through 200 cells/μl at 2.9 years. Based on a study of 212 seroconverters and 1,129 seroprevalant infections within the MACS cohort, the occurrence of clinical AIDS-defining conditions follows T-cell homeostasis failure by 1.7 years (24, 48), or approximately 4.2 years from the emergence of X4 viruses.
FIG. 6.
Schematic illustration of proposed consistent patterns in development of HIV disease in moderate progressors. (a) Clinical phases of HIV infection as well as typical patterns of CD4+ and CD3+ T cells and plasma viral RNA loads. The initial transient increase in viral RNA is estimated from the literature, whereas the later RNA increase and the CD3+ and CD4+ T-cell declines are based on the data from this study and that of Rinaldo et al. (65). (b) Patterns of viral sequence evolution within the asymptomatic period of infection identified in this study. Circle diameters represent the mean viral population diversities at increasing intervals following seroconversion. Vertical displacement of the circles represents the extent of viral population divergence from the founder strain. Shading represents the proportion of the viral population comprised of viruses with an X4 genotype. Dotted vertical lines represent (from left) the end of acute infection, peak viral diversity, stabilization of divergence from the founder strain, and the development of AIDS. (c) Characteristic changes in viral evolution in the three periods of the asymptomatic phase identified in this study (↑, increasing; ↓, decreasing; ↔, stable).
Several caveats should be noted. It is not clear whether the patterns and sequence of events observed can be generalized to people with other rates (i.e., very rapid or very slow) of disease progression or over shorter observation periods (23, 45, 46, 67, 76). Seven of the participants we studied were typical progressors, whereas two (participants 9 and 11), were initially chosen as examples of nonprogressors but subsequently did progress. Second, the length of the intervals between the noted events is variable within the individuals we studied; the durations estimated here are not meant to set limits and are mentioned only as a frame of reference for the evaluation of additional HIV-infected individuals. Third, we drew our inferences about viral evolutionary patterns on only 650 bp of env; other genes and gene regions evolve more slowly and may not exhibit the same patterns.
Despite these concerns, recognition of the aforementioned patterns of HIV-1 evolution suggests a way to reconcile conflicting reports about the relationship between the rate of HIV-1 disease progression and the degree and rate of increase in viral diversity (15, 16, 23, 26, 27, 44–46, 49, 51, 52, 60, 73, 82, 83). Because only extensive sampling can detect the complex pattern of viral evolution we describe, viral diversification could appear spuriously slow if viral populations are assessed infrequently, e.g., only early and late in infection (thereby missing the point of peak diversity) or in people with highly atypical disease pathogenesis (e.g., the slowest and the most rapid progressors). A previous study of env sequences by Shankarappa et al. (71) suggested the existence of a diversity peak with continuing divergence from the founder strain, although sampling was less frequent and potentially compromised by resampling (44).
In this study, X4 viruses were detected in the blood of all nine participants and grew to predominance in six. This represents a significantly higher frequency of detection of X4 viruses than in approximately 50% of the individuals observed in previous studies of people progressing to AIDS (36, 70, 78). Previous studies have used phenotypic assays to detect X4 viruses, as opposed to the genotypic analysis done here. Both methodologies can miss detection of X4 viruses on occasion. However, we also noted that the representation of X4 viruses peaked and then diminished over time, which could lead to falsely low estimates of the proportion of people who have circulating X4 viruses if they were sampled subsequent to such a decline. Our higher detection rate by sequence analysis could also reflect the fact that PBMC DNA harbors defective and latent proviruses for extended periods (12, 61, 63, 69) and thus may provide a record of past transient replication of X4 viruses. Evidence for the transience of X4 viruses has also been seen in previous studies (30, 51). We have also detected transient representation of X4 viruses as well as a similar pattern of viral evolutionary dynamics in the two other patients we have studied throughout infection: a gay man with rapidly progressing disease (43, 45) and a perinatally infected child (40a). Overall, our results support the view that X4 viruses are likely to occur in significantly more than 50% of people who develop AIDS. It should be noted that X4 viruses have often been linked to AIDS in HIV-1 subtype A, B, D, and E infections (18, 37, 59), have been found with significant frequency in early HIV-1 subtype C infections (79), but are seen at low frequency in cases of late-stage subtype C infections (59). Thus, it has not been established that our findings apply to infections with all HIV-1 subtypes.
Interestingly, the timing of evolution and rapidity of outgrowth of X4 viruses relative to the point of peak viral diversity appeared to differ by mutation. X4 viruses with a mutation at position 306 (participants 5 and 8) increased rapidly, reaching a peak level shortly after initial detection. In contrast, viruses with a mutation at position 320 (participants 1, 2, and 3) reached peak levels more gradually, and the appearance of the position 319 mutation was most variable and tended to be found at low levels (participants 3, 6, 7, and 8 but not 9) and in individuals with other X4-specific mutations. Thus, although these data are preliminary, the specific X4 mutation may be important to viral growth properties and in disease progression.
These observations suggest an important role for X4 viruses in T-cell destruction late in the progression of HIV-1 infection in the participants studied. This is plausible in view of the preferential expression of CXCR4 on naive T cells (9) and on thymocytes (50), which are needed to replenish memory T cells (6). Among other recent findings suggesting a differential impact of R5 and X4 viruses on T-cell populations (5, 8, 29, 33, 75, 81) is the mediation of apoptosis of CD8+ cells by X4 Env (29), a mechanism that may apply to CXCR4-expressing naive CD4+ cells as well.
Our data also suggest that the degree and rapidity of immune reconstitution (1) that occurs under HAART may differ according to the phase at which HAART is initiated, with greater reconstitution possible prior to some of the milestones we described. For example, it may be that people treated with HAART before X4 viruses are detected will be able to restore lost immunological functioning more effectively than those whose treatment is begun later. In addition, given the serious long-term side effects of HAART (10, 28), the ability to predict AIDS onset ∼4 years in advance based on detection of X4 viruses (57) may help refine therapeutic strategies.
ACKNOWLEDGMENTS
We thank Lisa M. Frenkel, Patricia d’Souza, and Thera Mulvania for helpful discussions; Dan Littman for cell lines; Elvia Ramirez, LuAnn Borowski, Cindy Kleeberger, and Ming Ding for assistance; David Swofford for permission to use the computer program PAUP*, version 4.0d59, to estimate evolutionary distances under the general time-reversible model; and MACS participants for their dedication to the cohort study.
This work was supported by Public Health Service awards AI37984, AI32885, AI34783, AI35042, RR00772, AI35039, AI35040, AI35041, and AI27757, the latter while R.S. was supported by a New Investigator award from the University of Washington Center for AIDS Research.
REFERENCES
- 1.Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debré P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 2.Bagnarelli P, Mazzola F, Menzo S, Montroni M, Butini L, Clementi M. Host-specific modulation of the selective constraints driving human immunodeficiency virus type 1 env gene evolution. J Virol. 1999;73:3764–3777. doi: 10.1128/jvi.73.5.3764-3777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E, Doms R, Fenyo E, Korber B, Littman D, Moore J, Sattentau Q, Schuitemaker H, Sodroski J, Weiss R. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 4.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 5.Berkowitz R D, Alexander S, Bare C, Linquist-Steppes V, Bogan M, Moreno M E, Gibson L, Wieder E, Kosek J, Stoddart C, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vitro. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Beckerman K P, Schall T J, McCune J M. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T cell differentiation. J Immunol. 1998;161:3702–3710. [PubMed] [Google Scholar]
- 7.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr A, Samaras K, Chisholm D J, Cooper D A. Pathogenesis of HIV-1-protease inhibitor-associated peripheral lipodystrophy, hyperlipidaemia, and insulin resistance. Lancet. 1998;351:1881–1883. doi: 10.1016/S0140-6736(98)03391-1. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morbid Mortal Weekly Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- 12.Chun T-W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 13.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delwart E L, Busch M P, Kalish M L, Mosley J W, Mullins J I. Rapid molecular epidemiology of HIV transmission. AIDS Res Hum Retrovir. 1995;11:1181–1193. doi: 10.1089/aid.1995.11.1081. [DOI] [PubMed] [Google Scholar]
- 15.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delwart E L, Shpaer E G, McCutchan F E, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 18.De Wolf F, Hogervorst E, Goudsmit J, Fenyo E M, Rubsamen-Waigmann H, Holmes H, Galvao-Castro B, Karita E, Wasi C, Sempala S D K, Baan E, Zorgdrager F, Lukashov V, Osmanov S, Kuiken C, Cornelissen M WHO Network for HIV Isolation and Characterization. Syncytium-inducing and non-syncytium-inducing capacity of HIV-1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res Hum Retrovir. 1994;10:1387–1401. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 19.Erlich H A, Gelfand D, Sninsky J J. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP—Phylogeny inference package, version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- 21.Folks T M, Powell D, Lightfoote M, Koenig S, Fauci A S, Benn S, Rabson A, Daugherty D, Gendelman H E, Hoggan M D, et al. Biological and biochemical characterization of a cloned Leu-3− cell surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain (V3) of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganeshan S, Dickover R E, Korber B T, Bryson Y J, Wolinsky S M. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gange S J, Muñoz A, Chmiel J S, Donnenberg A D, Kirstein L M, Detels R, Margolick J B. Identification of inflections in T-cell counts among HIV-1-infected individuals and relationship with progression to clinical AIDS. Proc Natl Acad Sci USA. 1998;95:10848–10853. doi: 10.1073/pnas.95.18.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgi J V, Cheng H L, Margolick J B, Bauer K D, Ferbas J, Waxdal M, Schmid I, Hultin L E, Jackson A L, Park L, et al. Quality control in the flow cytometric measurement of T-lymphocyte subsets: the Multicenter AIDS Cohort Study experience. The Multicenter AIDS Cohort Study Group. Clin Immunol Immunopathol. 1990;55:173–186. doi: 10.1016/0090-1229(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 26.Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immun Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 27.Haynes B F, Pantaleo G, Fauci A S. Toward an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- 28.Henry K, Melroe H, Huebsch J, Hermundson J, Levine C, Swensen L, Daley J. Severe premature coronary artery disease with protease inhibitors. Lancet. 1998;351:1328. doi: 10.1016/S0140-6736(05)79053-X. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 29.Herbein G, Mahlknecht U, Batliwalla F, Gregersen P, Pappas T, Butler J, O’Brien W A, Verdin E. Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature. 1998;395:189–194. doi: 10.1038/26026. [DOI] [PubMed] [Google Scholar]
- 30.Ida S, Gatanaga H, Shioda T, Nagai Y, Kobayashi N, Shimada K, Kimura S, Iwamoto A, Oka S. HIV type 1 V3 variation dynamics in vivo: long-term persistence of non-syncytium-inducing genotypes and transient presence of syncytium-inducing genotypes during the course of progressive AIDS. AIDS Res Hum Retrovir. 1997;13:1597–1609. doi: 10.1089/aid.1997.13.1597. [DOI] [PubMed] [Google Scholar]
- 31.Japour A J, Fiscus S A, Arduino J M, Mayers D L, Reichelderfer P S, Kurizkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurriaans S, Van Gemen B, Weverling G J, Van Strijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection: virus load and virus phenotype independent determinants of clinical course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 33.Kaneshima H, Su L, Bonyhadi M L, Connor R I, Ho D D, McCune J M. Rapid-high, syncytium-inducing isolates of human immunodeficiency virus type 1 induce cytopathicity in the human thymus of the SCID-hu mouse. J Virol. 1994;68:8188–8192. doi: 10.1128/jvi.68.12.8188-8192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaslow R A, Ostrow D G, Detels R, Phair J P, Polk B F, Rinaldo C R., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 36.Koot M, Keet I, Vos A, deGoede R, Roos M, Coutinho R, Miedema F, Schellekens P, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 37.Korber B, Hoelscher M, McCutchan F, Williamson C, von Sonnenburg F, Mullins J, Pletschette M, Weber J, van der Groen G, Osmanov S Workshop Report from the European Commission and the Joint United Nations Programme on HIV/AIDS. HIV-1 subtypes: implications for epidemiology, pathogenicity, vaccines and diagnostics. AIDS. 1997;11:UNAIDS17–UNAIDS36. [PubMed] [Google Scholar]
- 38.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuiken C L, Korber B. Epidemiological significance of intra- and inter-person variation of HIV-1. AIDS. 1994;8:S73–S83. [Google Scholar]
- 40.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 40a.Laco, G., et al. Unpublished data.
- 41.Leitner T, Kumar S, Albert J. Tempo and mode of nucleotide substitutions in gag and env gene fragments in human immunodeficiency virus type 1 populations with a known transmission history. J Virol. 1997;71:4761–4770. doi: 10.1128/jvi.71.6.4761-4770.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, S.-L., T. Mulvania, A. Rodrigo, B. Kosloff, X. He, D. Shriner, L. Corey, and J. I. Mullins. Emergence, evolutionary dynamics and selective advantage of human immunodeficiency virus type-1 chimeras in vivo. Submitted for publication.
- 44.Liu S-L, Rodrigo A G, Shankarappa R, Learn G H, Hsu L, Davidov O, Zhao L P, Mullins J I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 45.Liu S L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 48.Margolick J B, Muñoz A, Donnenberg A D, Park L P, Galai N, Giorgi J V, O’Gorman R G, Ferbas J for the Multicenter AIDS Cohort Study. Failure of T-cell homeostasis preceding AIDS in HIV-1 infection. Nat Med. 1995;1:674–680. doi: 10.1038/nm0795-674. [DOI] [PubMed] [Google Scholar]
- 49.Markham R B, Wang W C, Weisstein A E, Wang Z, Munoz A, Templeton A, Margolick J, Vlahov D, Quinn T, Farzadegan H, Yu X F. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci USA. 1998;95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCune J M, Loftus R, Schmidt D K, Carroll P, Webster D, Swor Y-L B, Francis I R, Gross B H, Grant R M. High prevalence of thymic tissue in adults with human immunodeficiency virus-1 infection. J Clin Investig. 1998;101:2301–2308. doi: 10.1172/JCI2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald R A, Mayers D L, Chung R C-Y, Wagner K F, Ratto-Kim S, Birx D L, Michael N L. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J Virol. 1997;71:1871–1879. doi: 10.1128/jvi.71.3.1871-1879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellors J W, Kingsley L A, Rinaldo C R J, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 54.Mellors J W, Muñoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 55.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 56.Muñoz A, Sabin C A, Phillips A N. The incubation period of AIDS. AIDS. 1997;11:S69–S76. [PubMed] [Google Scholar]
- 57.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowak M A, Anderson R M, McLean A R, Wolfs T F, Goudsmit J, May R M. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 59.Peeters M, Vincent R, Perret J L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 60.Pelletier E, Wain-Hobson S. AIDS is not caused by the extreme genetic variability of HIV. J NIH Res. 1996;8:45–48. [Google Scholar]
- 61.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 62.Poss M, Rodrigo A G, Gosink J J, Learn G H, de Vange Panteleeff D, Martin H L, Jr, Bwayo J, Kreiss J K, Overbaugh J. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Psallidopoulos M C, Schnittman S M, Thompson III L M, Baseler M, Fauci A S, Lane H C, Salzman N P. Integrated proviral human immunodeficiency virus type 1 is present in CD4+ peripheral blood lymphocytes in healthy seropositive individuals. J Virol. 1989;63:4626–4631. doi: 10.1128/jvi.63.11.4626-4631.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richman D D, Bozzette S A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 65.Rinaldo C R, Jr, Gupta P, Huang X, Fan Z, Mullins J I, Gange S, Farzadegan H, Shankarappa R, Muñoz A, Margolick J B. Anti-HIV-1 memory cytotoxic T lymphocyte responses associated with changes in CD4+ T cell numbers in the progression of HIV-1 infection. AIDS Res Hum Retrovir. 1998;14:1423–1433. doi: 10.1089/aid.1998.14.1423. [DOI] [PubMed] [Google Scholar]
- 66.Rodrigo A G, Goracke P C, Rowhanian K, Mullins J I. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retrovir. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 67.Salvatori F, Masiero S, Giaquinto C, Wade C M, Leigh Brown A J, Chieco-Bianchi L, De Rossi A. Evolution of human immunodeficiency virus type 1 in perinatally infected infants with rapid and slow progression to disease. J Virol. 1997;71:4694–4706. doi: 10.1128/jvi.71.6.4694-4706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schenker E L, Hultin L E, Bauer K D, Ferbas J, Margolick J B, Giorgi J V. Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry. 1993;14:307–317. doi: 10.1002/cyto.990140311. [DOI] [PubMed] [Google Scholar]
- 69.Schnittman S M, Psallidopoulos M C, Lane H C, Thompson L, Baseler M, Massari F, Fox C H, Salzman N P, Fauci A S. The reservoir for HIV-1 in human peripheral blood is a T cell that maintains expression of CD4. Science. 1989;245:305–308. doi: 10.1126/science.2665081. . (Erratum, 245: before 694, 1989.) [DOI] [PubMed] [Google Scholar]
- 70.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shankarappa R, Gupta P, Learn G H, Rodrigo A G, Rinaldo C R, Gorry M C, Mullins J I, Nara P L, Ehrlich G D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 71a.Shankarappa, R., A. G. Rodrigo, D. Upchurch, G. H. Learn, and J. I. Mullins. 20 September 1999, posting date. Mullins Molecular Retrovirology Lab. [Online.] http://ubik.microbiol.washington.edu/HIV/evolution1/. [15 October 1999, last date accessed.]
- 72.Shearer W T, Quinn T C, LaRussa P, Lew J F, Mofenson L, Almy S, Rich K, Handelsman E, Diaz C, Pagano M, Smeriglio V, Kalish L A the Women and Infants Transmission Study Group. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. New Engl J Med. 1997;336:1337–1342. doi: 10.1056/NEJM199705083361901. [DOI] [PubMed] [Google Scholar]
- 73.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan M K, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smouse P, Jong J C, Sokal R R. Multiple regression and correlation extensions of the Mantel’s test of matrix correspondence. Syst Zool. 1986;35:627–632. [Google Scholar]
- 75.Spina C A, Prince H E, Richman D D. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Investig. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strunnikova N, Ray S C, Livingson R A, Rubalcaba E, Viscidi R P. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J Virol. 1995;69:7548–7558. doi: 10.1128/jvi.69.12.7548-7558.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swofford D L, Olsen G J, Waddell P J, Hillis D M. Phylogenetic inference. In: Hillis D M, Moritz C, Mable B K, editors. Molecular systematics. 2nd ed. Sunderland, Mass: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- 78.Tersmette M, Gruters R A, de Wolf F, de Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tien P C, Chiu T, Latif A, Ray S, Batra M, Contag C H, Zejena L, Mbizvo M, Delwart E L, Mullins J I, Katzenstein D A. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:147–153. doi: 10.1097/00042560-199902010-00006. [DOI] [PubMed] [Google Scholar]
- 80.Waddle D M. Matrix correlation tests support a single origin for modern humans. Nature. 1994;368:452–454. doi: 10.1038/368452a0. [DOI] [PubMed] [Google Scholar]
- 81.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J ALIVE Study, Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, San Francisco City Cohort. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 82.Wolfs T F W, de Jong J J, van den Berg H, Tijnagel J M G H, Krone W J A, Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci USA. 1990;87:9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kuntsman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of HIV-1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]