Abstract
Glycoproteins gM and gN are conserved throughout the herpesviruses but are dispensable for viral replication in cell cultures. To assay for a function of these proteins in infection of an animal, deletion mutants of pseudorabies virus lacking gM or gN and corresponding revertants were analyzed for the ability to penetrate and propagate in the nervous systems of adult mice after intranasal inoculation. We demonstrate that neither of the two glycoproteins is required for infection of the nervous systems of mice by pseudorabies virus.
Pseudorabies virus (PrV) is an alphaherpesvirus which causes Aujeszky’s disease in pigs. The viral genome, of approximately 150 kbp, encodes at least 11 glycoproteins, 10 of which are present at the virion surface (reviewed in references 10 and 19). These virion glycoproteins are involved in the initial steps in virus infection, the attachment of virions to target cells and penetration by fusion between the virion envelope and the cellular cytoplasmic membrane. Thus, they play an important role in determining the host range of the virus. After replication in peripheral tissues, alphaherpesviruses are known to penetrate and propagate in the nervous system, where they infect neurons and glial cells and eventually establish a latent infection. PrV fatally infects most mammals except higher primates, including humans, and the pig is the only host animal able to survive a productive infection. The respiratory tract is the most frequent route of entry for PrV. Results obtained after intranasal inoculation of adult mice in our laboratory and by other investigators can be summarized as follows: wild-type PrV multiplies first in the respiratory epithelium (2, 10). Primary target cells also include few accessible trigeminal and sympathetic nerve endings in the nasal cavity, which can be infected by inoculated virus. However, most neurons of these two systems become infected by progeny viruses produced in the respiratory epithelium after primary replication. Few ganglionic cells in the olfactory epithelium are also permissive to PrV infection. From these infected neurons, rapid viral spread within sympathetic and trigeminal ganglia ensues, probably by local transfer of the virus between nonconnected neurons as well as by transneuronal transfer to connected neurons beyond the ganglion, leading to the death of the animal in approximately 50 h. Although the parasympathetic ganglion was not examined in these studies, it must have contained infected neurons since transneuronal transfer to second-order neurons of the parasympathetic pathway was observed. In mice, the olfactory route is poorly permissive to PrV and the infection does not propagate to the olfactory bulb. This differs from the situation in the pig (10, 15, 22). Although neurons are the first cells infected in the nervous system, virus spread to glial cells is soon observed.
In order to analyze the role of glycoproteins in viral neuroinvasion, PrV mutants singly lacking each of the known glycoproteins have been engineered. A glycoprotein gG deletion mutant exhibited a wild-type phenotype, and no obvious phenotypic alteration as a consequence of the gene deletion was observed (2). Glycoproteins gB, gD, and gH are essential for the propagation of the virus in cell cultures, and respective viral mutants require transcomplementing cells for productive replication (4, 23–25). After intranasal infection, complemented virions were able to perform one cycle of multiplication, giving rise to noncomplemented virions whose neurotropic properties were studied. Complemented viruses infected the respiratory epithelium and few olfactory, trigeminal, and sympathetic neurons. In the absence of gB or gH, infection did not spread, indicating that both glycoproteins are essential for the penetration of neurons from the respiratory epithelium and for local or transneuronal transfer between neurons (1, 4). In contrast, after primary infection by complemented virions, the presence of gD was not required for virus spread from the respiratory epithelium to neurons, between neurons by local and transneuronal transfer, and between neurons and glial cells (1, 21). This mimics the situation found in cell cultures (23, 24). Interestingly, glycoproteins gE and gI are dispensable for viral replication in cell cultures but are specifically required for transneuronal transfer of PrV between infected first-order neurons and several categories of connected neurons. For instance, after intranasal inoculation, a gE− PrV mutant is not transmitted to second-order neurons of the trigeminal, sympathetic, or parasympathetic route (3, 11, 15, 17, 21, 22). In rats, after inoculation into the posterior chamber of the eye, infection of the retina occurs normally but propagation of the mutant is restricted to a subset of neurons connected to ganglionic cells, e.g., those involved in circadian timing (6, 9, 10, 16, 18, 20, 26–28). The molecular basis for the restriction of transneuronal transfer is unclear. Glycoprotein gC is the major heparan sulfate binding envelope protein of alphaherpesviruses and thus is implicated in the primary attachment of virions to the host cell (reviewed in references 10 and 19). Despite this function in the initiation of infection, gC is not essential for the infectivity of PrV, presumably because attachment to cell surface components other than proteoglycans could also be mediated by gD (14). Penetration of and propagation in the nervous systems of adult mice by a gC deletion mutant are slower than with the wild type, and infected animals survive 24 h longer. However, they ultimately die with classical pseudorabies symptoms (5).
We have examined the roles of the nonessential structural glycoproteins gM and gN. Both proteins are conserved throughout the Herpesviridae, and they form a noncovalently linked complex (12). gM is N glycosylated and contains eight clusters of hydrophobic amino acids long enough to span the lipid bilayer. In a gM− PrV mutant, approximately 60% of the UL10 gene, which encodes gM, was deleted and replaced by a gG–β-galactosidase (gG–β-Gal) expression cassette (Fig. 1A). In cell cultures, gM− PrV replicates to yield approximately 50-fold-lower titers, and it is strongly attenuated in pigs (8). The product of the UL49.5 gene is O glycosylated in PrV and thus has been designated gN (13). A gN− PrV mutant which carries a 24-bp deletion within the UL49.5 gene and concomitant insertion of a gG–β-Gal expression cassette has been engineered (Fig. 1B). Both mutants and corresponding rescued viruses (gMR and gNR, in which the expression of gM and gN, respectively, was restored) were propagated on Vero cells. After the development of a complete cytopathic effect, cells were harvested and intracellular and extracellular viruses were concentrated and purified by centrifugation through a glycerol cushion (2). The genotypes of mutant viruses were verified by Southern blotting and PCR. Viral DNA was extracted from infected Vero cells after lysis in a solution containing 1% Nonidet P-40, 0.1% sodium deoxycholate, 10 mM Tris, and 1 mM EDTA (pH 8). After the removal of the nuclei by centrifugation, sodium dodecyl sulfate and proteinase K were added to the supernatant to final concentrations of 0.5% and 200 μg/ml, respectively, followed by an hour of incubation at 65°C. DNA was purified according to standard procedures (25a).
FIG. 1.
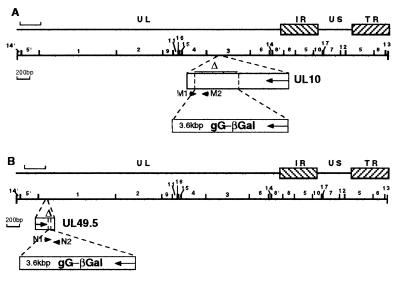
Construction of gM− and gN− mutants. The schematic diagram of the PrV genome consists of unique long (UL) and unique short (US) regions bracketed by repeats (TR, terminal repeat; IR, inverted repeat). The scales of the top lines are given by brackets which represent 8 kbp. A BamHI restriction map is given (BamHI fragments 1 and 3 include genes UL49.5 and UL10, respectively). The deletions (▵) of the gM (A) and gN (B) genes and the insertion of the gG-lacZ expression cassette are shown. The locations of primers M1, M2, N1, and N2, which were used for PCR amplifications, are given. The sequences of the primers are as follows: M1, 5′-GCC AGC AGG TAC TCG TCG TTG-3′; M2, 5′-CGG CCT TCT GCG TGC TCG TGG-3′; N1, 5′-GGC CAC GAC GAG CAC CGC CAG-3′; and N2, 5′-CTC GCA CAC ACC AGG ATG GTC-3′. M1 and M2 start, respectively, at positions 2474 and 2750 of the published sequence (GenBank accession no. X97257). N1 and N2 start, respectively, at positions 135 and 360 of the published sequence (GenBank accession no. U38547).
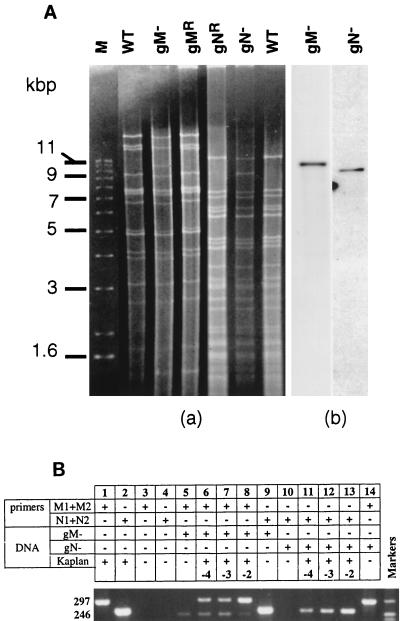
The genotypes of the viruses were first verified by Southern analysis (Fig. 2A). Replacement of part of the gM gene with a lacZ cassette introduced a new BamHI site within BamHI fragment 3, splitting this 17-kbp fragment into 9- and 11-kbp subfragments, the latter including the 3.6-kbp lacZ cassette. In the gN− mutant, the lacZ cassette introduced within the gN gene shifted PstI fragment 5 from 6 to 9.6 kbp. After digestion of the DNA with BamHI (wild type, gM−, or gMR) or PstI (wild type, gN−, or gNR), the fragments were separated on a 0.6% agarose gel containing ethidium bromide (Fig. 2A). As expected, the disappearance of a 17-kbp band and the appearance of a band of around 11 kbp was observed in gM− viruses, and wild-type and gMR profiles were similar. The 9-kbp subfragment probably comigrated with another BamHI fragment and could not be clearly seen in gM− bands. In the gN− bands, one band of 6 kbp was replaced by a band slightly larger than 9 kbp. Wild-type and gNR profiles were similar. Digestion products were also transferred to nylon membranes (Hybond N+; Amersham). Membranes were hybridized with a LacZ probe labelled with digoxigenin and developed according to the manufacturer’s instructions (Roche). The enhanced chemiluminescence autoradiograms are shown in Fig. 2A, gel b. The expected fragments from the gM− and gN− viruses hybridized with the LacZ probe and thus do include LacZ, which is absent from both the wild-type and the rescued viruses (not shown). Thus, the genotypes of the virus stocks were as expected. To further ascertain the absence of wild-type contamination from the recombinant virus stocks, we used a more sensitive PCR assay. Two sets of 21-mer primers were used for the amplification of a portion of the UL10 (M1 and M2) and UL49.5 (N1 and N2) genes. They were designed to yield amplification products of 297 and 246 bp, respectively, from wild-type PrV DNA. For each pair, one primer was localized within the deletion and the other was localized outside (Fig. 1). As expected, DNA of gM− PrV-infected cells yielded an amplification product only from the UL49.5-specific primers (Fig. 2B, lanes 5 and 9) whereas gN− PrV DNA was amplified by the UL10-specific primers only (Fig. 2B, lanes 10 and 14), indicating homogeneity of the virus stocks. The lack of contamination of mutant virus stocks by wild-type virus or revertants was further investigated by mixing dilutions of wild-type PrV DNA with mutant DNA. As shown in Fig. 2, wild-type PrV DNA was detectable by PCR at dilutions of up to 10−4 (lanes 6 to 8 and 11 to 13). Further dilutions did not result in detectable amplification. Thus, we conclude that the mutant virus stocks did not contain a detectable amount of wild-type or revertant virus.
FIG. 2.
Genotype assessment of the viral stocks. (A) Southern analysis of wild-type (WT) PrV, gM− and gN− mutants, and revertants. After extraction, viral DNA was digested with either BamHI (WT, gM−, and gMR) or PstI (gNR, gN−, and WT). Fragments were separated on a 0.6% agarose gel containing ethidium bromide (gel a), transferred to a nylon membrane, and hybridized with a LacZ probe (pGEM 3Zf; Promega) labeled with digoxigenin (gel b). Size markers (M) are on the left. (B) PCR amplifications were done with around 20 ng of wild-type or mutant DNA, 1 pmol of each primer, 2.5 U of Taq DNA polymerase (Appligene), 50 μM concentration of each deoxynucleoside triphosphate, and 10% dimethyl sulfoxide per reaction mixture. In addition to mutant DNA, 2, 20, and 200 pg of wild-type PrV DNA were added to samples 6 and 11, 7 and 12, and 8 and 13, respectively. Thirty cycles of amplification at 95, 50, and 72°C for 60, 40, and 120 s, respectively, were performed. PCR products were separated in 2% agarose gels containing ethidium bromide and were visualized in UV light. A pBR322 DNA-MspI digest was used for size markers (sizes of the bands, 217, 238, 242, and 307 nucleotides). Numbers at the left are molecular sizes of the amplified products, in nucleotides.
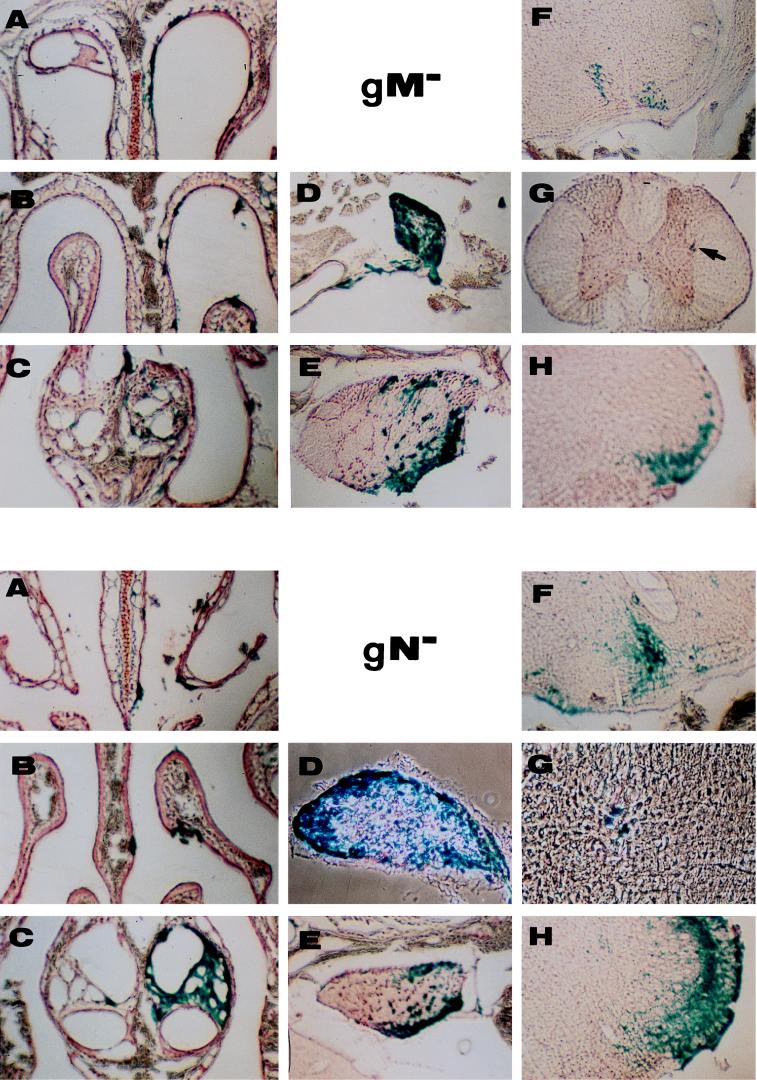
Intranasal inoculation of adult mice with the mutants was performed as described previously (2). Briefly, 3 μl of viral suspension containing 106 PFU of either the gN− or gM− mutant or corresponding rescued viruses (7, 13) was instilled in the right nostrils of 6-week-old Swiss mice with a Hamilton syringe connected to a catheter. Infected animals did not survive longer than those infected with wild-type or gG− PrV (48 to 52 h postinfection). All mice developed typical symptoms, e.g., hunched position and itching. To study viral penetration of and propagation in the nervous system, three mice per mutant virus and one mouse per corresponding rescued virus were infected and sacrificed when moribund. The head was skinned and the lower jaw and teeth were removed. The head was decalcified for 10 days in 0.1 M EDTA (pH 8.0) at 4°C. It was further incubated in 20% sucrose–phosphate-buffered saline for cryoconservation, embedded in Tissue Tek, frozen at −70°C, and cut into 30-μm sections. Sections were collected on gelatin-coated slides, incubated in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), counterstained with neutral red, and covered with a coverslip and Entelan. The superior cervical ganglia and spinal chord were dissected separately and incubated for 3 days at 4°C in 20% sucrose–phosphate-buffered saline containing X-Gal. They were then frozen, cut, and treated as the heads were treated. A Zeiss microscope with a 4× objective was used for observation. Sections from mice infected with rescued viruses which no longer contained the β-Gal gene were treated sequentially with polyclonal anti-PrV rabbit antibodies, biotinylated anti-rabbit antibodies, avidin, and biotinylated β-Gal. The eight mice yielded similar results, which can be summarized as follows: at the time of death, there was no obvious difference in the neuroinvasiveness of gM− and gN− mutants and rescued viruses. Infection was also similar to what was previously observed with the gG− mutant, which in every aspect was equivalent to wild-type PrV (2) (Fig. 3). Numerous foci of infection were seen in the respiratory and olfactory epithelia (Fig. 3A and B) and in the vomeronasal organ (Fig. 3C) on the inoculated side. On the other side, very few foci of infection were found, indicating that the inoculum remained mostly in the right nasal cavity. The mutants entered and propagated normally in the nervous system and showed signs of local transfer of the infection. For instance, the majority of sympathetic neurons in the superior cervical ganglia on the inoculated side were infected (Fig. 3D), including neurons which do not innervate the nasal cavity. The trigeminal ganglion on the inoculated side was heavily infected (Fig. 3E). Contralateral sympathetic and trigeminal ganglia were also infected, although less heavily than ipsilateral ganglia (not shown). By the time of death, the virus had infected synaptically connected neurons and was found in the superior salivary nucleus (parasympathetic pathway) (Fig. 3F), the intermediolateral nucleus (sympathetic pathway) (Fig. 3G), and the spinal trigeminal nucleus (Fig. 3H). Transfer was more efficient in the trigeminal pathway than in the parasympathetic pathway and was poorly efficient in the sympathetic pathway in which only a few infected second-order neurons were found 48 to 52 h postinfection (Fig. 3G), even though the upper half of the spinal cord was examined in its totality. Such differences in the timing of infection of second-order neurons was already observed with PrV strain Kaplan and its gG− mutant (2). Transfer was also more efficient with the gN− mutant than with the gM− mutant (compare the intensities of labelling in the spinal trigeminal nucleus, for instance), and this could be related to the fact that the deletion of glycoprotein gM affects viral multiplication more than the removal of gN (13). One reason could be that glycoprotein gM is found on the virion surface in the absence of gN whereas gN requires gM for virion localization (12).
FIG. 3.
Penetration and propagation of gM− and gN− mutants in the nervous systems of adult mice after intranasal inoculation. (A to C) The nasal cavity with the respiratory epithelium (A), the olfactory epithelium (B), and the vomeronasal organ (C); (D and E) ganglia containing first-order neurons of the sympathetic (D) and trigeminal (E) routes; (F to H) infected second-order neurons in the superior salivatory nucleus (parasympathetic pathway) (F), intermediolateral nucleus (sympathetic pathway) (arrow in gM− panel G) and spinal trigeminal nucleus (Sp5) (H). Magnifications, ×20 (gM− panels A to H and gN− panels A to C, E, F, and H), ×50 (gN− panel D), and ×100 (gN− panel G).
It is surprising that the deletion of proteins which are conserved among the herpesviruses did not dramatically modify the neuropathogenicity of the virus in mice after intranasal inoculation. Different results have been reported after intranasal infection of 6-week-old piglets with the gM− mutant (8). The mutant did not induce fever or any symptoms of Aujeszky’s disease, and nasal excretion of the virus was drastically reduced compared to excretion of wild-type PrV strain Kaplan. However, a direct comparison of the results is difficult since PrV strain Kaplan did not kill the piglets at a dose which would represent at least 104 50% lethal doses for mice.
A striking observation derived from our study of the neurovirulence and neuroinvasiveness of eight glycoprotein deletion mutants (some of them allowing a much longer survival time for the infected mice) is that the symptoms, which were certainly of nervous origin and rapidly led to the deaths of the infected animals, appeared only when peripheral ganglia were heavily infected. For example, at the time of death, the sympathetic and trigeminal ganglia were always massively invaded. Whether this is also true for the parasympathetic ganglion remains to be established because it is difficult to localize in mice. On the other hand, only a few isolated foci of infection in the brain were observed, which probably does not explain the severity of the symptoms.
This and previous studies have delineated the role of eight glycoproteins of PrV in neuroinvasiveness after intranasal infection of mice. Perhaps the most interesting finding was that gE and gI, two nonessential glycoproteins, are necessary for transneuronal transfer, at least in several chains of connected neurons. The roles of the other glycoproteins in neuroinvasiveness paralleled their functions in cell cultures: those proteins which are essential for cell-to-cell spread in cultures are also required for viral spread in the nervous system, whereas glycoproteins dispensable for viral replication in cultured cells are also nonessential for neuroinvasion. Whether these proteins function differently in different animal hosts, such as pigs, and which role they play in various virus-host systems remain to be analyzed.
Acknowledgments
We thank Richard Miselis for help in localizing several brain structures, Françoise Bras for suggesting the PCR experiment, Thérèse Bennardo for excellent technical assistance, and Sandie Munier for help with the Southern analysis.
This work was supported by the CNRS through the UPR A9053, the DFG (grant Me854/3-3), and the European Community (EEC contract no. BMH4-CT97-2573).
REFERENCES
- 1.Babic N, Mettenleiter T C, Flamand A, Ugolini G. Role of essential glycoproteins gII and gp50 in transneuronal transfer of pseudorabies virus from the hypoglossal nerves of mice. J Virol. 1993;67:4421–4426. doi: 10.1128/jvi.67.7.4421-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babic N, Mettenleiter T C, Ugolini G, Flamand A, Coulon P. Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology. 1994;204:616–625. doi: 10.1006/viro.1994.1576. [DOI] [PubMed] [Google Scholar]
- 3.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 4.Babic N, Klupp B G, Makoschey B, Karger A, Flamand A, Mettenleiter T C. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J Gen Virol. 1996;77:2277–2285. doi: 10.1099/0022-1317-77-9-2277. [DOI] [PubMed] [Google Scholar]
- 5.Babic, N., and A. Flamand. Unpublished results.
- 6.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkstra J M, Gerdts V, Klupp B G, Mettenleiter T C. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol. 1997;78:2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 9.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs L, Mulder W A M, Oirschot J T V, Gielkens A L J, Kimman T J. Deleting two amino acids in glycoprotein gI of pseudorabies virus decreases virulence and neurotropism for pigs, but does not affect immunogenicity. J Gen Virol. 1993;74:2201–2206. doi: 10.1099/0022-1317-74-10-2201. [DOI] [PubMed] [Google Scholar]
- 12.Jöns A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jöns A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karger A, Saalmüller A, Tufaro F, Banfield B W, Mettenleiter T C. Cell surface proteoglycans are not essential for infection by pseudorabies virus. J Virol. 1995;69:3482–3489. doi: 10.1128/jvi.69.6.3482-3489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimman T G, de Wind N, Oei-Lie N, Pol J M A, Berns A J M, Gielkens A L J. Contribution of single genes within the unique short region of Aujeszky’s disease virus (suid herpes type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 16.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE- and gI-deleted mutants of Aujeszky’s disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 18.Levine J D, Zhao X-S, Miselis R R. Direct and indirect retino-hypothalamic projections to the supraoptic nucleus in the female albino rat. J Comp Neurol. 1994;341:214–224. doi: 10.1002/cne.903410207. [DOI] [PubMed] [Google Scholar]
- 19.Mettenleiter T C. Pseudorabies (Aujeszky’s disease) virus: state of the art. Acta Vet Hung. 1993;42:153–177. [PubMed] [Google Scholar]
- 20.Moore R Y, Speh J C, Card J P. The retinohypothalamic tract originates from a distinct subset of retinal ganglion cells. J Comp Neurol. 1995;352:351–366. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 21.Mulder W, Pol J, Kimman T, Kok G, Priem J, Peeters B. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J Virol. 1996;70:2191–2200. doi: 10.1128/jvi.70.4.2191-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder W A M, Jacobs L, Priem J, Kok G L, Wagenaar F, Kimman T G, Pol J M A. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 23.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauh I, Weiland F, Fehler F, Keil G M, Mettenleiter T C. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J Virol. 1991;65:621–631. doi: 10.1128/jvi.65.2.621-631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sams J M, Jansen A S P, Mettenleiter T C, Loewy A D. Pseudorabies virus mutants as transneuronal markers. Brain Res. 1995;687:182–190. doi: 10.1016/0006-8993(95)00484-8. [DOI] [PubMed] [Google Scholar]
- 27.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]