Abstract
Using real-time fluorescence PCR, we quantitated the numbers of copies of latent varicella-zoster virus (VZV) and herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) genomes in 15 human trigeminal ganglia. Eight (53%) and 1 (7%) of 15 ganglia were PCR positive for HSV-1 or -2 glycoprotein G genes, with means of 2,902 ± 1,082 (standard error of the mean) or 109 genomes/105 cells, respectively. Eleven of 14 (79%) to 13 of 15 (87%) of the ganglia were PCR positive for VZV gene 29, 31, or 62. Pooling of the results for the three VZV genes yielded a mean of 258 ± 38 genomes/105 ganglion cells. These levels of latent viral genome loads have implications for virus distribution in and reactivation from human sensory ganglia.
Herpes simplex virus types 1 and 2 (HSV-1 and -2) and varicella-zoster virus (VZV) are alphaherpesviruses that infect, establish latency in, and subsequently reactivate from human sensory nerve ganglia (1, 36). Following reactivation from latent ganglia reservoirs, each of these herpesviruses may cause significant clinical disease in the individual and may spread to uninfected persons. Symptomatic VZV reactivation is an infrequent, usually once-in-a-lifetime event that results in zoster (shingles), while HSV-1 and -2 reactivation occurs frequently and results in numerous symptomatic and asymptomatic recurrences of oral and genital herpes.
Little is known regarding the mechanism underlying the particular patterns of latency and reactivation that distinguish HSV-1 and -2 infection from that with VZV. Abundant data from both human studies and animal models confirm that HSV-1 and -2 persist in sensory neurons but that satellite glial cells are spared from harboring latent HSV (7, 29, 30). Data regarding the site of VZV latency have been controversial, with various reports indicating it to be neurons, nonneuronal cells, or both (7, 9, 17, 22, 26). Moreover, estimates of the proportions of cells harboring HSV and VZV and the quantity of latent viral DNA in ganglia have varied widely (7, 17, 22, 25, 30). Recent animal studies show that latent viral genome levels in sensory ganglia influence the reactivation frequency of HSV-1 and -2, suggesting that the quantity of latent viral genome copies per ganglion—the latent viral load—may be a significant determinant of herpesvirus reactivation from the nervous system (21, 31, 32). To further clarify the nature and distribution of latent HSV and VZV genomes in human ganglia, we developed and used several sensitive and specific PCR assays.
Human tissue samples.
Human trigeminal ganglia were harvested within 24 h postmortem, frozen in dry ice, and stored at −70°C until DNA extraction. The general clinical histories and causes of death are summarized in Table 1.
TABLE 1.
Demographic data on subjects from whom trigeminal ganglia were recovered
| Subject | Gendera | Age (yr) | Immediate cause of death | Underlying and/or contributing disease |
|---|---|---|---|---|
| 669 | M | 65 | Alcohol abuse | None known |
| 671 | M | 27 | Motor vehicle accident | None known |
| 672 | M | 25 | Drug overdose | HIV infected |
| 673 | M | 34 | Gunshot wound | None known |
| 674 | F | 35 | Unknown | None known |
| 675 | M | 40 | Gunshot wounds | None known |
| 676 | M | 73 | Drug overdose | Systemic lupus erythmatosus |
| 677 | M | 45 | Unknown | None known |
| 678 | M | 18 | Drug overdose | None known |
| 679 | F | 43 | Drug overdose | None known |
| 680 | F | 62 | Drug overdose | None known |
| 681 | F | 30 | Respiratory failure | Aplastic anemia |
| 682 | M | 41 | Sepsis | Large granular lymphocytic leukemia |
| 683 | M | 23 | Pneumonia | Multiple myeloma, bone marrow transplant |
| 684 | M | 18 | Diffuse alveolar damage | Graft vs host disease, organ transplant recipient |
M, male; F, female.
DNA was extracted by the protocol described in the instructions for a Puregene DNA isolation kit (D-5000A; Gentra Systems, Minneapolis, Minn.) with a few modifications. Ganglia were pulverized to a fine powder on dry ice and incubated in cell lysis buffer and 10 mg of proteinase K per ml for 3 days to ensure complete homogenization. Following protein precipitation, DNA was ethanol precipitated and resuspended in water. DNA concentration was estimated by spectrophotometry, and purity was determined from the ratios of the optical density at 260 nm to that at 280 nm. On average, the ganglia yielded 478 ± 46 (mean ± standard error of the mean [SEM]) μg of DNA.
QF-PCR assays.
Quantitative fluorescent (QF) PCR was performed with a Prism 7700 sequence detector (PE Applied Biosystems, Foster City, Calif.) according to supplied guidelines for real-time DNA amplification. Real-time PCR relies on a quantitative increase in fluorescence due to cleavage of a 5′ reporter dye from a dually labeled fluorogenic probe oligonucleotide by the 5′→3′ nuclease activity of Taq DNA polymerase.
The genes encoding VZV glycoprotein B (gB; open reading frame [ORF] 31), ORF 62 (which encodes the major immediate early transactivator), ORF 29 (which encodes a putative early major DNA-binding protein), HSV-1 glycoprotein G (gG1), and HSV-2 glycoprotein G (gG2) were selected for quantification. The forward and reverse primers and probe for the VZV gB gene were described by Kimura et al. (18) (Table 2). The forward and reverse primers and probes for VZV ORF 29 and ORF 62 and for the gG genes of HSV-1 and HSV-2 were designed with the Primer Express program (PE Applied Biosystems) (Table 2) and synthesized by Bioserve Biotechnologies (Laurel, Md.). QF PCR was also performed with the primers and the probe (Table 2) provided with the Taqman β-actin reagent kit (PE Applied Biosystems) to normalize each of the ganglion extracts for amplifiable human DNA.
TABLE 2.
Probe and primer sequences for genes amplified by quantitative PCR
| Viral or cellular gene | Probea | Forward primer | Reverse primer |
|---|---|---|---|
| VZV | |||
| gB | 5′-(FAM)ATTACTGGAACCTGCAGCGCGGA(TAMRA)-3′ | 5′-GATGGTGCATACAGAGAACATTCC-3′ | 5′-CCGTTAAATGAGGCGTGACTAA-3′ |
| ORF 29 | 5′-(FAM)CCCGTGGAGCGCGTCGAAA(TAMRA)-3′ | 5′-CGTACACGTATTTTCAGTCCTCTTC-3′ | 5′-GGCTTAGACGTGGAGTTGACA-3′ |
| ORF 62 | 5′-(FAM)TCTCGACTGGCTGGGACTTGCG(TAMRA)-3′ | 5′-TCTTGTCGAGGAGGCTTCTG-3′ | 5′-TGTGTGTCCACCGGATGAT-3′ |
| HSV-1 gG1 | 5′-(FAM)CCCTGGACACCCTCTTCGTCGTCAG(TAMRA)-3′ | 5′-CTGTTCTCGTTCCTCACTGCCT-3′ | 5′-CAAAAACGATAAGGTGTGGATGAC-3′ |
| HSV-2 gG2 | 5′-(FAM)ACACATCCCCCTGTTCTGGTTCCTAACG(TAMRA)-3′ | 5′-CAAGCTCCCGCTAAGGACAT-3′ | 5′-GGTGCTGATGATAAAGAGGATATCTAGA-3′ |
| Human genomic β-actin | 5′-(FAM)ATGCCCX(TAMRA)CCCCCATGCCATCCTGCGTp-3′ | 5′-TCACCCACACTGTGCCCATCTACGA-3′ | 5′-CAGCGGAACCGCTCATTGCCCAATGG-3′ |
TAMRA, 6-carboxy-tetramethyl-rhodamine. X in the β-actin probe sequence is a linker arm nucleotide, and p is a phosphorylation site.
All PCRs were performed in triplicate on two separate occasions with a Taqman PCR kit (PE Applied Biosystems) in the absence of reverse transcriptase, so that only DNA was amplified. Each 50-μl PCR mixture contained 500 ng of human trigeminal ganglion DNA with final concentrations of each primer of 1,000 nM and of each probe sequence of 200 nM. Multiple human trigeminal DNA samples were run together on a plate in parallel with duplicate sets of DNA standards. Each primer set mixture was run on a separate plate. Standard curves for each targeted viral gene were generated by mixing serial 10-fold dilutions of plasmids containing 100 to 106 copies of the desired genes. Additional dilutions within that range were analyzed for plasmids bearing the genes for gB, ORF 62, and gG1 to better delineate the sensitivity limits at the low ends of the standard curves. Uninfected BALB/c mouse trigeminal DNA (500 ng) was added to all plasmid standards to compensate for the potential inhibition of amplification reactions by added DNA. For VZV gB, the gB gene contained in the pUC19 vector, a gift of Liyanage Perera, was used. For ORF 29 and ORF 62, their respective EcoRI-B and EcoRI-A fragments from the VZV strain Ellen contained in the pGEM vector were used. Plasmids containing the HSV-1 and HSV-2 gG genes were obtained from Mark Challberg and Philip Krause, respectively. In addition, no template control reaction mixtures containing the appropriate probe and primer system without DNA were run on all plates in triplicate. PCR mixtures were subjected to 2 min at 50°C (reaction of AmpErase uracil-N-glycosylase), 10 min at 95°C (activation of AmpliTaq Gold), and 55 cycles of 15 s at 95°C and 1 min at 60°C. For β-actin amplification, 40 cycles were performed.
For each reaction, real-time fluorescence values were measured as a function of the quantity of a reporter dye (6-carboxy-fluorescein [FAM]) released during amplification. A threshold cycle (Ct) value for each sample was determined as the number of the first cycle at which the measured fluorescence exceeded the threshold limit (10 times the standard deviation of the baseline). Ct values observed for human trigeminal DNA samples were used to calculate the viral genome copy number for each sequence amplified based on the standard curves for plasmids containing test sequence.
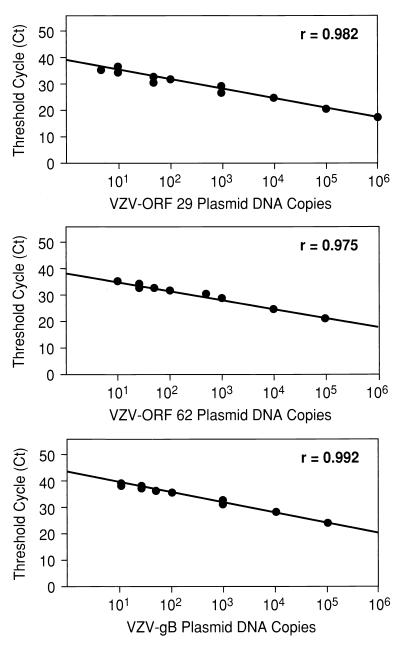
Standard curves for three VZV-containing plasmids are shown in Fig. 1 as examples. No cross-reactivity was observed between any of the viral DNA assays. The limits of sensitivity of the QF PCRs for all of the viral genes were estimated as the lowest plasmid dilutions that yielded comparable Ct values in replicate samples: they were all <10 copies/500 ng of input DNA. The human β-actin standards were not evaluated below 100 copies/500 ng of DNA. All standard curves were fit by linear regression, with correlation coefficients of >0.92.
FIG. 1.
Standard curves for QF-PCR assays for VZV genes encoding ORFs 29 and 62 and gB. Shown are the Ct values at which a given input of a VZV DNA-containing plasmid was detected by PCR. Plasmid concentrations tested ranged from 100 to 106 copies per reaction mixture. The Spearman rank coefficient of correlation (r) for lines fitting each standard curve are given.
Ct values for plasmid dilutions in these standard curves were compared with Ct values of human trigeminal ganglia samples to estimate the number of copies of each viral gene present in the approximately 500 ng of extracted DNA analyzed for each reaction. To more accurately estimate the amount of human genomic DNA in each trigeminal ganglion extract, we quantitated the number of copies of β-actin genes in that sample volume and normalized the number of DNA copies observed to the number of copies per 2 × 105 β-actin copies, i.e., per 105 cells. Because the mean number of copies of β-actin quantified in 500 ng of DNA was 6.4 × 104 ± 0.8 × 104, we calculated that each cell contained 15.6 pg of DNA.
VZV and HSV latent DNA load.
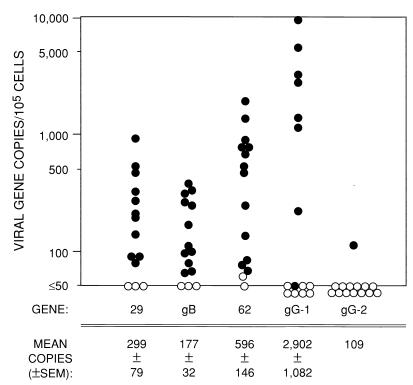
The numbers of copies of the various VZV and HSV genes per 105 cells are displayed in Fig. 2 along with tabulations of their mean numbers (± SEM) for ganglia that yielded positive results (≥10 copies/500 ng of DNA). By these criteria, the proportions (and percentages) of ganglia positive for each viral gene were as follows: 11 of 14 were positive (79%) for the ORF 29 gene, 12 of 15 were positive (80%) for the gB gene, 13 of 15 were positive (87%) for the ORF 62 gene, 8 of 15 were positive (53%) for the gG1 gene, and 1 of 14 were (7%) positive for the gG2 gene.
FIG. 2.
Copies of VZV, HSV-1, and HSV-2 genes in human trigeminal ganglia. The mean ± SEM number of copies per 105 cells for each gene in the PCR-positive ganglia are tabulated below the figure. The filled circles show the gene copy numbers for samples that exceeded the assay limit of detection (≥10 copies/500 ng of total DNA). Open circles represent values for samples for which gene copy numbers may have been extrapolated from the QF-PCR results but were below the threshold of reliable DNA detection (<10 copies/500 ng of DNA). The inability to define a precise threshold for positive ganglia on the per-105-cell basis used here reflects the variability in β-actin gene number per 500 ng of total DNA from the individual ganglia.
By the Spearman rank test, the numbers of copies of all three VZV genes correlated highly significantly with each other (P < 0.001). There were no statistical associations between HSV gene copy numbers and VZV gene numbers. Moreover, the mean number of VZV ORF 62 genes, 2.7 ± 0.4 times the numbers of the gB and ORF 29 genes, was not dissimilar from the expected ratio of 2.0 for this diploid viral gene. With the numbers obtained for gB and ORF 29 and one-half of the number obtained for the diploid ORF 62 gene being pooled, the QF-PCR assay revealed that PCR-positive ganglia contained a mean of 258 ± 38 VZV genome copies/105 cells. This value was significantly less than the 2,902 ± 1,082 HSV-1 genomes per 105 cells in positive ganglia (P = 0.02; Wilcoxon rank sum test) but comparable to the copy number of genomes in the one ganglion containing detectable HSV-2 DNA, 109 genomes/105 cells. Latent viral DNA loads were not grossly different for ganglia obtained from individuals known to be immunocompromised and those presumed to be immunocompetent.
Implications of the data.
The detection of VZV DNA in 79 to 87% of ganglia is in concordance with the very high proportion of American adults who are seropositive for VZV and with data obtained from prior standard nonquantitative PCR assays and in situ hybridization (1, 13, 27). Slightly more than half of the ganglia were HSV-1 DNA positive, also commensurate with the seroprevalence of 50 to 70% reported for this virus for young American adults and with prior molecular studies of human ganglia (7, 11, 36). HSV-2 DNA was detected in only 1 of 15 ganglia, reflecting the relatively low rate of facial infection with this virus (36).
Extrapolations of our data permit estimates of the total copy numbers and distributions of these three viruses in human trigeminal ganglia. We recovered a mean of 478 μg of DNA per ganglion. Based on our estimate of 15.6 pg of DNA/cell, the average trigeminal ganglion contained upward of 3 × 107 cells. Thus, we estimate that, on average, each ganglion latently infected with VSV, HSV-1, or HSV-2 contained 7.7 × 104, 8.7 × 105, or 3.2 × 104 genomes, respectively. Our estimate for latent HSV-1 load in human ganglia (Fig. 2) is similar to that reported by Efstathiou et al. (11) (1,000 to 10,000 copies), but the estimate for VZV load is higher than the 6 to 31 copies/105 cells reported by Mahalingam et al. using the less precise PCR and Southern hybridization methods (26).
Ball et al. reported that human trigeminal ganglia contain an average of 8.1 × 104 neurons (2). Since HSV-1 and -2 persist exclusively in neurons (5, 7, 8, 29, 30, 34, 35), we can project that each latently infected neuron contains at least 11 copies of HSV-1 DNA, assuming all neurons are infected. While in situ hybridization studies for HSV-1 latency-associated transcripts suggested that only 1 to 4% of neurons are positive (7, 8, 10, 34, 35), the more recent PCR and in situ-PCR analyses demonstrated that 3 to 10 times that percentage are positive (6–8, 29–32, 34, 35). These data indicate that latently infected trigeminal neurons contain an average of 28 or more HSV-1 genomes, a copy number similar to those of Epstein-Barr virus episomes carried in B cell lines (12, 28) and papillomavirus episomes in human cervical cancer cells (3, 4, 15, 16).
The low copy number of detectable HSV-2 DNA in trigeminal ganglia may parallel the very low frequency with which this virus recurs following initial infection in the mouth (20). Since the latent viral load correlates well with recurrence rates in infected animals, if further studies confirm the low HSV-2 copy number in human ganglia, these data suggest that the same is also true for humans (21, 24, 31, 32). If so, some obstacle must impede infection and establishment of latency of HSV-2 in human trigeminal ganglia.
Although our initial data suggested that VZV persists only in nonneuronal cells (7), recent data compel us to conclude that both neurons and nonneuronal cells are infected by VZV both during productive infection (7, 22) and during latency (7, 9, 14, 17, 22, 25). Were VZV to persist exclusively in neurons, the average of 7.7 × 104 genome copies we detected could be distributed among nearly all of the estimated 8.1 × 104 neurons; however, they are not distributed in this way (17, 22). While we found no published estimates of the numbers of satellite and other nonneuronal cell populations in human ganglia, sensory neurons are very large (50 to 100 μm in diameter) and are encircled and contacted by about 10 nucleated satellite cells each in most thin (6 to 8-μm) histologic sections (unpublished data and references 6, 10, and 19), which implies that there are at least 100 satellite cells surrounding every neuron or at least 8 × 106 satellite cells per human trigeminal ganglion.
If we were to assume that VZV persisted in the same proportions of neurons and nonneuronal cells and at roughly the same copy number per cell as that of HSV-1 (≥28), fewer than 1 in 1,000 total cells would prove positive. Lungu et al. (22, 23) estimated that 5 to 30% of both neurons and satellite cells are positive for VZV DNA or VZV proteins, an estimate that is far higher than that permitted by these data. Kennedy et al. (17), however, estimated by in situ PCR that 2% of neurons (∼1,600 cells according to the estimate of Ball et al. [2]) and 0.1% of satellite cells (∼8,000 cells according to our estimate) are VZV DNA positive. If the estimate of Kennedy et al. is correct, the 7.7 × 104 VZV genomes could persist in this number of cells at a density of eight copies/cell, not dissimilar from that for HSV-1. We are currently testing the validity of these extrapolations from our QF-PCR data by analyzing sections microdissected from human ganglia (33).
The present data provide refined estimates of the proportion of human trigeminal ganglia containing VZV, HSV-1, and HSV-2 DNAs and the latent DNA loads for each of these neurotropic viruses. Moreover, they have implications regarding the distribution of these three viruses in human ganglia and the pathogenesis of reactivation infections associated with them.
Acknowledgments
We thank Jeffrey Cohen, Philip Krause, and Nancy Tresser for advice and assistance in this project, Peter Kennedy for additional comments on the manuscript, and Uri Lopatin for help with statistical analyses.
REFERENCES
- 1.Arvin A M. Varicella-zoster virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2547–2585. [Google Scholar]
- 2.Ball M J, Nuttall K, Warren K G. Neuronal and lymphocytic populations in human trigeminal ganglia: implications for aging and for latent virus. Neuropathol Appl Neurobiol. 1982;8:177–187. doi: 10.1111/j.1365-2990.1982.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 3.Berumen J, Casas L, Segura E, Amezcua J L, Garcia-Carranca A. Genome amplification of human papillomavirus types 16 and 18 in cervical carcinomas is related to the retention of E1/E2 genes. Int J Cancer. 1994;56:640–645. doi: 10.1002/ijc.2910560506. [DOI] [PubMed] [Google Scholar]
- 4.Brown D R, Bryan J T, Cramer H, Katz B P, Handy V, Fife K H. Detection of multiple human papillomavirus types in condylomata acuminata from immunosuppressed patients. J Infect Dis. 1994;170:759–765. doi: 10.1093/infdis/170.4.759. [DOI] [PubMed] [Google Scholar]
- 5.Croen K D, Ostrove J M, Dragovic L, Straus S E. Characterization of herpes simplex virus type 2 latency-associated transcription in human sacral ganglia and in cell culture. J Infect Dis. 1991;163:23–28. doi: 10.1093/infdis/163.1.23. [DOI] [PubMed] [Google Scholar]
- 6.Croen K D, Ostrove J M, Dragovic L J, Smialek J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense” transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 7.Croen K D, Ostrove J M, Dragovic L J, Straus S E. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc Natl Acad Sci USA. 1988;85:9773–9777. doi: 10.1073/pnas.85.24.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deatly A, Lumsden A. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci USA. 1984;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dueland A N, Ranneberg-Nilsen T, Degre M. Detection of latent varicella zoster virus DNA and human gene sequences in human trigeminal ganglia by in situ amplification combined with in situ hybridization. Arch Virol. 1995;140:2055–2066. doi: 10.1007/BF01322692. [DOI] [PubMed] [Google Scholar]
- 10.Ecob-Prince M S, Preston C M, Rixon F J, Hassan K, Kennedy P G E. Neurons containing latency-associated transcripts are numerous and widespread in dorsal root ganglia following footpad inoculation of mice with herpes simplex virus type 1 mutant in 1814. J Gen Virol. 1993;74:985–994. doi: 10.1099/0022-1317-74-6-985. [DOI] [PubMed] [Google Scholar]
- 11.Efstathiou S, Minson A C, Field H J, Anderson J R, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986;57:446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernberg I, Andersson-Anvret M, Klein G. Relationship between amount of Epstein-Barr virus-determined nuclear antigen per cell and number of EBV-DNA copies per cell. Nature. 1977;226:269–271. doi: 10.1038/266269a0. [DOI] [PubMed] [Google Scholar]
- 13.Furuta Y, Takasu T, Fukuda S, Sato-Matsumura K C, Inuyama Y, Hondo R, Nagashima K. Detection of varicella-zoster virus DNA in human geniculate ganglia by polymerase chain reaction. J Infect Dis. 1992;166:1157–1159. doi: 10.1093/infdis/166.5.1157. [DOI] [PubMed] [Google Scholar]
- 14.Gilden D H, Vafai A, Shtram Y, Becker Y, Devlin M, Wellish M. Varicella-zoster virus DNA in human sensory ganglia. Nature. 1983;306:478–480. doi: 10.1038/306478a0. [DOI] [PubMed] [Google Scholar]
- 15.Ikenberg H, Runge M, Goppinger A, Pfleiderer A. Human papillomavirus DNA in invasive carcinoma of the vagina. Obstet Gynecol. 1990;76:432–438. [PubMed] [Google Scholar]
- 16.Jeon S, Allen-Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy P G, Grinfeld E, Gow J W. Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc Natl Acad Sci USA. 1998;95:4658–4662. doi: 10.1073/pnas.95.8.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura H, Wang Y, Pesnicak L, Cohen J I, Hooks J J, Straus S E, Williams R K. Recombinant varicella-zoster virus glycoproteins E and I: immunologic responses and clearance of virus in a guinea pig model of chronic uveitis. J Infect Dis. 1998;178:310–317. doi: 10.1086/515638. [DOI] [PubMed] [Google Scholar]
- 19.Krause P R, Croen K D, Straus S E, Ostrove J M. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J Virol. 1988;62:4819–4823. doi: 10.1128/jvi.62.12.4819-4823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafferty W E, Coombs R W, Benedetti J, Critchlow C, Corey L. Recurrences after oral and genital herpes simplex virus infection. Influence of site of infection and viral type. N Engl J Med. 1987;316:1444–1449. doi: 10.1056/NEJM198706043162304. [DOI] [PubMed] [Google Scholar]
- 21.Lekstrom-Himes J A, Pesnicak L, Straus S E. The quantity of latent viral DNA correlates with the relative rates at which herpes simplex virus types 1 and 2 cause recurrent genital herpes outbreaks. J Virol. 1998;72:2760–2764. doi: 10.1128/jvi.72.4.2760-2764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lungu O, Annunziato P W, Gershon A, Staugaitis S M, Josefson D, LaRussa P, Silverstein S J. Reactivated and latent varicella-zoster virus in human dorsal root ganglia. Proc Natl Acad Sci USA. 1995;92:10980–10984. doi: 10.1073/pnas.92.24.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lungu O, Panagiotidis C A, Annunziato P W, Gershon A A, Silverstein S J. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc Natl Acad Sci USA. 1998;95:7080–7085. doi: 10.1073/pnas.95.12.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggioncalda J, Mehta A, Su Y H, Fraser N W, Block T M. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology. 1996;225:72–81. doi: 10.1006/viro.1996.0576. [DOI] [PubMed] [Google Scholar]
- 25.Mahalingam R, Wellish M, Cohrs R, Debrus S, Piette J, Rentier B, Gilden D. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc Natl Acad Sci USA. 1996;93:2122–2124. doi: 10.1073/pnas.93.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahalingam R, Wellish M, Lederer D, Forghani B, Cohrs R, Gilden D. Quantitation of latent varicella-zoster virus DNA in human trigeminal ganglia by polymerase chain reaction. J Virol. 1993;67:2381–2384. doi: 10.1128/jvi.67.4.2381-2384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahalingam R, Wellish M C, Dueland A N, Cohrs R J, Gilden D H. Localization of herpes simplex virus and varicella zoster virus DNA in human ganglia. Ann Neurol. 1992;31:444–448. doi: 10.1002/ana.410310417. [DOI] [PubMed] [Google Scholar]
- 28.Nonoyama M, Pagano J S. Homology between Epstein-Barr virus DNA and viral DNA from Burkitt’s lymphoma and nasopharyngeal carcinoma determined by DNA-DNA reassociation kinetics. Nature. 1973;242:44–47. doi: 10.1038/242044a0. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan R, Poliani P L, Levine M, Glorioso J C, Fink D J. Detection of herpes simplex virus type 1 latency-associated transcript expression in trigeminal ganglia by in situ reverse transcriptase PCR. J Virol. 1996;70:6519–6523. doi: 10.1128/jvi.70.9.6519-6523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawtell N M. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J Virol. 1997;71:5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawtell N M. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J Virol. 1998;72:6888–6892. doi: 10.1128/jvi.72.8.6888-6892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawtell N M, Poon D K, Tansky C S, Thompson R L. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J Virol. 1998;72:5343–5350. doi: 10.1128/jvi.72.7.5343-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simone N L, Bonner R F, Gillespie J W, Emmert-Buck M R, Liotta L A. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 34.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 35.Tenser R B, Dawson M, Ressel S J, Dunstan M E. Detection of herpes simplex virus mRNA in latently infected trigeminal ganglion neurons by in situ hybridization. Ann Neurol. 1982;11:285–291. doi: 10.1002/ana.410110309. [DOI] [PubMed] [Google Scholar]
- 36.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2297–2342. [Google Scholar]