Abstract
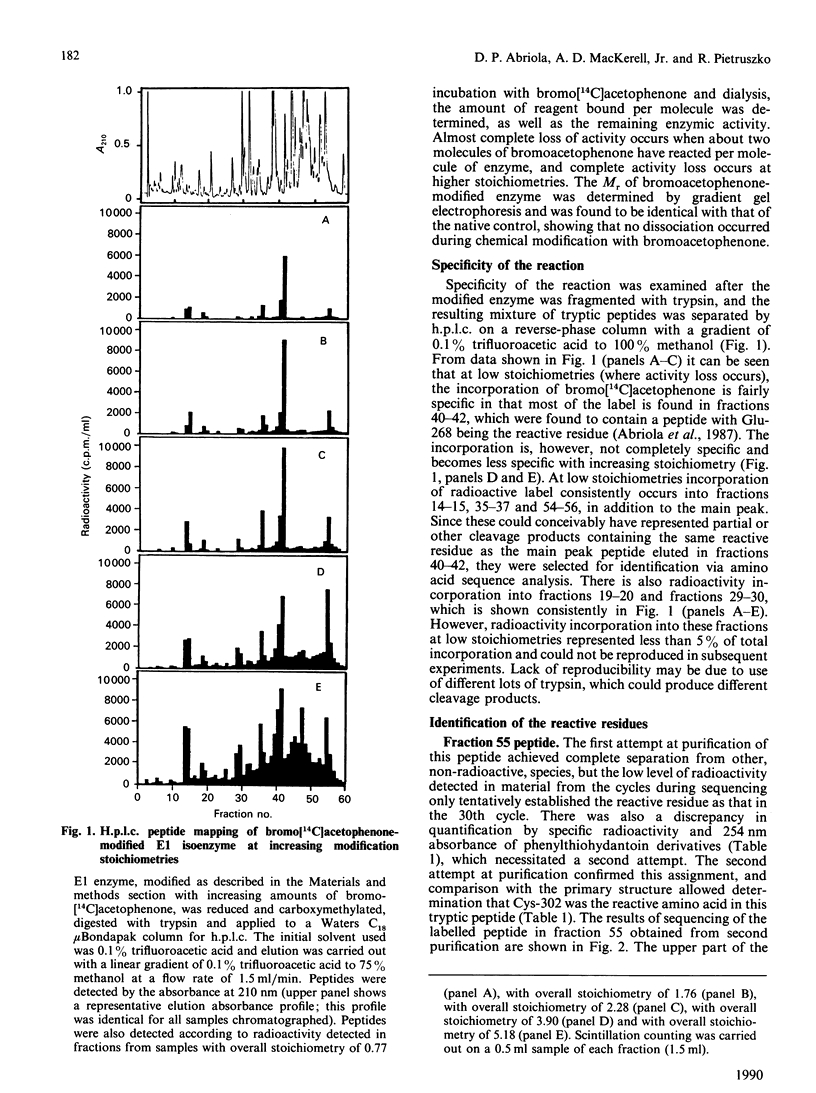
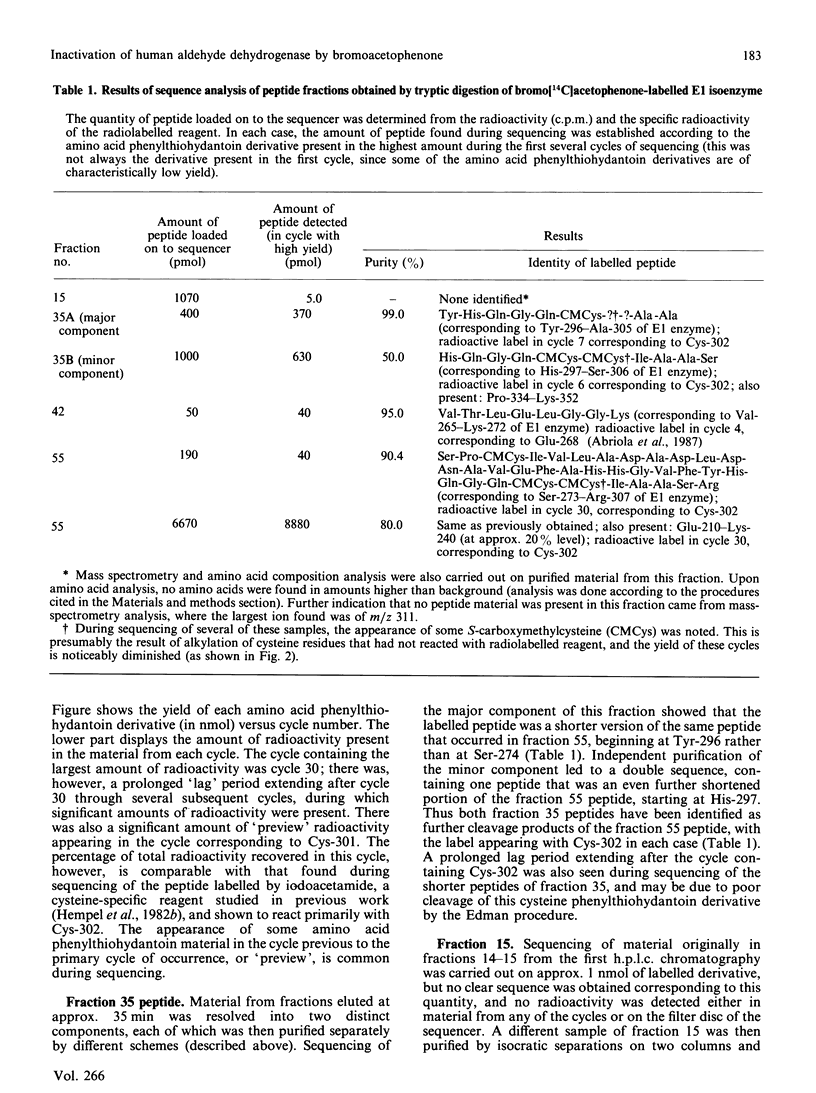
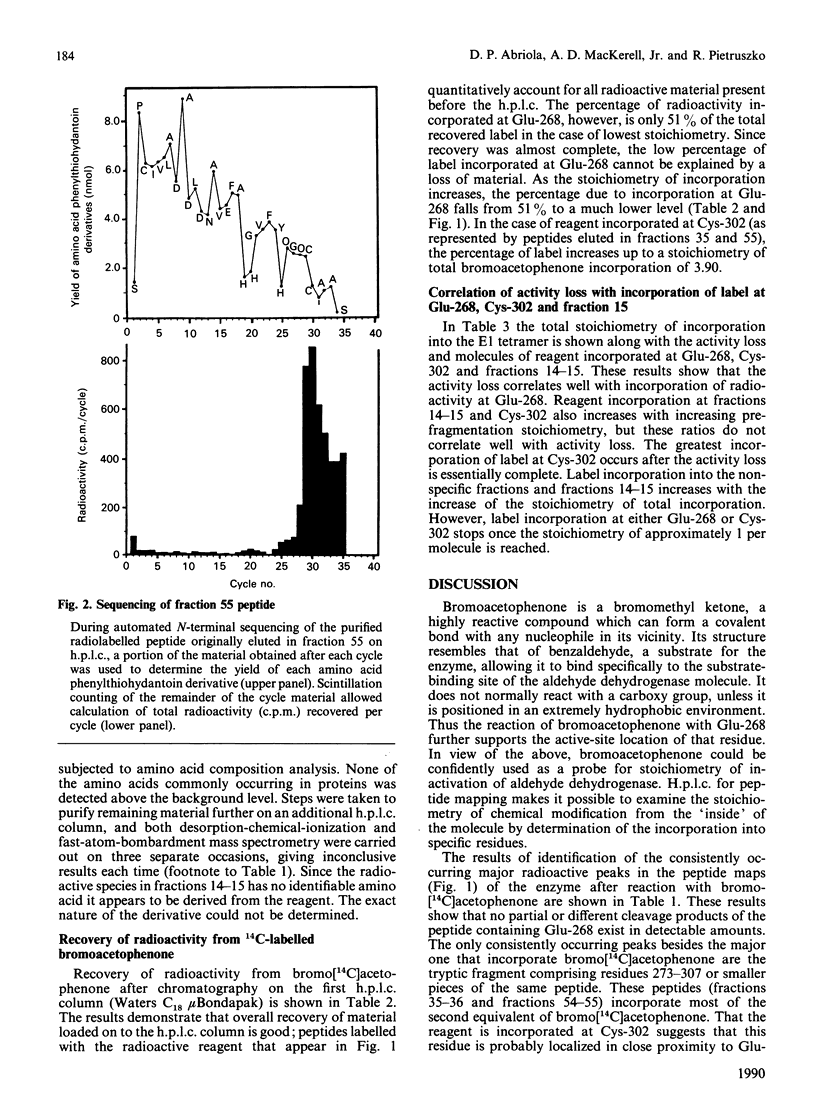
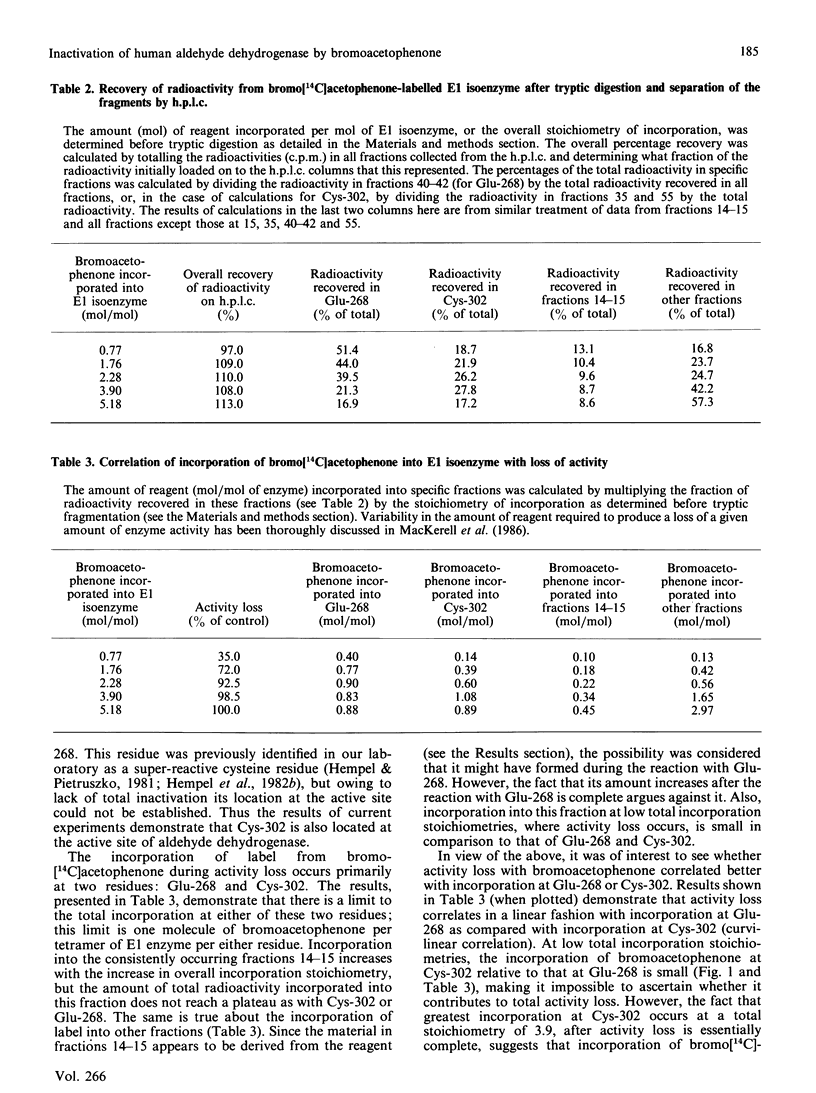
Bromoacetophenone (2-bromo-1-phenylethanone) has been characterized as an affinity reagent for human aldehyde dehydrogenase (EC 1.2.1.3) [MacKerell, MacWright & Pietruszko (1986) Biochemistry 25, 5182-5189], and has been shown to react specifically with the Glu-268 residue [Abriola, Fields, Stein, MacKerell & Pietruszko (1987) Biochemistry 26, 5679-5684] with an apparent inactivation stoichiometry of two molecules of bromoacetophenone per molecule of enzyme. The specificity of bromoacetophenone for reaction with Glu-268, however, is not absolute, owing to the extreme reactivity of this reagent. When bromo[14C]acetophenone was used to label the human cytoplasmic E1 isoenzyme radioactively and tryptic fragmentation was carried out, peptides besides that containing Glu-268 were found to have reacted with reagent. These peptides were purified by h.p.l.c. and analysed by sequencing and scintillation counting to quantify radioactive label in the material from each cycle of sequencing. Reaction of bromoacetophenone with the aldehyde dehydrogenase molecule during enzyme activity loss occurs with two residues, Glu-268 and Cys-302. The activity loss, however, appears to be proportional to incorporation of label at Glu-268. The large part of incorporation of label at Cys-302 occurs after the activity loss is essentially complete. With both Glu-268 and Cys-302, however, the incorporation of label stops after one molecule of bromoacetophenone has reacted with each residue. Reaction with other residues continues after activity loss is complete.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abriola D. P., Fields R., Stein S., MacKerell A. D., Jr, Pietruszko R. Active site of human liver aldehyde dehydrogenase. Biochemistry. 1987 Sep 8;26(18):5679–5684. doi: 10.1021/bi00392a015. [DOI] [PubMed] [Google Scholar]
- Ambroziak W., Kosley L. L., Pietruszko R. Human aldehyde dehydrogenase: coenzyme binding studies. Biochemistry. 1989 Jun 27;28(13):5367–5373. doi: 10.1021/bi00439a008. [DOI] [PubMed] [Google Scholar]
- Dickinson F. M., Hart G. J., Kitson T. M. The use of pH-gradient ion-exchange chromatography to separate sheep liver cytoplasmic aldehyde dehydrogenase from mitochondrial enzyme contamination, and observations on the interaction between the pure cytoplasmic enzyme and disulfiram. Biochem J. 1981 Dec 1;199(3):573–579. doi: 10.1042/bj1990573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. J., Koleske A. J., Lindahl R., Pitot H. C. Phenobarbital-inducible aldehyde dehydrogenase in the rat. cDNA sequence and regulation of the mRNA by phenobarbital in responsive rats. J Biol Chem. 1989 Aug 5;264(22):13057–13065. [PubMed] [Google Scholar]
- Eckfeldt J. H., Yonetani T. Kinetics and mechanism of the F1 isozyme of horse liver aldehyde dehydrogenase. Arch Biochem Biophys. 1976 Mar;173(1):273–281. doi: 10.1016/0003-9861(76)90260-5. [DOI] [PubMed] [Google Scholar]
- Farrés J., Guan K. L., Weiner H. Primary structures of rat and bovine liver mitochondrial aldehyde dehydrogenases deduced from cDNA sequences. Eur J Biochem. 1989 Mar 1;180(1):67–74. doi: 10.1111/j.1432-1033.1989.tb14616.x. [DOI] [PubMed] [Google Scholar]
- Greenfield N. J., Pietruszko R. Two aldehyde dehydrogenases from human liver. Isolation via affinity chromatography and characterization of the isozymes. Biochim Biophys Acta. 1977 Jul 8;483(1):35–45. doi: 10.1016/0005-2744(77)90005-5. [DOI] [PubMed] [Google Scholar]
- Hart G. J., Dickinson F. M. The coenzyme-binding characteristics of highly purified preparations of sheep liver cytoplasmic aldehyde dehydrogenase. Biochem J. 1983 May 1;211(2):363–371. doi: 10.1042/bj2110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel J. D., Pietruszko R. Selective chemical modification of human liver aldehyde dehydrogenases E1 and E2 by iodoacetamide. J Biol Chem. 1981 Nov 10;256(21):10889–10896. [PubMed] [Google Scholar]
- Hempel J. D., Reed D. M., Pietruszko R. Human aldehyde dehydrogenase: improved purification procedure and comparison of homogeneous isoenzymes E1 and E2. Alcohol Clin Exp Res. 1982 Summer;6(3):417–425. doi: 10.1111/j.1530-0277.1982.tb05001.x. [DOI] [PubMed] [Google Scholar]
- Hempel J., Kaiser R., Jörnvall H. Mitochondrial aldehyde dehydrogenase from human liver. Primary structure, differences in relation to the cytosolic enzyme, and functional correlations. Eur J Biochem. 1985 Nov 15;153(1):13–28. doi: 10.1111/j.1432-1033.1985.tb09260.x. [DOI] [PubMed] [Google Scholar]
- Hempel J., Pietruszko R., Fietzek P., Jörnvall H. Identification of a segment containing a reactive cysteine residue in human liver cytoplasmic aldehyde dehydrogenase (isoenzyme E1). Biochemistry. 1982 Dec 21;21(26):6834–6838. doi: 10.1021/bi00269a032. [DOI] [PubMed] [Google Scholar]
- Hempel J., von Bahr-Lindström H., Jörnvall H. Aldehyde dehydrogenase from human liver. Primary structure of the cytoplasmic isoenzyme. Eur J Biochem. 1984 May 15;141(1):21–35. doi: 10.1111/j.1432-1033.1984.tb08150.x. [DOI] [PubMed] [Google Scholar]
- Johansson J., von Bahr-Lindström H., Jeck R., Woenckhaus C., Jörnvall H. Mitochondrial aldehyde dehydrogenase from horse liver. Correlations of the same species variants for both the cytosolic and the mitochondrial forms of an enzyme. Eur J Biochem. 1988 Mar 15;172(3):527–533. doi: 10.1111/j.1432-1033.1988.tb13920.x. [DOI] [PubMed] [Google Scholar]
- Jones D. E., Jr, Brennan M. D., Hempel J., Lindahl R. Cloning and complete nucleotide sequence of a full-length cDNA encoding a catalytically functional tumor-associated aldehyde dehydrogenase. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1782–1786. doi: 10.1073/pnas.85.6.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Meulenberg C. H., Kingma J., Witholt B. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J Biol Chem. 1989 Apr 5;264(10):5442–5451. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MacKerell A. D., Jr, MacWright R. S., Pietruszko R. Bromoacetophenone as an affinity reagent for human liver aldehyde dehydrogenase. Biochemistry. 1986 Sep 9;25(18):5182–5189. doi: 10.1021/bi00366a030. [DOI] [PubMed] [Google Scholar]
- MacQuarrie R. A., Bernhard S. A. Subunit conformation and catalytic function in rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1971 Jan 28;55(2):181–192. doi: 10.1016/0022-2836(71)90190-2. [DOI] [PubMed] [Google Scholar]
- Meltzer N. M., Tous G. I., Gruber S., Stein S. Gas-phase hydrolysis of proteins and peptides. Anal Biochem. 1987 Feb 1;160(2):356–361. doi: 10.1016/0003-2697(87)90060-1. [DOI] [PubMed] [Google Scholar]
- Pickett M., Gwynne D. I., Buxton F. P., Elliott R., Davies R. W., Lockington R. A., Scazzocchio C., Sealy-Lewis H. M. Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene. 1987;51(2-3):217–226. doi: 10.1016/0378-1119(87)90310-6. [DOI] [PubMed] [Google Scholar]
- Pietruszko R., MacKerell A. D., Jr Stoichiometry of chemical modification of human aldehyde dehydrogenase: evidence for "quarter of the sites" reactivity. Prog Clin Biol Res. 1987;232:37–52. [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Roth M. Automated amino acid analysis with sensitive fluorescence detection. J Clin Chem Clin Biochem. 1976 Jul;14(7):361–364. doi: 10.1515/cclm.1976.14.1-12.361. [DOI] [PubMed] [Google Scholar]
- Roth M. Fluorescence reaction for amino acids. Anal Chem. 1971 Jun;43(7):880–882. doi: 10.1021/ac60302a020. [DOI] [PubMed] [Google Scholar]


