Abstract
Hepatitis delta virus (HDV) replication requires both the cellular RNA polymerase and one virus-encoded protein, small delta antigen (S-HDAg). S-HDAg has been shown to be a phosphoprotein, but its phosphorylation status is not yet clear. In this study, we employed three methods to address this question. A special two-dimensional gel electrophoresis, namely, nonequilibrium pH gradient electrophoresis, was used to separate the very basic S-HDAg. By carefully adjusting the pH of solubilization solution, the ampholyte composition, and the appropriate electrophoresis time periods, we were able to clearly resolve S-HDAg into two phosphorylated isoforms and one unphosphorylated form. In contrast, the viral large delta antigen (L-HDAg) can only be separated into one phosphorylated and one unphosphorylated form. By metabolic 32P labeling, both immunoprecipitated S-HDAg and L-HDAg were found to incorporate radioactive phosphate. The extent of S-HDAg phosphorylation was increased upon 12-O-tetradecanoylphorbol-13-acetate treatment, while that of L-HDAg was not affected. Finally, phosphoamino acid analysis identified serine and threonine as the phospho residues in the labeled S-HDAg and only serine in the L-HDAg. Therefore, HDV S- and L-HDAgs differ in their phosphorylation patterns, which may account for their distinct biological functions.
Hepatitis delta virus (HDV) is the smallest member of negative-strand RNA viruses and possesses a circular genome of 1.7 kb (19, 21, 29). Replication of HDV is entirely through RNA intermediates and proposed to proceed by a double rolling-circle mechanism (19, 29). Hepatitis delta antigen (HDAg) is the only known protein encoded by HDV and exists as two major forms: small HDAg (S-HDAg; 195 amino acids) and large HDAg (L-HDAg; 214 amino acids) (29, 30, 31). Both forms of HDAg are translated from the same initiation codon of one open reading frame (ORF), but L-HDAg contains an additional 19 amino acids at the C terminus of S-HDAg (31) because of RNA editing of the original termination codon during replication. Hence, they have different biological functions: S-HDAg is essential for viral replication (18); however, L-HDAg exerts a dominant-negative role in HDV replication and is involved in viral assembly (5, 10).
L-HDAg was first shown as a nuclear phosphoprotein when expressed in mammalian cells, and a previous result indicated that L-HDAg is phosphorylated only at the serine residue(s) (6). In a subsequent study, both L- and S-HDAg were found to be phosphorylated in insect cells (16). S-HDAg was further demonstrated to be a phosphoprotein in mammalian cells (35), and the phosphorylation level of S-HDAg is reported to be downregulated by casein kinase (CK II)- and protein kinase C (PKC)-specific inhibitors, whereas that of L-HDAg is downregulated only by CK II inhibitor (35). A recent study (2) tried to distinguish the phosphorylated from the unphosphorylated delta antigens by using an NEPHGE (nonequilibrium pH gradient electrophoresis)-sodium dodecyl sulfate (SDS) system instead of the conventional isoelectric focusing (IEF)-SDS system. The results of that study revealed ca. 20 to 40% of L-HDAg to be phosphorylated. However, in contrast to previous studies, hardly any phosphorylated S-HDAg was detected (2).
The different results concerning the phosphorylation status of S-HDAg may be reconciled by at least two factors. First, because of its nonequilibrium nature, separation of the isoforms of basic protein by NEPHGE has been empirical. Careful optimization of running conditions is required to achieve the best resolution. Second, protein phosphorylation is subjected to modulation by many signals; thus, different cells and culture conditions may influence the extent of S-HDAg phosphorylation. In this study, we tried to address the question by optimizing the conditions for NEPHGE by carefully adjusting many parameters in order to separate the phosphorylated forms of both HDAgs from the unphosphorylated forms.
Separation of HDAg isoforms of different pI by fine-tuning the NEPHGE.
Because the IEF allows a clear separation of proteins and their isoforms of different pIs, it is usually used in separating the unphosphorylated from the phosphorylated forms of a protein (32, 34). However, it has been noted that the standard IEF would not give satisfactory resolution in separating very basic or acidic proteins (23, 33). According to the predicted pI values of S-HDAg and L-HDAg (10.2 and 9.9, respectively), both HDAgs seemed to be very basic, and it was difficult to reach pH equilibrium in conventional IEF. Therefore, we adapted an alternative approach, namely, NEPHGE, which was developed to resolve very basic protein by using a nonequilibrium condition rather than true focusing (1, 23, 33). To investigate S- and L-HDAgs in the NEPHGE system, two plasmids, pCDAg-S and pCDAg-L, which express S- and L-HDAg, respectively, were cotransfected into HuH-7 cells. pCDAg-S contained the S-HDAg ORF of HDV (nucleotides 46 to 781) under the control of human cytomegalovirus immediate-early promoter. To construct pCDAg-L, the site-directed mutagenesis (Transformer Site-Directed Mutagenesis Kit; Clontech Laboratories, Inc.) was employed to change the stop codon (UAG) in the S-HDAg ORF of pCDAg-S to UAA (encoding Trp). On day 3 posttransfection, cells were washed twice with cold TBS (150 mM NaCl, 20 mM Tris; pH 7.5) and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl; 50 mM Tris, pH 8.0; 5 mM EDTA; 0.2% NP-40; 1% Triton X-100; 0.1% SDS). The cell lysates were centrifuged at 12,000 rpm for 20 min to remove debris. For preclearing, the supernatant was incubated with a 1/10 volume of normal mouse serum (50 mg/ml; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at 4°C and then with 25 μl of protein G-agarose beads (Boehringer Mannheim) for another 1 h at 4°C. After centrifugation at 12,000 rpm for 1 min, precleared supernatant was reacted for 1 h at 4°C with a mouse monoclonal anti-HDAg antibody (5 μg/ml), D9-3, followed by another 1 h at 4°C with 25 μl of protein G-agarose. D9-3 was raised in BALB/c mice by inoculation of S-HDAg expressed in Escherichia coli. After centrifugation at 12,000 rpm for 1 min, the pellet was washed with 1 ml of RIPA buffer once, 1 ml of high-salt buffer (25 mM HEPES, pH 7.5; 1% Triton X-100; 1% deoxycholate; 0.1% SDS; 500 mM NaCl; 5 mM EDTA) twice, and then low-salt buffer (25 mM HEPES, pH 7.5; 0.2% Triton X-100; 1 mM EDTA) once.
The immune complex was resuspended in 50 μl of urea–NP-40 solubilizer (9 M urea, 4% NP-40; 1.6% Pharmalyte [pH 3.5 to 10; Amersham Pharmacia Biotech AB], 0.4% Servalyte [pH 9 to 11; Serva FeinBiochemica GmbH and Co.], and 1% dithiothreitol [adjusted to pH 3.0]) and held at room temperature for 10 min. After dissolution, the supernatant was loaded onto the well of one cylindrical gel (12 cm [length] by 4 mm [diameter]) and covered with 100 μl of 4 M urea overlay solution. The gel was made with 3.3% acrylamide (30% acrylamide and 1.8% bisacrylamide) containing 9 M urea, 2% NP-40, 1.6% Pharmalyte (pH 3.5 to 10) and 0.4% Servalyte (pH 9 to 11). Finally, the gels were installed vertically in electrophoresis apparatus (Desaga GmbH, D-6900 Heidelberg, Germany) with 20 mM NaOH in the lower chamber and 10 mM H3PO4 in the upper chamber. Lysozyme and trypsinogen (Sigma, St. Louis, Mo.) were used as the reference for pI markers and run in parallel cylindrical gels.
In NEPHGE, proteins migrate in nonequilibrium pH gradient; therefore, it is important to find the critical window (defined by the running volt-hour) at which the proteins are best focused (23, 33). Proteins did not enter gels completely if the electrophoresis time period was insufficient. On the other hand, prolonged period of electrophoresis made proteins in acidic ends migrate backwards off the gel and caused the collapse of the pH gradient in the basic ends. Therefore, outside the critical window, proteins will clump together, spread into a broader pattern, or intermingle with others, and this results in poor separation (23). In order to fine-tune the NEPHGE, gels were electrophoresed at four different volt-hour levels: 600, 800, 1,300, and 1,600 V · h, respectively. Gels were electrophoresed at 400 V for 1 h and then run at 800 V at different periods of time with the current reversed. After electrophoresis, the cylindrical gels were extruded from the glass tubes by syringes, and each one was soaked in 10 ml of equilibration buffer (10% glycerol, 4.9 mM dithiothreitol, 2% SDS, 0.125 M Tris; adjusted to pH 6.8) for 10 min. Then, the tube gel was laid on top of the second-dimension gel. The second dimension was the standard discontinuous SDS-polyacrylamide gel electrophoresis (PAGE) employing a 20-by-20-by-0.15-cm gel (12% separation gel and 5% stacking gel) with a beveled plate to assemble the glass-plate sandwich (BRL Vertical Gel Electrophoresis System, model V16-2). This plate provided a wider space to accommodate the thick cylindrical gel from the NEPHGE. After equilibration, the extruded tube gel was laid on top of the stacking gel and sealed with 0.5% agarose. Electrophoresis was carried out overnight (70 V, 16 to 18 h), and gels were electrotransferred onto nitrocellulose membrane (Hybond-c Super; Amersham Pharmacia Biotech). Both HDAgs were detected by Western blot probing with human polyclonal anti-HDAg antibody.
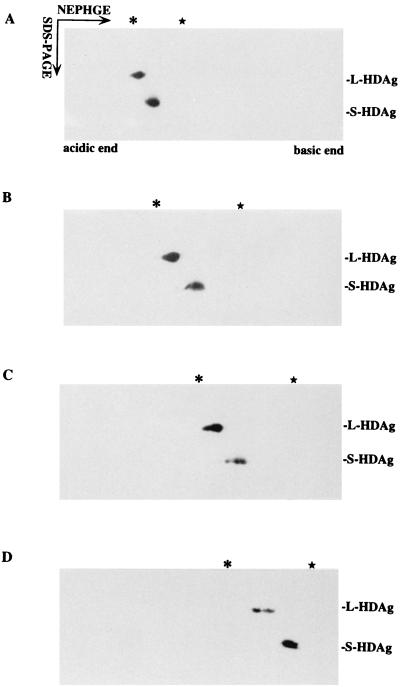
Based upon the migration of the molecular weight marker and recognition of HDAgs by specific antibody in a subsequent Western blot, we identified both HDAgs in the NEPHGE-SDS gel (Fig. 1). In the shorter electrophoresis periods (600 and 800 V · h), both the S- and L-HDAgs migrated as a large, clumped spot without separation, probably due to insufficient time for electrophoresis. Because of a higher pI, the S-HDAg is noted moving farther right than the L-HDAg (Fig. 1B). Notably, in the appropriate volt-hour (in the case of 1,300 V · h), the L-HDAg separates into two spots (Fig. 1C), and the S-HDAg resolves into three spots. However, with a longer electrophoresis period, the L-HDAg is still separated into two spots (Fig. 1D), but the S-HDAg now becomes broader again. This clumped signal may result from the S-HDAg isoforms residing in the collapsed pH gradient at the basic end of gel after prolonged electrophoresis. The results suggested that the electrophoresis period needs to be experimentally titrated, so different pI isoforms of one basic protein can be resolved.
FIG. 1.
Effect of different electrophoresis time periods (NEPHGE) in separating isoforms of S- and L-HDAgs in a two-dimensional system. L-HDAg- and S-HDAg-expressing constructs (pCDAg-L and pCDAg-S) were cotransfected into HuH-7 cells. On day 3 posttransfection, cell lysates were immunoprecipitated with the anti-HDAg monoclonal antibody D9-3. The immune complex was resuspended in 50 μl of solubilizer and electrophoresed in the NEPHGE gel for different periods of time: 600 (A), 800 (B), 1,300 (C), and 1,600 (D) V · h. Electrophoresis in the first dimension migrated from the acidic to the basic end of the NEPHGE gel and in the second dimension separated proteins by their different molecular weights. The isoforms of both HDAgs were detected by Western blot with polyclonal anti-HDAg antibody. Lysozyme and trypsinogen (Sigma) were used as the references for pI markers and were run in parallel NEPHGE experiments. The positions of these markers were identified after Coomassie blue staining. The asterisk above each panel indicates the position of trypsinogen with a known pI value of 9.3, and the star indicates the position of lysozyme with a pI value of 10.5 to 11 as pH markers.
Effect of protein dephosphorylation on pI isoforms of both HDAgs.
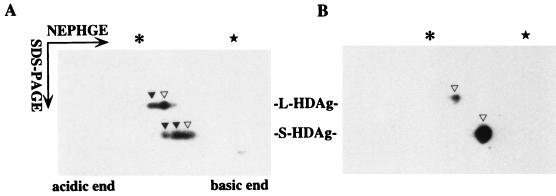
If the observed pI isoforms of both HDAgs were generated by differential phosphorylation, then the stepwise decrease in pI value should be caused by the incorporation of phosphates. The incorporation with one phosphate into the protein is noted to decrease its pI, which varies from 0.04 to 0.46 pI units (11, 12, 32, 34). Phosphorylation on more residues will further reduce its pI and generate more isoforms. However, treatment with phosphatase will remove the phosphates, and then the dephosphorylated protein should become a single species with the same pI. To test this, immunoprecipitated HDAgs were treated with alkaline phosphatase before the NEPHGE–SDS-PAGE and detected by Western blot as described above. The immunoprecipitated HDAgs were resuspended in 90 μl 1× dephosphorylation buffer (50 mM Tris, 0.1 mM EDTA; pH 8.8) for 10 min at 30°C. Then, 10 μl of calf intestinal alkaline phosphatase (Boehringer Mannheim) was added into reaction buffer (100 U/ml) for 1 h at 37°C, followed by another 1 h at 42°C. The sample was washed with 1 ml of high-salt buffer once and 1 ml of low-salt buffer once and then dissolved in the solubilizer for NEPHGE–SDS-PAGE. In the untreated samples, the L-HDAgs are separated into two spots and the S-HDAgs are separated into three spots (Fig. 2A). After alkaline phosphatase treatment, both the L- and S- HDAgs were reduced to only one spot (Fig. 2B). This spot migrated to the same distance as the isoform with the highest pI in untreated HDAgs (Fig. 2A, indicated by open triangles). It supported that the isoform with the highest pI value is the unphosphorylated HDAg. Those isoforms with lower pI values were phosphorylated S- or L-HDAgs, which could be dephosphorylated by alkaline phosphatase treatment (Fig. 2A, indicated by solid triangles).
FIG. 2.
Effect of alkaline phosphatase treatment on the mobility of delta antigen isoforms. The immunoprecipitated HDAgs were prepared as described in Fig. 1. Prior to resuspension in solubilizer, the immune complex was treated with (B) or without (A) alkaline phosphatase and then subjected to NEPHGE for 1,300 V · h, followed by SDS-PAGE. The open and solid triangles represent unphosphorylated and phosphorylated isoforms of both HDAgs, respectively, as identified by Western blot.
These data demonstrated that the L-HDAg contains two isoforms, one unphosphorylated and the other perhaps monophosphorylated. For S-HDAgs, other than the unphosphorylated one, there exist two obvious phosphorylated isoforms (probably mono- and diphosphorylated). However, there could be small amounts of S-HDAg with higher phosphorylation, since minor spots with even lower pIs sometimes were seen (data not shown). Although there was another posttranslational modification for L-HDAg farnesylation (13, 14, 24), farnesylated L-HDAg could not be separated from unfarnesylated isoforms, presuming that farnesylation does not change the pI value (2). By establishing this two-dimensional system to separate phosphorylated isoforms of HDAgs, we concluded that there are two major phosphorylation forms for S-HDAg but only one for L-HDAg.
Metabolic labeling of both HDAgs.
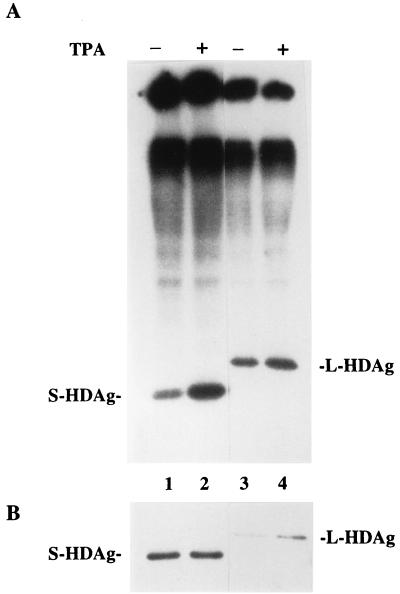
The results described above suggested that the S-HDAg is phosphorylated differently from that of L-HDAg. To further investigate this, the in vivo-labeling experiment was performed. On day 3 posttransfection, cells were washed twice with TBS and starved in 0.8 ml of phosphate-free DMEM (Gibco BRL/Life Technologies) for 2 h. Then, 0.5 mCi of [32P]orthophosphate (PBS-13; Amersham Pharmacia Biotech) was added into medium for 4 h to label the cellular phosphoprotein. Cells were then harvested and lysed for immunoprecipitation as described above. The L-HDAg was shown to be a phosphoprotein in previous reports (2, 6, 35, 36). It was also labeled in our experiments (Fig. 3A, lane 3). For S-HDAg, there was a major phospholabeled protein of the expected molecular weight being immunoprecipitated (Fig. 3A, lane 1). This labeled protein presumably represented the S-HDAg, since it was not detected in the cells transfected with L-HDAg expressing plasmid (Fig. 3A, lane 3). The metabolically labeled L-HDAg and S-HDAg detected in Fig. 3A were confirmed by a Western blot of the same sample (Fig. 3B, lanes 1 and 3).
FIG. 3.
In vivo phosphate labeling of both HDAgs and the effect of TPA stimulation on HDAg phosphorylation. S-HDAg (lanes 1 and 2) and L-HDAg (lanes 3 and 4) were transiently expressed in HuH-7 cells and metabolically labeled with [32P]orthophosphate on day 3 posttransfection. After immunoprecipitation with monoclonal anti-HDAg antibody, seven-eighths of the immunoprecipitates was analyzed by SDS-PAGE and autoradiography (A), and the remainder was detected by Western blot analysis with human polyclonal anti-HDAg antibody (B). Where indicated (lanes 2 and 4), 100 ng of TPA per ml was added to the labeled cells 15 min before harvest (+). The positions of S-HDAg and L-HDAg were marked.
It has been demonstrated that PKC inhibitor, H7, reduces the phosphorylation level of S-HDAg but not that of L-HDAg (35). As the phorbol ester TPA (12-O-tetradecanoylphorbol-13-acetate) is known to activate PKC, mimicking the physiological activator diacylglycerol, to mediate TPA-induced biological effects (22). Therefore, metabolically labeled S-HDAg or L-HDAg in transfected cells were exposed to 100 ng of TPA (Sigma) per ml for 15 min before harvest to observe any effect. As shown in Fig. 3A, the phosphorylation level of S-HDAg increased by ca. threefold after TPA treatment (Fig. 3A, comparing lanes 1 and 2). The parallel immunoprecipitates analyzed by Western blot showed that the amount of precipitated S-HDAg was equivalent with or without TPA treatment (Fig. 3B, lanes 1 and 2). Therefore, TPA stimulates the phosphorylation level instead of the expression level of S-HDAg. However, it was noted that the phosphorylation of L-HDAg was not affected by TPA treatment (Fig. 3A, lanes 3 and 4). The stimulation of S-HDAg phosphorylation may be through a TPA signaling pathway, and the phosphorylation pathway of S-HDAg at least partly differs from that of L-HDAg.
S-HDAg is phosphorylated at both serine and threonine residues.
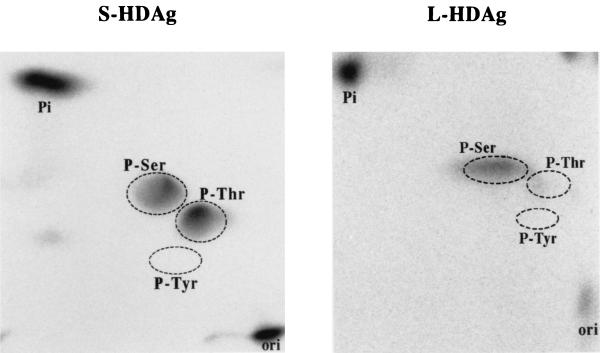
We have shown that phosphorylation of S-HDAg may be different from that of the L-HDAgs, in terms of the number of phosphorylation residues and possible kinases. We then tried to identify the phosphorylated residues of the S-HDAg by performing two-dimensional phosphoamino acid analysis (PAA). The 32P-labeled S-HDAg in Fig. 3A, which was separated by SDS–12% PAGE, was electrotransferred onto a polyvinylidene difluoride (PVDF) membrane and identified by autoradiography. The labeled S-HDAg was excised from the membrane and acid hydrolyzed in 300 μl of 6 N HCl at 110°C for 90 min. After a lyophilizing and resuspending step in 10 μl of pH 1.9 buffer (0.58 M formic acid and 1.36 M glacial acetic acid), the sample was applied to a thin-layer chromatography (TLC) plate (20 by 20 cm; Merck). Electrophoresis was carried out in pH 1.9 buffer at 1,200 V for 20 min for the first dimension and the run for second dimension in pH 3.5 buffer (0.87 M glacial acetic acid, 0.5% pyridine, 0.5 mM EDTA) at 1,600 V for 25 min (Hunter Thin-Layer Peptide Mapping System, model HTLE-7000; C.B.S. Scientific Co.). After electrophoresis, the plate was dried in a 65°C oven for 20 to 30 min, and phosphoamino acid standards (Sigma) were visualized by spraying them with 0.25% ninhydrin solution in acetone. The labeled signals were subjected to autoradiography by a radioanalytic imaging system (Fujix BAS 1000). As shown in Fig. 4, the hydrolyzed, 32P-labeled amino acids from S-HDAg were found comigrating with both authentic phosphoserine and phosphothreonine standards, but not with phosphotyrosine, indicating that S-HDAg is phosphorylated at both serine and threonine residues. We also carried out PAA to determine the phosphorylated residue(s) of L-HDAg as a control, and the data obtained were consistent with the former report (6) that L-HDAg is phosphorylated at only the serine residue (Fig. 4). Since the phosphorylated residues of S-HDAg were identified as both serine and threonine, there were at least two phosphorylation sites in S-HDAg, thus corresponding with previous data that showed in an NEPHGE-SDS system that S-HDAg contained two major phospho isoforms.
FIG. 4.
PAA of both HDAgs. The labeled immune complex as described in Fig. 3 was separated by SDS-PAGE and then electrotransferred onto PVDF membrane. The membrane slice containing labeled delta antigens was cut out and acid hydrolyzed in 6 N HCl. The phosphorylated amino acids were then separated by two-dimensional electrophoresis on TLC plates. The regions circled with dashed lines indicated the authentic phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) forms identified by use of ninhydrin. Phospho residues of S-HDAg were shown by autoradiography in the left panel, and that of L-HDAg is shown in the right panel.
For HDV delta antigens, it is generally accepted that the L-HDAg is a phosphoprotein. However, the phosphorylation of S-HDAg has not yet been settled. Though S-HDAg could be labeled by [32P]orthophosphate in both mammalian and insect cells (15, 35), the data from a previous two-dimensional gel analysis of S-HDAg showed very little phosphorylation (2). To reconcile the difference in the S-HDAgs, we tried to determine whether the resolution of NEPHGE could be improved by careful adjustments. By optimizing several parameters for NEPHGE, especially the running time for electrophoresis, we finally succeeded in separating the phosphorylated forms of S-HDAg expressed in transfected cells. It has been noted that the separation by NEPHGE was achieved long before approaching pH gradient equilibrium and that more basic proteins need shorter periods of electrophoresis for separation (23, 33). So the optimum electrophoresis time is very important. As shown in former reports, NEPHGE was able to resolve the phospho isoform of L-HDAg at 1,600 V · h (2). We obtained the same results (Fig. 1D). However, this electrophoresis time period was not suitable for separating the phospho isoforms for the S-HDAg (Fig. 1D). Probably the S-HDAg is a more basic protein than L-HDAg and should be run in a shorter time period before the collapse of the basic end in NEPHGE. Actually, our results showed that 1,300 V · h is optimal for resolving different phospho isoforms of S-HDAg (Fig. 1C). However, with time periods shorter than 1,300 V · h, the resolution failed again (Fig. 1A and B). Therefore, the electrophoresis time period is critical for the successful separation of S-HDAg. Our results explained the poor separation of phospho isoforms of S-HDAg by NEPHGE obtained in the previous study, which employed an inappropriate condition (2).
The phosphorylation of S-HDAg apparently differed from that of L-HDAg. There were two major phospho isoforms and one unphosphorylated form for S-HDAg. (We noted that, after a longer exposure, there were minor phospho isoforms with lower pIs.) Phosphoamino acid analysis also showed that S-HDAg is phosphorylated at both serine and threonine. This further confirmed that more than one phosphorylation occurred on S-HDAg. L-HDAg has one phosphorylated form and is phosphorylated only at serine residues. In metabolically labeling experiments, the phosphorylation level of S-HDAg was enhanced three times after TPA stimulation, but that of L-HDAg was not affected. These findings indicated that the phosphorylation pathways for both HDAgs are quite different. Although these two HDAgs shared the same ORF, except that L-HDAg has an extra 19 amino acids at the C terminus; however, they have different protein conformations (13, 14). Their phosphorylation properties may account for such a difference.
Our results only indicated the S-HDAg to be a phosphoprotein but did not yet identify the exact phosphorylation sites. There are 10 serine residues and 5 threonine in S-HDAg coding sequences of the specific HDV strain used in this study (Fig. 5, strain I). Among these 10 serine residues, three (Ser2, Ser4, and Ser123) have been studied for their biochemical and biological properties related to phosphorylation by site-directed mutagenesis (35). Both CK II and PKC have been demonstrated to modulate the function and phosphorylation of S-HDAg, and Ser2 was proposed as one of the candidates for S-HDAg phosphorylation affected by CK II. As for Ser4 and Ser123, both were not affected by CK II inhibitors, even though they also resided in the predicted consensus sequences for CK II. A comparison of deduced primary sequences of HDAgs among HDV variants revealed that, in addition to Ser2 and Ser123, there is another completely conserved serine residue, Ser177 (Fig. 5, including all three genotypes), which resides in a consensus sequence conforming the motif of mitogen-activated protein (MAP) kinase substrate (PESP) (25, 26). Since PKC has been demonstrated to be involved in the phosphorylation of S-HDAg, the activation of PKC could lead to the phosphorylation of MAP kinase (3, 7). Therefore, the effect of PKC on S-HDAg may mediate the phosphorylation of Ser177 through PKC-MAP kinase pathway. Though Ser2 and Ser177 may be the phosphorylated residues for S-HDAg, this speculation awaits further investigation.
FIG. 5.
Conserved serine and threonine residues among HDV strains. The boldface characters in the left lane represented geographically distinct HDV isolates: A, American (21); I, Italian (30); N, Nauru (8); F, French (27); L, Lebanon (20); T, Taiwan (9); C, Central African (28); J, Japan-2 (16); E, Ethiopia (37); S, Somalia (37); J′, Japan-1 (17); P, Peru (4). The numbers indicate completely or highly conserved Ser and Thr residues.
Besides serine, we discovered that threonine is also a phosphorylation acceptor in S-HDAg. Amino acid 95 is the only completely conserved Ser or Thr residue among all the HDV variants (Fig. 5; amino acid 95 is threonine in the strain we used). Thr134 and Thr182 are highly but not completely conserved (Fig. 5, amino acid 134 as Ala in the S and P isolates and amino acid 182 as Arg in the S isolate and as His in the E and L isolates, respectively). The sequences encompassing Ser95/Thr95 and Thr134 were similar, with predicted consensus sequence for CK II or PKC (25). Phosphorylations in S-HDAg might be more likely to occur on these highly conserved serine and threonine residues (Ser2 and Ser177 and Thr95, -134, and -182). However, the functional involvement of the less-conserved ones (Ser6, -83, -116, -159, -170, and -180 and Thr37 and -76) could not be completely excluded.
The biological function of S-HDAg is to assist the replication of HDV RNA, and the phosphorylation of S-HDAg may modulate this replication (35). In our preliminary data, phosphorylated S-HDAg could only be detected inside the cells but not in viral particles (unpublished data). It appears that the phosphorylation of S-HDAg is relevant for HDV RNA replication.
Phosphoamino acid analysis indicated that L-HDAg is phosphorylated only at the serine residue. Completely conserved serines (Ser2, Ser123, and Ser177) and highly conserved serines (Ser4, Ser207, and Ser210) have been studied by using site-directed mutagenesis, and none of their mutants seemed to affect the biological and biochemical significance for L-HDAg (2, 35, 36). Besides, CK II inhibitor decreased the phosphorylation level of L-HDAg, but it does not affect both particle assembly and the trans-dominant function of L-HDAg (36). However, mutation at the farnesylation site (Cys211) has been reported to eliminate the phosphorylation on L-HDAg, though its biological significance has not yet been investigated (2). Therefore, the function of L-HDAg phosphorylation is still not clear.
Despite the speculation on the possible phosphorylation sites in S- or L-HDAg, a more solid basis is required. It is necessary to identify the exact amino acid residues to be phosphorylated by biochemical procedures, such as two-dimensional peptide mapping or liquid chromatography-mass spectrometry. With this information, we can address the biological significance of each phosphorylation of delta antigens in the viral life cycle. Finally, the results also help in searching for the kinases responsible for phosphorylating the delta antigens.
Acknowledgments
This work was supported by grants (NSC88-2314-B002-028) from National Science Council, Executive Yuan, Taiwan.
REFERENCES
- 1.Anderson L. Two-dimensional electrophoresis: operation of the ISO-DALT system. 2nd ed. Rockville, Md: Large Scale Biology Press; 1991. [Google Scholar]
- 2.Bichko V, Barik S, Taylor J. Phosphorylation of the hepatitis delta virus antigens. J Virol. 1997;71:512–518. doi: 10.1128/jvi.71.1.512-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabedo H, Felipo V, Minana M D, Grisolia S. H7, an inhibitor of protein kinase C, prevents serum-induced phosphorylation of Raf and MAP kinase in neuroblastoma cells. Neurosci Lett. 1996;214:13–16. doi: 10.1016/0304-3940(96)12867-6. [DOI] [PubMed] [Google Scholar]
- 4.Casey J L, Brown T L, Colan E J, Wignall F S, Gerin J L. A genotype of hepatitis D virus that occurs in northern South America. Proc Natl Acad Sci USA. 1993;90:9016–9020. doi: 10.1073/pnas.90.19.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang F-L, Chen P-J, Tu S-J, Wang C-J, Chen D-S. The large isoform of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang M-F, Baker S C, Soe L H, Kamahora T, Keck J G, Makino S, Govindarajan S, Lai M M. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J Virol. 1988;62:2403–2410. doi: 10.1128/jvi.62.7.2403-2410.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y Y, Kim S J, Park T K, Kang S S, Ha M J, Mushinski J F, Chun J S. Modulation of MAP kinase signaling and growth characteristics by the overexpression of protein kinase C in NIH 3T3 cells. Biochem Mol Biol Int. 1998;45:1139–1148. doi: 10.1080/15216549800203362. [DOI] [PubMed] [Google Scholar]
- 8.Chao Y-C, Chang M-F, Gust I, Lai M M. Sequence conservation and divergence of hepatitis delta virus RNA. Virology. 1990;178:384–392. doi: 10.1016/0042-6822(90)90335-o. [DOI] [PubMed] [Google Scholar]
- 9.Chao Y-C, Lee C-M, Tang H-S, Govindarajan S, Lai M M. Molecular cloning and characterization of an isolate of hepatitis delta virus from Taiwan. Hepatology. 1991;13:345–352. [PubMed] [Google Scholar]
- 10.Chen P-J, Chang F-L, Wang C-J, Lin C-J, Sung S-Y, Chen D-S. Functional study of hepatitis delta virus large antigen in packaging and replication inhibition: role of the amino-terminal leucine zipper. J Virol. 1992;66:2853–2859. doi: 10.1128/jvi.66.5.2853-2859.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan R, Milburn S C, Hershey J W B. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 12.Grgacic E V L, Lin B, Gazina E V, Snooks M J L, Anderson D A. Normal phosphorylation of duck hepatitis B virus L protein is dispensable for infectivity. J Gen Virol. 1998;79:2743–2751. doi: 10.1099/0022-1317-79-11-2743. [DOI] [PubMed] [Google Scholar]
- 13.Hwang S B, Lai M M. A unique conformation at the carboxyl terminus of the small hepatitis delta antigen revealed by a specific monoclonal antibody. Virology. 1993;193:924–931. doi: 10.1006/viro.1993.1201. [DOI] [PubMed] [Google Scholar]
- 14.Hwang S B, Lai M M. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J Virol. 1994;68:2958–2964. doi: 10.1128/jvi.68.5.2958-2964.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang S B, Lee C-Z, Lai M M. Hepatitis delta antigen expressed by recombinant baculoviruses: comparison of biochemical properties and post-translational modifications between the large and small isoforms. Virology. 1992;190:413–422. doi: 10.1016/0042-6822(92)91227-l. [DOI] [PubMed] [Google Scholar]
- 16.Imazeki F, Omata M, Ohto M. Heterogeneity and evolution rates of delta virus RNA sequences. J Virol. 1990;64:5594–5599. doi: 10.1128/jvi.64.11.5594-5599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imazeki F, Omata M, Ohto M. Complete nucleotide sequence of hepatitis delta virus RNA in Japan. Nucleic Acids Res. 1991;19:5439. doi: 10.1093/nar/19.19.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 20.Lee C-M, Bih F-Y, Chao Y-C, Govindarajan S, Lai M M. Evolution of hepatitis delta virus RNA during chronic infection. Virology. 1992;188:165–173. doi: 10.1016/0042-6822(92)90756-f. [DOI] [PubMed] [Google Scholar]
- 21.Makino S, Chang M F, Shieh C K, Kamahora T, Vannier D M, Govindarajan S, Lai M M. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature (London) 1987;329:343–346. doi: 10.1038/329343a0. [DOI] [PubMed] [Google Scholar]
- 22.Newton A C. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 23.O’Farrell P Z, Goodman H M, O’Farrell P H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 24.Otto J, Casey P. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J Biol Chem. 1996;271:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 25.Pinna L A, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 26.Rothmann K, Schnolzer M, Radziwill G, Hildt E, Moelling K, Heinz S. Host cell-virus cross talk: phosphorylation of a hepatitis B virus envelope protein mediates intracellular signaling. J Virol. 1998;72:10138–10147. doi: 10.1128/jvi.72.12.10138-10147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saldanha J A, Thomas H C, Monjardino J P. Cloning and sequencing of RNA of hepatitis delta virus isolated from human serum. J Gen Virol. 1990;71:1603–1606. doi: 10.1099/0022-1317-71-7-1603. [DOI] [PubMed] [Google Scholar]
- 28.Tang J R, Hantz O, Vitvitski L, Lamelin J P, Parana R, Cova L, Lesbordes J L, Trepo C. Discovery of a novel point mutation changing the HDAg expression of a hepatitis delta virus isolate from Central African Republic. J Gen Virol. 1993;74:1827–1835. doi: 10.1099/0022-1317-74-9-1827. [DOI] [PubMed] [Google Scholar]
- 29.Wang K S, Choo Q L, Weiner A J, Ou J H, Najarian R C, Thayer R M, Mullenbach G T, Denniston K J, Gerin J L, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature (London) 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 30.Wang K S, Choo Q L, Weiner A J, Ou J H, Najarian R C, Thayer R M, Mullenbach G T, Denniston K J, Gerin J L, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature (London) 1987;328:456. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 31.Weiner A J, Choo Q-L, Wang K-S, Govindarajan S, Redeker A G, Gerlin J L, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24δ and p27δ. J Virol. 1988;62:594–599. doi: 10.1128/jvi.62.2.594-599.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch W J. Phorbol ester, calcium ionophore, or serum added to quiescent rat embryo fibroblast cells all result in the elevated phosphorylation of two 28,000-dalton mammalian stress proteins. J Biol Chem. 1985;260:3058–3062. [PubMed] [Google Scholar]
- 33.Willard K E, Giometti C S, Anderson L, Onnor T E O, Anderson N G. Analytical techniques for cell fractions. A two-dimensional electrophoretic analysis of basic proteins using phosphatidyl choline/urea solubilization. Anal Biochem. 1979;100:289–298. doi: 10.1016/0003-2697(79)90232-x. [DOI] [PubMed] [Google Scholar]
- 34.Woppmann A, Patschinsky T, Bringmann P, Godt F, Luhrmann R. Characterisation of human and murine snRNP proteins by two-dimensional gel electrophoresis and phosphopeptide analysis of U1-specific 70K protein variants. Nucleic Acids Res. 1990;18:4427–4438. doi: 10.1093/nar/18.15.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh T-S, Lo S-J, Chen P-J, Wu Lee Y-H. Casein kinase II and protein kinase C modulate hepatitis delta virus RNA replication but not empty viral particle assembly. J Virol. 1996;70:6190–6198. doi: 10.1128/jvi.70.9.6190-6198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh T-S, Wu Lee Y-H. Assembly of hepatitis delta virus particles: package of multimeric hepatitis delta virus genomic RNA and role of phosphorylation. Virology. 1998;249:12–20. doi: 10.1006/viro.1998.9310. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y Y, Tsega E, Hansson B G. Phylogenetic analysis of hepatitis D viruses indicating a new genotype I subgroup among African isolates. J Clin Microbiol. 1996;34:3023–3030. doi: 10.1128/jcm.34.12.3023-3030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]