Abstract
The herpesvirus saimiri strain C488 genome contains five genes for small nuclear RNAs, termed herpesvirus saimiri URNAs (or HSURs). Using a cosmid-based approach, all HSURs were precisely deleted from the genome. The mutant virus replicated at levels that were similar to those of wild-type viruses in OMK cells. Although the HSURs are expressed in wild-type virus-transformed human T-cell lines, the deletion does not affect viral transformation in cell culture.
The rhadinovirus prototype, herpesvirus saimiri strain A11 (2), contains seven genes for virus-encoded small nuclear RNAs that have been termed herpesvirus saimiri URNAs (or HSURs 1 to 7) (1, 16, 20, 25). The HSURs assemble into ribonucleoprotein particles (17) and can interact with cellular factors involved in mRNA destabilization (11, 12, 22). These 32- or 70-kDa proteins physiologically bind to AU-rich elements in the 3′ untranslated regions of cellular mRNAs, which leads to the rapid degradation of the respective mRNAs. HSURs may also be involved in the regulation of cytokine transcription in T cells, as cytokine mRNA levels for interleukin-4 but not interleukin-2 were reduced in tumor cells (6). It was conceived that the abundant HSURs bind and compete for the cellular destabilization factors, thus increasing the half-life of cellular mRNAs expressing growth factors or proto-oncogenes. This led to the hypothesis that the viral URNAs might have a relevant function in the transformation of T cells. The herpesvirus saimiri strain subgroup C viruses, especially strain C488, are distinguished by the ability to transform human T cells to permanent antigen-independent growth (3). In the left terminal region of the coding low G+C content DNA (L-DNA), strain C488 contains five of the HSUR genes. According to their homology to strain A11 HSURs, they represent HSURs 1, 2, 5, 4, and 7 (Fig. 1a). As in subgroup A strains, a gene with strong homology to the family of dihydrofolate reductases (DHFR) is located in the same region of the viral genome (24). The herpesvirus saimiri transformation-associated protein StpC and a tyrosine kinase interacting protein (Tip) (4) located at the very left end of the L-DNA have been found to be essential for transformation (7).
FIG. 1.
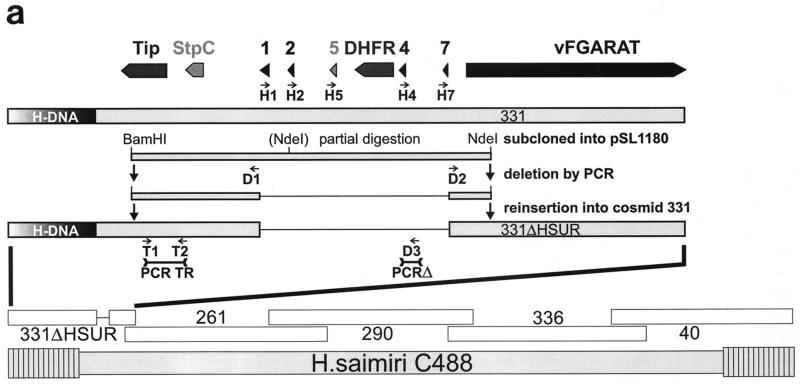
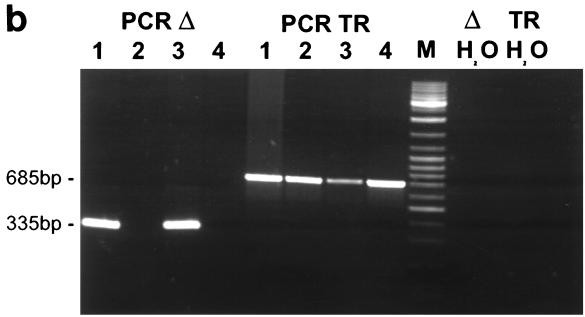
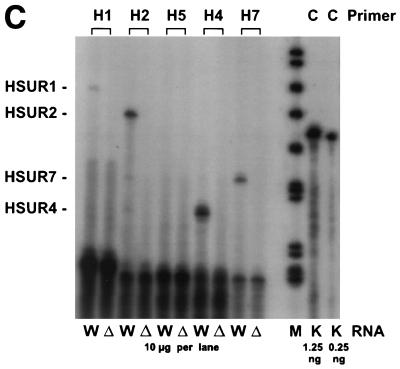
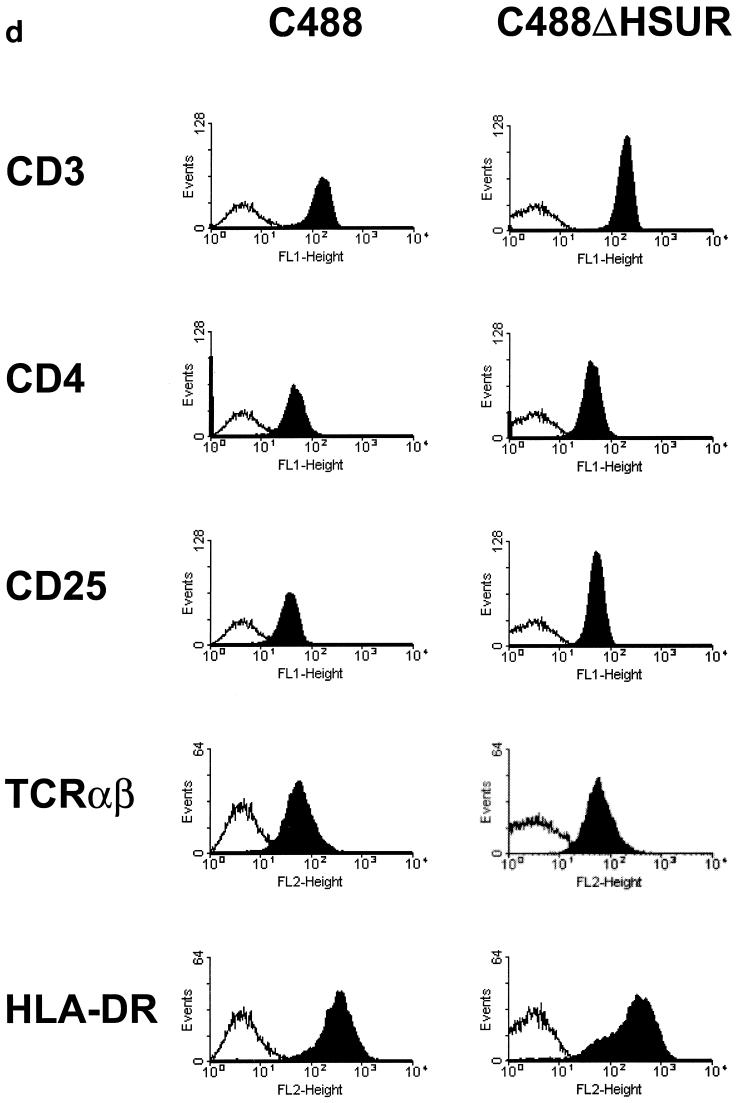
(a) Construction of the mutant virus C488ΔHSUR from overlapping cosmids. The HSUR genes were precisely deleted from a subcloned fragment by PCR mutagenesis with the indicated oligonucleotide primers. The fragment was reinserted to generate cosmid 331ΔHSUR. StpC, herpesvirus saimiri subgroup C transforming protein; Tip, tyrosine kinase interacting protein; HSURs are numbered H1, H2, H4, H5, and H7, according to the most homologous HSURs of herpesvirus saimiri A11; vFGARAT, herpesvirus saimiri open reading frame 3 with homology to formylglycineamide-ribonucleotidyl-aminotransferase; D1 and D2, oligonucleotides for the PCR-based deletion of HSURs; H1, H2, H5, H4, and H7, oligonucleotides used for primer extension analysis (results shown in panel c); T1, T2, H4, and D3, oligonucleotides used for PCR (the results shown in panel b); 331, 331ΔHSUR, 261, 290, 336, and 40, overlapping subgenomic cosmid clones used to generate recombinant virus. (b) Recombinant virus was generated by transfection of NotI-linearized cosmids into permissive OMK cells. PCR TR with oligonucleotides T1 and T2 was specific for the StpC-Tip transformation-associated genomic region; the PCR TR product size was 685 bp; PCRΔ with oligonucleotides H4 and D3 was specific for the deleted HSUR-DHFR region; PCRΔ product size, 335 bp. Lanes: 1 and 2, C488- and C488ΔHSUR-transformed T-cell lines from one donor, respectively; 3 and 4, as in lanes 1 and 2, from a second donor. Δ H2O and TR H2O show negative control reactions without template for PCRΔ and PCR TR, respectively. (c) Primer extension analysis with 10 μg of total RNA from C488 (W)- and C488ΔHSUR (Δ)-transformed human CBL (as in panel b, lanes 1 and 2). A 1.2-kb kanamycin mRNA control (K) and suitable control primer (C) supplied with the kit show the expected product of 82 bp. (d) Flow cytometry analysis of transformed human T cells after 4 months of continuous culture. The following antibodies (Becton Dickinson) were used on 300,000 transformed cells/assay: αCD3 (Leu-4), αTCR-αβ-1, αCD4 (Leu-3a), αCD8 (Leu-2a), and αHLA-DR. A typical pair of C488-and C488ΔHSUR-transformed T-cell lines from the same donor are shown.
Previous deletion analysis of herpesvirus saimiri strains could not reveal the presumed transformation-associated function of the HSURs. There are published reports describing deletions in strain A11 where all HSURs are deleted, but the gene encoding the herpesvirus saimiri transforming protein of strain A11 (StpA) was deleted as well. For a herpesvirus saimiri subgroup C isolate that is less efficient in the sustained transformation of human T cells, strain C484-77, it was reported that HSURs 1 and 2 are not essential in short-term replication assays with human T cells (19). In these experiments, a C484 mutant (2RNA) with a deletion of HSURs 1 and 2 was still able to exert effects on the growth of human T cells similar to those of wild-type C484. HSURs 1 and 4 had been detected in those transient transformation assays with herpesvirus saimiri strain C484, but the homologs of HSURs 2 and 3 were absent or at very low abundance (19). However, these detected transcripts could also be ascribed to lytic viral replication that may occur in the early infection of human T cells. The authors (19) further showed that a large part of the left end of the viral genome, including all HSURs and DHFR, was dispensable for the viral replication of this strain. The putative function of the HSURs in transformation could not be addressed with this deletion virus (5RNA), as it had no transforming phenotype due to the absence of the StpC and Tip genes that are essential for T-cell transformation (7, 19). Thus, it could not be decided if the HSURs contribute to T-cell transformation by herpesvirus saimiri.
In this study, all HSURs and the interspersed DHFR were precisely deleted from the strain C488 genome by a cosmid-based approach. First, a 6.6-kb fragment from a partial BamHI-NdeI digestion from the left terminal cosmid, 331, was subcloned into pSL1180 (Pharmacia, Freiburg, Germany), and then the HSUR genes were deleted by PCR with the indicated oligonucleotide primers (D1, TACGTAGTAAACACGCAAATGCACAAG, and D2, GCTGACTTCTTTATAAGTTATGGTG). After confirmatory sequencing, the fragment was reinserted into cosmid 331 via the BamHI and NdeI sites to generate cosmid 331ΔHSUR (Fig. 1a). In the cosmid vector pWE15, the cloned viral DNA fragments are flanked by adjacent NotI restriction sites. Herpesvirus saimiri strain C488 does not contain NotI restriction sites. Recombinant virus C488ΔHSUR was generated by the transfection of NotI-linearized cosmids into permissive OMK cells (ATCC CRL1556) and characterized by PCR. Each 50-μl reaction mixture contained 2 μl of template DNA, 0.2 μM deoxynucleoside triphosphate, 10 μM of each primer, and 2.5 U of AmpliTaq-Gold in 1× AmpliTaq buffer (PE Biosystems, Weiterstadt, Germany); the polymerase was activated at 96°C for 10 min, followed by 40 cycles of 96°C for 10 s, 58°C for 20 s, and 70°C for 45 s, followed by a final extension step of 70°C for 1 min. For template preparation, virus was pelleted from 1 ml of a tissue culture supernatant of lytically infected OMK cells and lysed in 100 μl of PCR buffer containing 100 μg of proteinase K per ml for 1 h at 56°C followed by 15 min at 96°C; alternatively, 0.5 million transformed human T cells were lysed in 200 μl of the same buffer. Primer pairs were specific for the StpC-Tip transformation-associated genomic region (for PCR TR: the primers were T1, GTAGTAAACTAAGAGCAAAGCAAGC, and T2, GTACAAGCTGTTCAAGTTTGTTAGC; product size, 685 bp) and the deleted HSUR-DHFR region (for PCRΔ, the primers were H4, TGGTTTGGGCTACCCCAGAG, and D3, GCAATGTCTTACAATTCTACCAGC; product size, 335 bp) (Fig. 1b). The replication kinetics of recombinant virus C488ΔHSUR were compared to those of wild-type strain C488. Confluent OMK cells were infected in a 25-cm2 flask (105 cells) with 104 PFU of virus at day 0. Until lysis was complete, infectious particles in the supernatant were titrated on subsequent days by limiting dilution on OMK cells in 48-well plates. Both the wild-type strain C488 and the mutant virus C488ΔHSUR replicated with similar kinetics to an endpoint titer of 106 infectious particles per ml in OMK cells. Transformation assays in vitro showed that the mutant virus C488ΔHSUR transformed human umbilical cord blood lymphocytes (CBL) to permanently growing T cell-lines as efficiently as did the wild-type strain C488. For the transformation assays, 3 × 106 to 5 × 106 cells were infected with 1 ml of virus-containing supernatant from completely lysed OMK cells (equivalent to 106 infectious particles). Eight of eight attempts, each with cells from different donors, were successful with both C488 and C488ΔHSUR. None of the uninfected controls yielded a transformed T-cell line.
The expression of viral genes in herpesvirus saimiri-transformed human cells is highly restricted. The bicistronic mRNA carrying the StpC and Tip genes which are essential for viral transformation and the mRNA needed for a putative viral superantigen have been detected in strain C488-transformed human T cells (10, 15). In contrast, transcription of various viral genes is detectable from transformed marmoset or rabbit cell lines, where limited viral replication occurs. Primer extension analysis was performed on total RNA from C488- and C488ΔHSUR-transformed human CBL with the Primer Extension System with avian mycloblastosis virus reverse transcriptase (Promega) and the oligonucleotide primers H1 (GGTTACTTTATATTTACACCC), H2 (TATATACACCCAGTGCTTTC), H5 (TAGCTGTTATTGAGCAACAC), H4 (TGGTTTGGGCTACCCCAGAG), and H7 (CCTAAATTTGCCTAAGTACC). Briefly, 10 pM primers and 0.25 μg of a size marker were labeled with T4 polynucleotide kinase and [γ-32P]ATP. Primer extension was performed with 100 fM labeled primer and 20 μg of template RNA. Extension products were separated on a denaturing 8% polyacrylamide gel containing 6 M urea. Gels were dried on filter paper, exposed to phosphorimaging screens, and scanned with a BAS2000 reader (Fuji). A 1.2-kb kanamycin mRNA control and a suitable primer supplied with the kit showed the expected product of 82 bp. The primer extension analysis showed that significant levels of HSURs 1, 2, 4, and 7 are detectable from human T cells permanently transformed with strain C488 (Fig. 1c), whereas HSUR 5 transcripts were not detectable. This is in agreement with the hypothesis that HSUR 5 is usually not expressed, as it lacks the proper promoter sequences. HSUR transcripts were not detectable from the C488ΔHSUR-transformed cells (Fig. 1c). This shows that although HSURs are expressed in transformed human T-cell lines, the deletion does not affect viral transformation. The C488ΔHSUR- and C488-transformed pairs of T-cell lines were further compared by flow cytometry analysis. Surface marker analysis showed no significant differences. The phenotype was similar to those of mature activated T cells, with similar expression levels of CD3, TCRαβ, CD4 or CD8, and CD25 and a strong expression level of major histocompatibility complex class II (Fig. 1d).
These results are not compatible with the previous assumption that HSURs may act as cofactors for lymphocyte transformation. Although expressed in C488 wild-type virus-transformed human T cells, the absence of all HSURs in the deletion virus did not result in any significant loss of transforming capacity. No phenotypical differences were apparent between human T cells transformed by the C488 wild-type virus and those transformed by the C488ΔHSUR mutant virus. Small RNAs are also carried by other gammaherpesviruses, namely the Epstein-Barr virus (EBV)-encoded small RNAs (EBERs) (e.g., EBV and human herpesvirus 4) (18) and the related herpesvirus papio (13), and the viral tRNAs of murine herpesvirus 68 (MHV-68) (5). Although they have been shown to interact with cellular ribonucleoproteins and are useful for the detection of latent infection, no specific function has been assigned to the EBERs. The MHV-68 tRNAs are not a substrate of aminoacyl tRNA synthases (5) and are dispensable for viral replication and infection (23). In conclusion, a biological function remains to be assigned for the HSURs, as it does for the EBV-encoded EBERs and MHV-68-encoded tRNAs. It may be speculated that HUSRs can induce some modulation of transcription or translation in infected cells. The ubiquitously expressed HuR protein was shown to be a 32-kDa nuclear shuttle protein with homology to the Drosophila ELAV protein. It is important in the nucleocytoplasmatic transport of AU-rich elements containing mRNAs that carry a subset of early response genes (8, 9, 14, 21). The presumed interaction of HSURs and HuR may change the subcellular distribution of HuR and could result in an altered translation or degradation of mRNAs encoding proto-oncoproteins or cytokines. Changes in early-response protein expression may be favorable for the establishment of a persistent viral infection in the natural host. Thus, the HSURs are not required for in vitro transformation of T cells or presumably for oncogenesis but may be relevant in the persistently infected animal host.
REFERENCES
- 1.Albrecht J C, Fleckenstein B. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 1992;20:1810. doi: 10.1093/nar/20.7.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesinger B, Müller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesinger B, Tsygankov A Y, Fickenscher H, Emmrich F, Fleckenstein B, Bolen J B, Bröker B M. The product of the herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p561ck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 5.Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 6.Chou C S, Medveczky M M, Geck P, Vercelli D, Medveczky P G. Expression of IL-2 and IL-4 in T lymphocytes transformed by herpesvirus saimiri. Virology. 1995;208:418–426. doi: 10.1006/viro.1995.1172. [DOI] [PubMed] [Google Scholar]
- 7.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan X C, Steitz J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fickenscher H, Biesinger B, Knappe A, Wittmann S, Fleckenstein B. Regulation of the herpesvirus saimiri oncogene stpC, similar to that of T-cell activation genes, in growth-transformed human T lymphocytes. J Virol. 1996;70:6012–6019. doi: 10.1128/jvi.70.9.6012-6019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geck P, Medveczky M M, Chou C S, Brown A, Cus J, Medveczky P G. Herpesvirus saimiri small RNA and interleukin-4 mRNA AUUUA repeats compete for sequence-specific factors including a novel 70K protein. J Gen Virol. 1994;75:2293–2301. doi: 10.1099/0022-1317-75-9-2293. [DOI] [PubMed] [Google Scholar]
- 12.Geck P, Whitaker S A, Medveczky M M, Medveczky P G. Expression of collagenlike sequences by a tumor virus, herpesvirus saimiri. J Virol. 1990;64:3509–3515. doi: 10.1128/jvi.64.7.3509-3515.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe J G, Shu M-D. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J Virol. 1988;62:2790–2798. doi: 10.1128/jvi.62.8.2790-2798.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene J D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knappe A, Hiller C, Thurau M, Wittmann S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S I, Murthy S C, Trimble J J, Desrosiers R C, Steitz J A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee S I, Steitz J A. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J Virol. 1990;64:3905–3915. doi: 10.1128/jvi.64.8.3905-3915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner M R, Andrews N C, Miller G, Steitz J A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medveczky M M, Geck P, Sullivan J L, Serbousek D, Djeu J Y, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 20.Murthy S, Kamine J, Desrosiers R C. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986;5:1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myer V E, Lee S I, Steitz J A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc Natl Acad Sci USA. 1992;89:1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simas J P, Bowden R J, Paige V, Efstathiou S. Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J Gen Virol. 1998;79:149–153. doi: 10.1099/0022-1317-79-1-149. [DOI] [PubMed] [Google Scholar]
- 24.Trimble J J, Murthy S C, Bakker A, Grassmann R, Desrosiers R C. A gene for dihydrofolate reductase in a herpesvirus. Science. 1988;239:1145–1147. doi: 10.1126/science.2830673. [DOI] [PubMed] [Google Scholar]
- 25.Wassarman D A, Lee S I, Steitz J A. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 1989;17:1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]