In the original publication [1], there was a mistake shown in Tables 1–8 and Figures 3–5 and 7 as published. The pre/post-categorization information of four patients in our proteomic dataset was incorrectly labeled, requiring the proteomic analysis to be repeated. We repeated our analyses depending on the newly constructed, corrected, and normalized dataset. The newly obtained results are given in the corrected Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7 and Table 8 and Figure 3, Figure 4 and Figure 5 and Figure 7 below.
Table 1.
Accuracy rates: Five supervised learning models with or without feature selection. Color changes from red to green display performance results from the lowest (red) to the highest values (green).
| ML-ACC | Without FS | LASSO FS | mRMR FS |
|---|---|---|---|
| SVM | 57.860 | 78.674 | 91.515 |
| LR | 67.633 | 85.341 | 92.708 |
| KNN | 62.197 | 53.068 | 88.466 |
| RF | 73.826 | 88.466 | 89.659 |
| AdaBoost | 88.409 | 89.072 | 88.447 |
Table 2.
Performance results (i.e., ACC%) using only LASSO-based feature selection and weighting methods. Color changes from red to green display performance results from the lowest (red) to the highest values (green). The bold value indicates the best result.
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 5 | 11 | 93.314 | 92.083 | 82.386 | 91.496 | 91.477 |
| 4 | 26 | 89.640 | 93.314 | 62.197 | 93.939 | 90.890 |
| 3 | 44 | 89.053 | 96.363 | 46.345 | 92.121 | 93.939 |
| 2 | 90 | 85.985 | 92.064 | 60.379 | 91.496 | 93.901 |
| 1 | 197 | 76.269 | 85.966 | 54.868 | 87.841 | 87.822 |
Table 3.
Performance results (i.e., ACC%) using only mRMR-based feature selection and weighting methods. Color changes from red to green display performance results from the lowest (red) to the highest (green) values. The bold value indicates the best result.
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 5 | 5 | 86.004 | 88.428 | 87.235 | 87.841 | 87.197 |
| 4 | 7 | 90.890 | 92.708 | 90.265 | 91.496 | 91.496 |
| 3 | 8 | 95.152 | 96.364 | 92.708 | 90.871 | 93.920 |
| 2 | 11 | 92.708 | 96.364 | 92.689 | 90.284 | 94.508 |
| 1 | 34 | 92.102 | 92.102 | 87.254 | 92.121 | 90.871 |
Table 4.
Mean performance results (i.e., ACC %, CV = 5) determined using both LASSO and mRMR-based feature selection with weighting methods. Color changes from red to green display performance results from the lowest (red) to the highest values (green). The bold value indicates the best result.
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 2 | 87.216 | 87.216 | 89.659 | 87.841 | 85.379 |
| 14 | 2 | 87.216 | 87.216 | 89.659 | 87.841 | 85.379 |
| 13 | 4 | 90.265 | 90.265 | 90.284 | 92.727 | 89.034 |
| 12 | 6 | 93.920 | 92.708 | 91.477 | 92.102 | 93.314 |
| 11 | 6 | 93.920 | 92.708 | 91.477 | 92.102 | 93.314 |
| 10 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 9 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 8 | 8 | 95.152 | 96.364 | 92.708 | 90.265 | 93.920 |
| 7 | 10 | 93.333 | 94.546 | 90.284 | 92.102 | 90.284 |
| 6 | 12 | 93.939 | 95.152 | 90.909 | 91.496 | 90.246 |
| 5 | 17 | 96.345 | 95.114 | 88.447 | 91.496 | 90.871 |
| 4 | 32 | 93.295 | 95.739 | 68.921 | 90.890 | 89.640 |
| 3 | 52 | 92.121 | 95.151 | 49.962 | 93.333 | 92.670 |
| 2 | 113 | 87.216 | 92.670 | 60.379 | 91.496 | 95.152 |
| 1 | 218 | 78.087 | 85.966 | 55.492 | 89.053 | 93.314 |
Table 5.
The standard deviation of performance results (i.e., ACC %, CV = 5) determined using both LASSO and mRMR-based feature selection with weighting methods. Color changes from red to green display performance results from the lowest (red) to the highest values (green).
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 2 | 5.175 | 4.408 | 4.072 | 4.232 | 2.191 |
| 14 | 2 | 5.175 | 4.408 | 4.072 | 4.232 | 2.191 |
| 13 | 4 | 4.423 | 4.821 | 4.425 | 4.924 | 2.384 |
| 12 | 6 | 5.060 | 5.269 | 3.509 | 4.088 | 3.515 |
| 11 | 6 | 5.060 | 5.269 | 3.509 | 4.088 | 3.515 |
| 10 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 9 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 8 | 8 | 3.636 | 4.454 | 5.269 | 3.496 | 4.272 |
| 7 | 10 | 4.848 | 5.555 | 4.425 | 4.088 | 4.425 |
| 6 | 12 | 4.285 | 3.636 | 6.357 | 3.995 | 3.526 |
| 5 | 17 | 2.966 | 2.444 | 3.476 | 4.827 | 5.400 |
| 4 | 32 | 6.177 | 4.105 | 6.384 | 4.259 | 1.442 |
| 3 | 52 | 4.535 | 2.424 | 7.329 | 4.020 | 2.469 |
| 2 | 113 | 4.807 | 1.557 | 4.954 | 4.430 | 4.924 |
| 1 | 218 | 4.720 | 3.133 | 5.559 | 3.031 | 3.515 |
Table 6.
Performance results without employing feature selection and feature weighting. Color changes from red to green display performance results from the lowest (red) to the highest values (green).
| ML | ACC% | AUC | F1 | PRE | REC | SPEC |
|---|---|---|---|---|---|---|
| SVM | 57.860 | 0.415 | 0.518 | 0.698 | 0.515 | 0.690 |
| LR | 67.633 | 0.755 | 0.676 | 0.681 | 0.681 | 0.673 |
| KNN | 62.197 | 0.647 | 0.581 | 0.662 | 0.527 | 0.722 |
| RF | 73.826 | 0.808 | 0.744 | 0.768 | 0.746 | 0.737 |
| AdaBoost | 88.409 | 0.951 | 0.886 | 0.882 | 0.893 | 0.873 |
Table 7.
Performance results employing LASSO and mRMR-based feature selection with weighting operation. Color changes from red to green display performance results from the lowest (red) to the highest values (green).
| ML | ACC% | AUC | F1 | PRE | REC | SPEC |
|---|---|---|---|---|---|---|
| SVM | 95.152 | 0.989 | 0.949 | 0.975 | 0.928 | 0.976 |
| LR | 96.364 | 0.987 | 0.964 | 0.963 | 0.965 | 0.965 |
| KNN | 92.708 | 0.965 | 0.930 | 0.929 | 0.932 | 0.923 |
| RF | 90.265 | 0.978 | 0.902 | 0.885 | 0.928 | 0.876 |
| AdaBoost | 93.920 | 0.979 | 0.941 | 0.941 | 0.942 | 0.935 |
The best ACC% is 96.364, which was obtained with the Logistic Regression Model, and the minimum weight is 10. Selected Number of Features: 8. Best Feature (Biomarker) Set is as follows: ‘K2C5’, ‘MIC-1’, ‘CSPG3’, ‘GFAP’, ‘Proteinase-3’, ‘STRATIFIN’, ‘Cystatin M’, and ‘Keratin-1’ [59].
Table 8.
Overview of the identified proteomic biomarkers illustrating the biological relevance to glioma.
| Entrez Gene Symbol | Target Full Name | Biological Relevance to Glioma |
|---|---|---|
| K2C5 | Keratin, type II cytoskeletal 5 | Yes, evolving biomarker/target [61] |
| Keratin-1 | Keratin, type II cytoskeletal 1 | Yes, evolving biomarker/target [61] |
| STRATIFIN (SFN) |
14-3-3 protein sigma | Yes, tumor suppressor gene expression pattern correlates with glioma grade and prognosis [62] |
| MIC-1 (GDF15) | Growth/differentiation factor 15 | Yes, biomarker, novel immune checkpoint [63] |
| GFAP | Glial fibrillary acidic protein | Yes, evolving biomarker/target [64] |
| CSPG3 (NCAN) | Neurocan core protein | Yes, glycoproteomic profiles of GBM subtypes, differential expression versus control tissue [65] |
| Cystatin M (CST6) | Cystatin M | Yes, cell type-specific expression in normal brain and epigenetic silencing in glioma [66] |
| Proteinase-3 (PRTN3) |
Proteinase-3 | Yes, evolving role, may relate to pyroptosis, oxidative stress and immune response [59] |
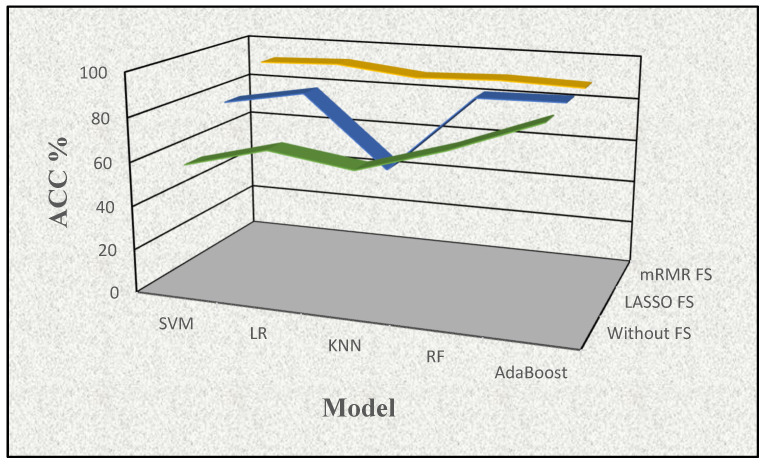
Figure 3.
The visualization of the effects of the feature selection procedures with accuracy (ACC%) determined by a supervised learning method in conjunction with the feature selection approach (mRMR FS (yellow), LASSO FS (blue), and no FS (green)).
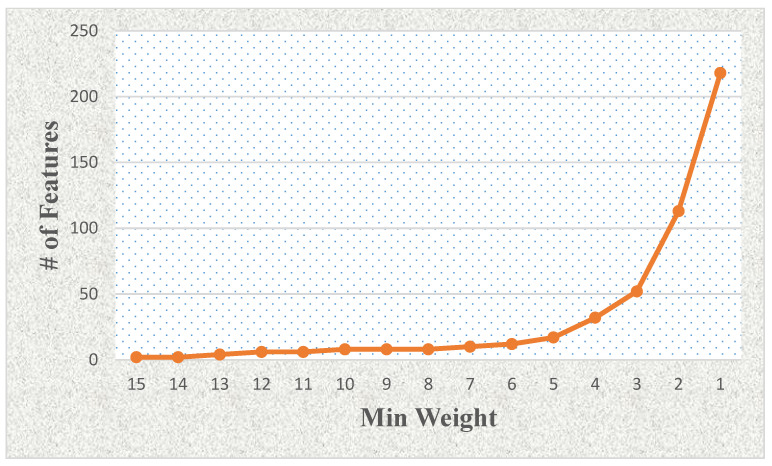
Figure 4.
The effects of the number of features related to the minimum weight value using LASSO and mRMR-based feature selection with weighting methods.
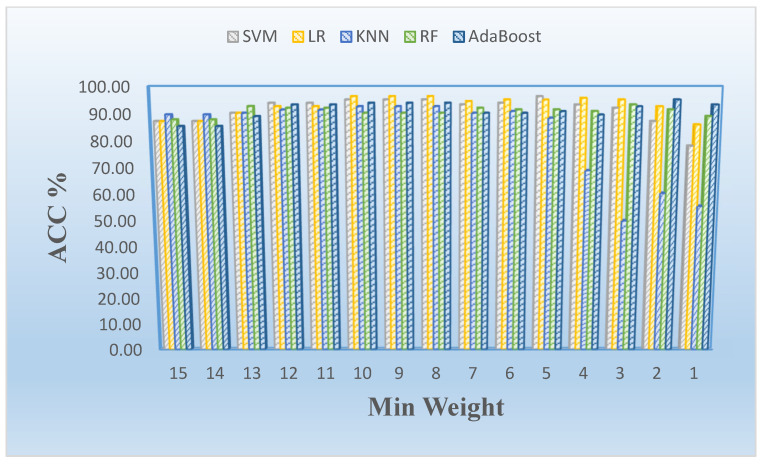
Figure 5.
Mean accuracy rate (ACC) vs. minimum weight stratified by model employed in analysis.
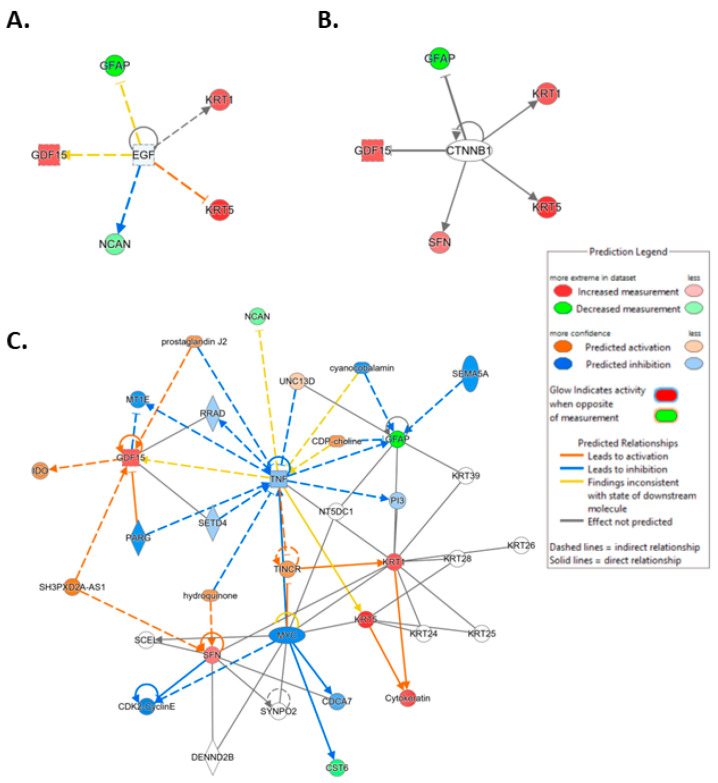
Figure 7.
Ingenuity pathway analysis (IPA) carried out on April 5, 2023, illustrating linkage of the identified protein features to the top 2 upstream mediators (Supplementary Data Table S2). (A) Epidermal growth factor (EGF) (p-value of overlap 2.53 × 10−7). (B) Catenin beta 1 (CTNNB1) (p-value of overlap 2.41 × 10−6). (C) IPA-generated merged network for the 8 ML-identified proteins using the disease classification brain cancer.
References
-
59.
Zeng, S.; Li, W.; Ouyang, H.; Xie, Y.; Feng, X.; Huang, L. A Novel Prognostic Pyroptosis-Related Gene Signature Correlates to Oxidative Stress and Immune-Related Features in Gliomas. Oxid. Med. Cell. Longev. 2023, 2023, 4256116. https://doi.org/10.1155/2023/4256116.
The repeat analysis has resulted in superior results as compared to the previous analysis with the best ACC% now 96.364, which was obtained with the Logistic Regression Model, and the minimum weight of 10. The selected number of features is now 8. Best Feature (Biomarker) Set is as follows: ‘K2C5’, ‘MIC-1’, ‘CSPG3’, ‘GFAP’, ‘STRATIFIN’, ‘Cystatin M’, ‘Keratin-1’ and ‘Proteinase-3’. All places in the manuscript text where the new results have resulted in a numerical change e.g., ACC, AUC, have been corrected to reflect the new findings in the updated tables. With this correction, the order of some references has been adjusted accordingly. The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Reference
- 1.Tasci E., Jagasia S., Zhuge Y., Sproull M., Cooley Zgela T., Mackey M., Camphausen K., Krauze A.V. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers. 2023;15:2672. doi: 10.3390/cancers15102672. [DOI] [PMC free article] [PubMed] [Google Scholar]