Abstract
Although the optimum substrate for lipoamidase (lipoyl-X hydrolase) has not yet been determined, it is known that lipoamidase activity, as determined by hydrolysis of the synthetic substrate lipoyl 4-aminobenzoate (LPAB), is widely distributed in pig brain tissues, i.e. in the cerebrum, cerebellum and medulla. Over 95% of the enzyme activity is present in the membrane subfractions, indicating that brain lipoamidase is an integral membrane protein enzyme. To elucidate the chemical nature and the optimum substrate of the abundant lipoamidase in the brain, we isolated it from the membrane subfractions. After an 8-step purification procedure, brain lipoamidase was purified 601-fold and identified as a 140 kDa glycoprotein by SDS/PAGE. A mechanistic study to determine Km and Vmax, values was carried out using various synthetic compounds. Lipoyl-lysine, which is generally believed to be a naturally occurring substrate of lipoamidase, was first compared with biotinyl-lysine, because these two vitamins have reactive sulphur atoms and are similar in molecular mass and structure. The Km for lipoyl-lysine was 333 microM, whereas biotinyl-lysine was not hydrolysed. Stringent specificity for the lipoyl moiety is demonstrated, as expected. Dipeptides of amino acid-lysine structures were studied, and dipeptides of aspartyl- and glutamyl-lysine hydrolysis occurred at high Km (3 mM) values. Thus lysine in the moiety is not very effective as an optimum substrate. The chemical bond structures of the amide bond (lipoyl-lysine) and peptide bond (aspartyl-lysine) were hydrolysed. Next, the ester bond compound was tested, and it was observed that lipolylmethyl ester was hydrolysed at high specificity. These findings indicate that this enzyme has broad specificities with respect to bond structure; it therefore is a unique hydrolase having stringent specificity for lipoic acid and relatively broad specificity for the chemical bond and the X moiety. Various inhibitors were tested; a few reagents, such as organic mercurials, di-isopropylfluorophosphate, 1,10-phenanthroline, sodium azide and angiotensin-converting enzyme inhibitor exhibited some inhibition (not more than 60%). Thus the active centre of this enzyme is a complex type. Although ATP is not hydrolysed and the lowest Km value is exhibited by the synthetic substrate reduced from LPAB (12 microM), some other compounds may still be expected to be hydrolysed by this unique and abundant brain lipoamidase.
Full text
PDF


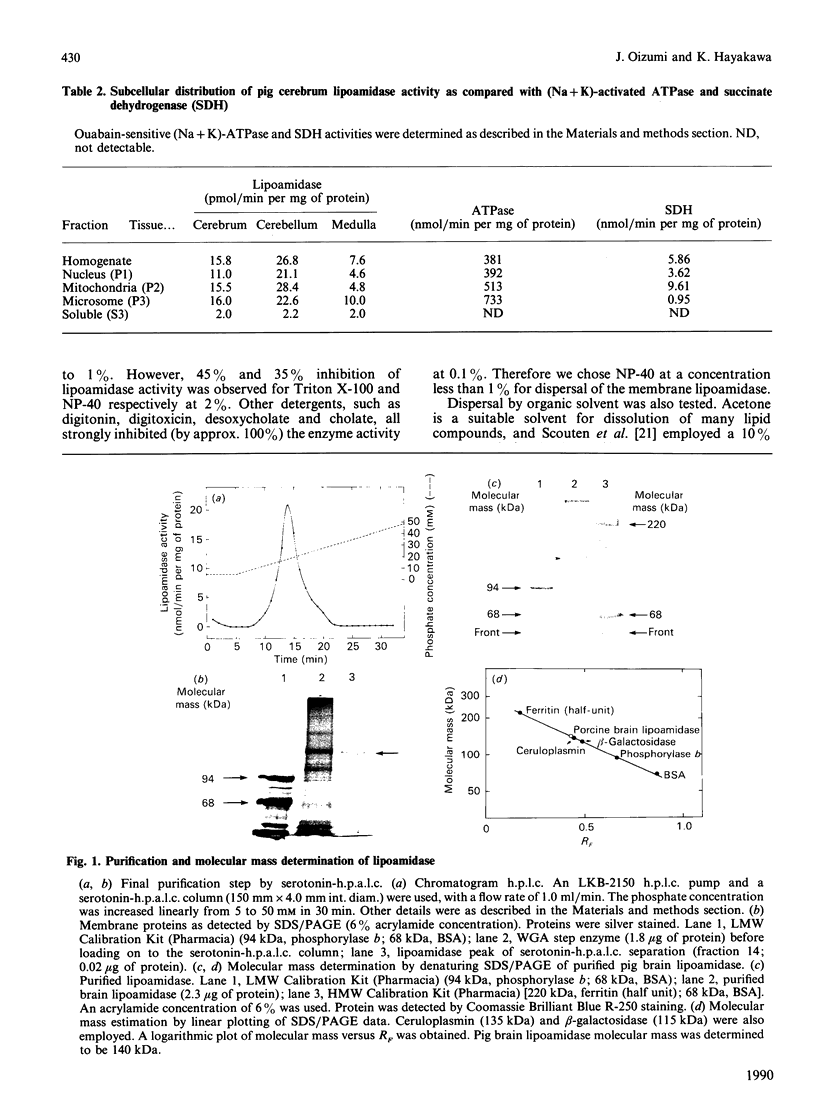
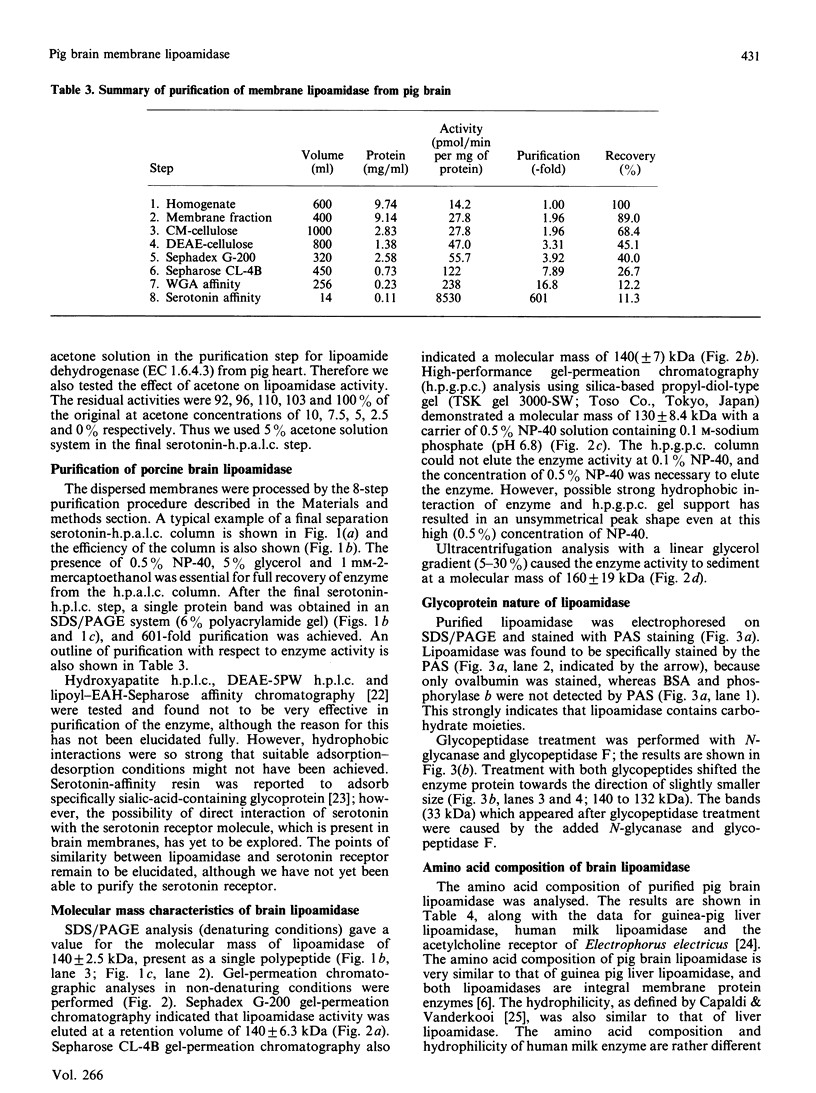
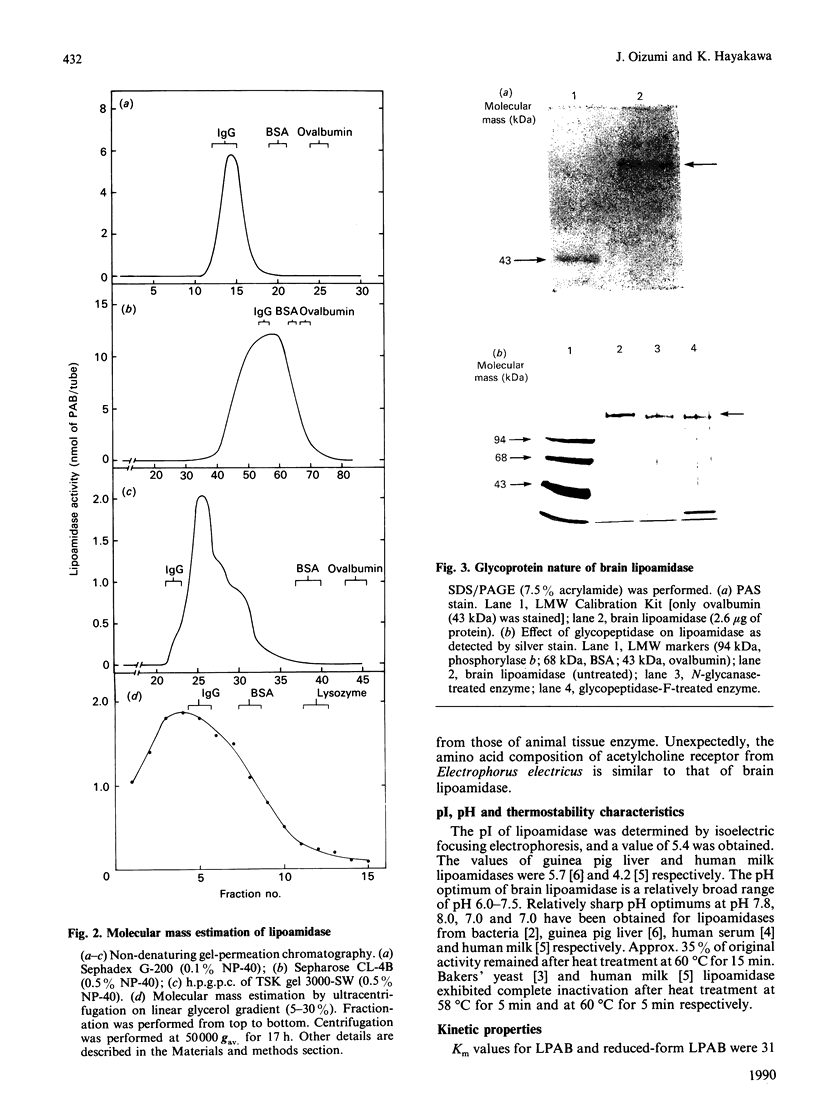

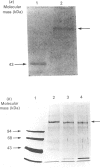
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALBERS R. W., RODRIGUEZDE LORES, DEROBERTIS E. SODIUM-POTASSIUM-ACTIVATED ATPASE AND POTASSIUM-ACTIVATED P-NITROPHENYLPHOSPHATASE: A COMPARISON OF THEIR SUBCELLULAR LOCALIZATIONS IN RAT BRAIN. Proc Natl Acad Sci U S A. 1965 Mar;53:557–564. doi: 10.1073/pnas.53.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARRIGONI O., SINGER T. P. Limitations of the phenazine methosulphate assay for succinic and related dehydrogenases. Nature. 1962 Mar 31;193:1256–1258. doi: 10.1038/1931256a0. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R. S., Potter L. T., Smith D. S. Postsynaptic membranes in the electric tissue of Narcine: IV. Isolation and characterization of the nicotinic receptor protein. Tissue Cell. 1977;9(4):623–644. doi: 10.1016/0040-8166(77)90031-3. [DOI] [PubMed] [Google Scholar]
- Clayton B. E., Dobbs R. H., Patrick A. D. Leigh's subacute necrotizing encephalopathy: clinical and biochemical study, with special reference to therapy with lipoate. Arch Dis Child. 1967 Oct;42(225):467–478. doi: 10.1136/adc.42.225.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellamora-Ortiz G. M., Ortiz C. H., Maia J. C., Panek A. D. Partial purification and characterization of the interconvertible forms of trehalase from Saccharomyces cerevisiae. Arch Biochem Biophys. 1986 Nov 15;251(1):205–214. doi: 10.1016/0003-9861(86)90067-6. [DOI] [PubMed] [Google Scholar]
- Harmon F. R. Purification of antibodies against biotin of lipoic acid-Sepharose. Anal Biochem. 1980 Mar 15;103(1):58–63. doi: 10.1016/0003-2697(80)90236-5. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Oizumi J. Determination of lipoamidase activity by liquid chromatography with fluorimetric detection. J Chromatogr. 1987 Dec 25;423:304–307. doi: 10.1016/0378-4347(87)80355-9. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Oizumi J. Determination of lipoyllysine derived from enzymes by liquid chromatography. J Chromatogr. 1989 May 5;490(1):33–41. doi: 10.1016/s0378-4347(00)82758-9. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Oizumi J. Human serum lipoamidase. Enzyme. 1988;40(1):30–36. doi: 10.1159/000469138. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Oizumi J. Isolation and characterization of human breast milk lipoamidase. Biochim Biophys Acta. 1988 Dec 2;957(3):345–351. doi: 10.1016/0167-4838(88)90224-5. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matalon R., Stumpf D. A., Michals K., Hart R. D., Parks J. K., Goodman S. I. Lipoamide dehydrogenase deficiency with primary lactic acidosis: favorable response to treatment with oral lipoic acid. J Pediatr. 1984 Jan;104(1):65–69. doi: 10.1016/s0022-3476(84)80591-0. [DOI] [PubMed] [Google Scholar]
- Oizumi J., Hayakawa K. Biotinidase and lipoamidase in guinea pig livers. Biochim Biophys Acta. 1989 Jun 27;991(3):410–414. doi: 10.1016/0304-4165(89)90066-4. [DOI] [PubMed] [Google Scholar]
- Prick M., Gabreëls F., Renier W., Trijbels F., Jaspar H., Lamers K., Kok J. Pyruvate dehydrogenase deficiency restricted to brain. Neurology. 1981 Apr;31(4):398–404. doi: 10.1212/wnl.31.4.398. [DOI] [PubMed] [Google Scholar]
- REED L. J., KOIKE M., LEVITCH M. E., LEACH F. R. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem. 1958 May;232(1):143–158. [PubMed] [Google Scholar]
- Reynolds S. F., Blass J. A possible mechanism for selective cerebellar damage in partial pyruvate dehydrogenase deficiency. Neurology. 1976 Jul;26(7):625–628. doi: 10.1212/wnl.26.7.625. [DOI] [PubMed] [Google Scholar]
- SEAMAN G. R. Purification of an enzyme from yeast which liberates protein-bound thioctic acid. J Biol Chem. 1959 Jan;234(1):161–164. [PubMed] [Google Scholar]
- SUZUKI K., REED L. J. LIPOAMIDASE. J Biol Chem. 1963 Dec;238:4021–4025. [PubMed] [Google Scholar]
- Scouten W. H., Torok F., Gitomer W. Purification of lipoamide dehydrogenase by affinity chromatography on propyllipoamide-glass columns. Biochim Biophys Acta. 1973 Jun 6;309(2):521–524. doi: 10.1016/0005-2744(73)90053-3. [DOI] [PubMed] [Google Scholar]
- Sturgeon R. J., Sturgeon C. M. Affinity chromatography of sialoglycoproteins, utilising the interaction of serotonin with n-acetylneuraminic acid and its derivatives. Carbohydr Res. 1982 May 16;103(2):213–219. doi: 10.1016/s0008-6215(00)80684-9. [DOI] [PubMed] [Google Scholar]
- Tashima Y. Removal of protein interference in the Fiske-Subbarow method by sodium dodecyl sulfate. Anal Biochem. 1975 Dec;69(2):410–414. doi: 10.1016/0003-2697(75)90143-8. [DOI] [PubMed] [Google Scholar]