Abstract
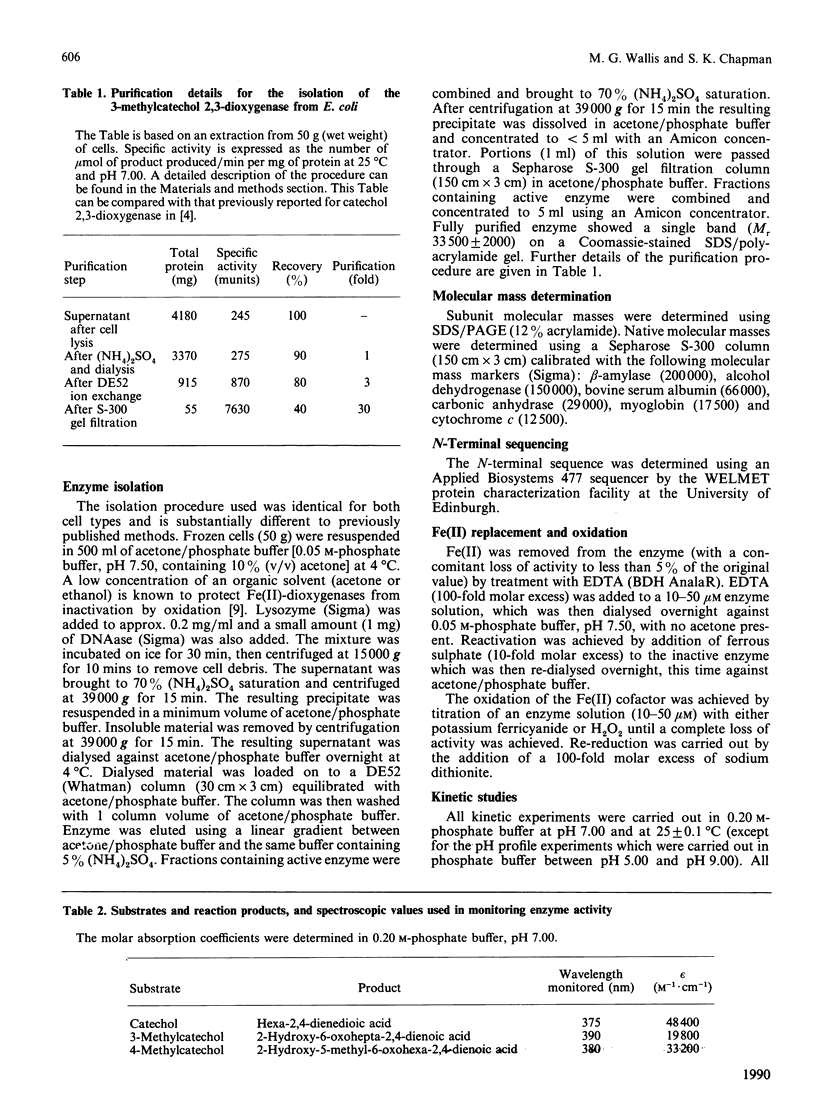
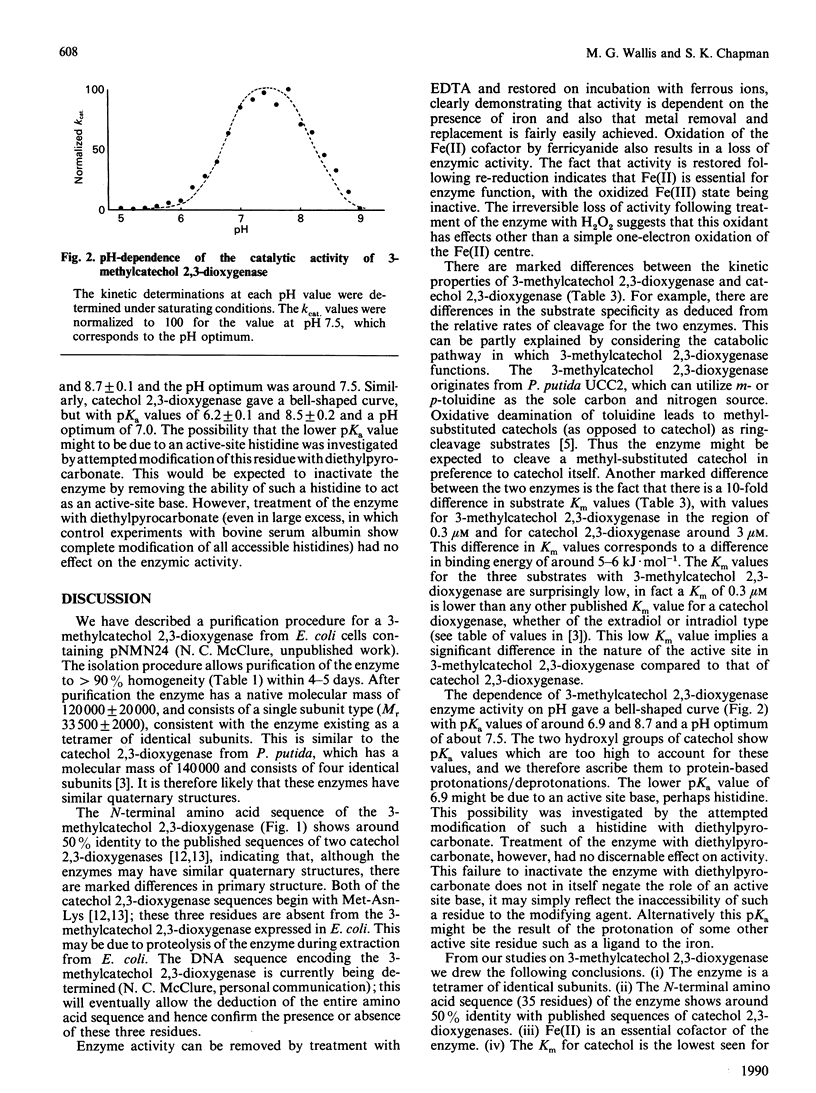
A purification procedure has been developed for an extradiol dioxygenase expressed in Escherichia coli, which was originally derived from a Pseudomonas putida strain able to grow on toluidine. Physical and kinetic properties of the enzyme have been investigated. The enzyme has a subunit Mr of 33,500 +/- 2000 by SDS/polyacrylamide-gel electrophoresis. Gel filtration indicates a molecular mass under non-denaturing conditions of 120,000 +/- 20,000. The N-terminal sequence (35 residues) of the enzyme has been determined and exhibits 50% identity with other extradiol dioxygenases. Fe(II) is a cofactor of the enzyme, as it is for other extradiol dioxygenases. The reactivity of this enzyme towards catechol and methyl-substituted catechols is somewhat different from that seen for other catechol 2,3-dioxygenases, with 3-methylcatechol cleaved at a higher rate than catechol or 4-methylcatechol. Km values for these substrates with this enzyme are all around 0.3 microM. The enzyme exhibits a bell-shaped pH profile with pKa values of 6.9 +/- 0.1 and 8.7 +/- 0.1. These results are compared with those found for other extradiol dioxygenases.
Full text
PDF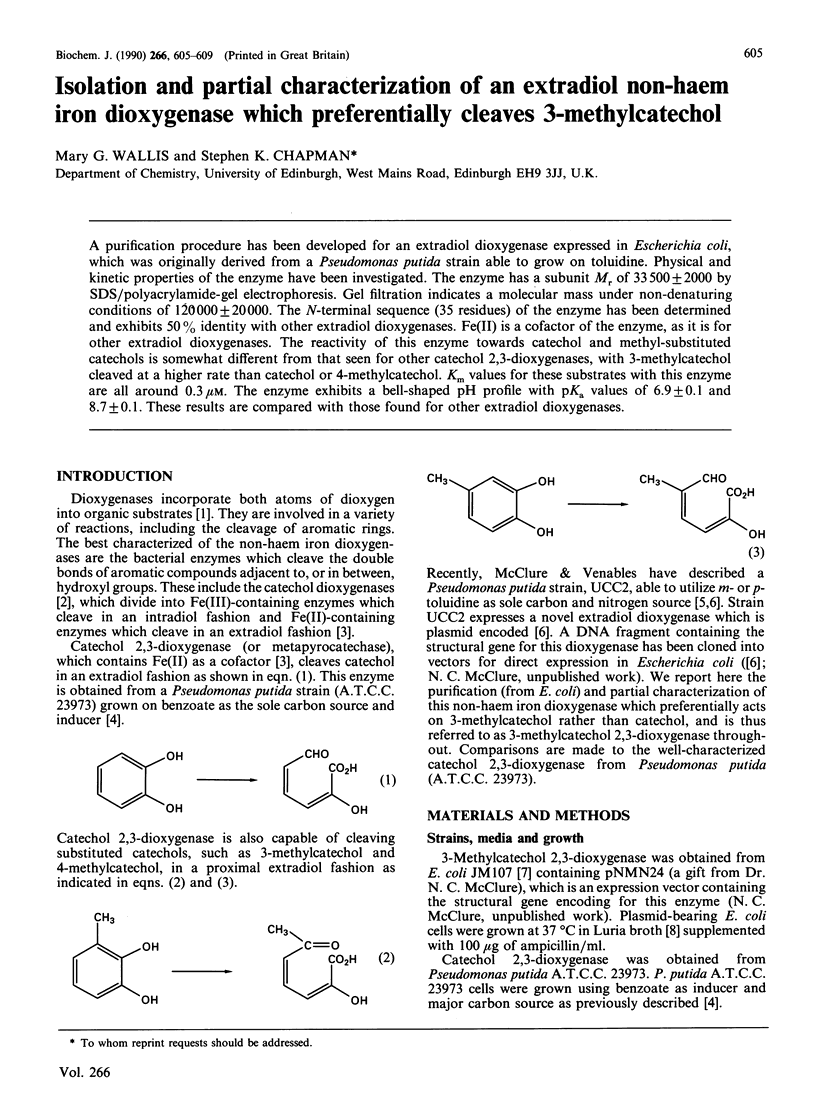


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ghosal D., You I. S., Gunsalus I. C. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene. 1987;55(1):19–28. doi: 10.1016/0378-1119(87)90244-7. [DOI] [PubMed] [Google Scholar]
- McClure N. C., Venables W. A. Adaptation of Pseudomonas putida mt-2 to growth on aromatic amines. J Gen Microbiol. 1986 Aug;132(8):2209–2218. doi: 10.1099/00221287-132-8-2209. [DOI] [PubMed] [Google Scholar]
- Miles E. W. Modification of histidyl residues in proteins by diethylpyrocarbonate. Methods Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- NOZAKI M., KAGAMIYAMA H., HAYAISHI O. METAPYROCATECHASE. I. PURIFICATION, CRYSTALLIZATION AND SOME PROPERTIES. Biochem Z. 1963;338:582–590. [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- Nozaki M., Kotani S., Ono K., Seno S. Metapyrocatechase. 3. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970 Nov 11;220(2):213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


