Abstract
Background
Hepatocellular carcinoma (HCC) is the most common primary liver cancer. Chronic hepatitis and liver cirrhosis lead to accumulation of genetic alterations driving HCC pathogenesis. This study is designed to explore genomic landscape of HCC in Egyptian patients by whole exome sequencing.
Methods
Whole exome sequencing using Ion Torrent was done on 13 HCC patients, who underwent surgical intervention (7 patients underwent living donor liver transplantation (LDLT) and 6 patients had surgical resection}.
Results
Mutational signature was mostly S1, S5, S6, and S12 in HCC. Analysis of highly mutated genes in both HCC and Non-HCC revealed the presence of highly mutated genes in HCC (AHNAK2, MUC6, MUC16, TTN, ZNF17, FLG, MUC12, OBSCN, PDE4DIP, MUC5b, and HYDIN). Among the 26 significantly mutated HCC genes—identified across 10 genome sequencing studies—in addition to TCGA, APOB and RP1L1 showed the highest number of mutations in both HCC and Non-HCC tissues. Tier 1, Tier 2 variants in TCGA SMGs in HCC and Non-HCC (TP53, PIK3CA, CDKN2A, and BAP1). Cancer Genome Landscape analysis revealed Tier 1 and Tier 2 variants in HCC (MSH2) and in Non-HCC (KMT2D and ATM). For KEGG analysis, the significantly annotated clusters in HCC were Notch signaling, Wnt signaling, PI3K-AKT pathway, Hippo signaling, Apelin signaling, Hedgehog (Hh) signaling, and MAPK signaling, in addition to ECM-receptor interaction, focal adhesion, and calcium signaling. Tier 1 and Tier 2 variants KIT, KMT2D, NOTCH1, KMT2C, PIK3CA, KIT, SMARCA4, ATM, PTEN, MSH2, and PTCH1 were low frequency variants in both HCC and Non-HCC.
Conclusion
Our results are in accordance with previous studies in HCC regarding highly mutated genes, TCGA and specifically enriched pathways in HCC. Analysis for clinical interpretation of variants revealed the presence of Tier 1 and Tier 2 variants that represent potential clinically actionable targets. The use of sequencing techniques to detect structural variants and novel techniques as single cell sequencing together with multiomics transcriptomics, metagenomics will integrate the molecular pathogenesis of HCC in Egyptian patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-01965-w.
Keywords: Hepatocelleular carcinoma (HCC), Living Donor Liver Transplantation (LDLT), Whole exome sequencing, Mutational signature
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, ranking the third leading cause of cancer related mortalities [1]. Chronic hepatitis and liver cirrhosis lead to accumulation of genetic alterations driving HCC pathogenesis. In Egypt, HCC constitutes a significant public health problem, where it is responsible for 33.63% and 13.54% of all cancers in males and females, respectively. There is a strong link between HCC and the hepatitis C virus (HCV) epidemic affecting 10–15% of the Egyptian population, which is reported as the highest prevalence of HCV in the world [2]. The molecular mechanisms driving HCC tumorigenesis are extremely complex and an understanding of these driver mutations is essential for prevention, as well as diagnostic, prognostic, and therapeutic purposes [3, 4]. However, these key genetic drivers remain unstudied, and an understanding of the underlying molecular mechanisms process in the development of HCC in Egyptian population is necessary, of which this study adds to relevance.
According to the European Association for Study of the Liver (EASL) guidelines, one of the unmet needs in HCC research is to develop new tools for early detection including the assessment of liquid biopsy (blood sample) [5]. In addition, without a liver biopsy, assessment of the genomic profile can be addressed by a noninvasive liquid biopsy which provides actionable genomic information without the risk of complications. Using a liquid biopsy, ctDNA can be extracted to comprehensively profile the tumor genome better than conventional sampling methods [6, 7].
Hepatocellular carcinoma is highly heterogenous malignancy and recent studies found that HCC commonly presents with mutations in the TERT promoter, TP53, CTNNB1, AXIN1, LAMA2, ARID1A, WWP1, and RPS6KA3 genes. Somatic mutations that are HCC-associated vary extensively among individuals and even within a single tumor [8]. Among the causes of genomic intratumor heterogeneity (ITH) including somatic mutations, epigenetic changes, and large-scale genomic alterations, somatic alterations represent the most abundant and studied mutations among cancers. Interplay between mutated driver genes is a major determinant of the carcinogenic process [9].
Several studies have attempted to identify driver mutations within the protein coding regions or the exomes in various types of cancers. Although the exomes represent 1% of human genome, it covers 85% of the disease-causing mutations. For this reason, sequencing of the whole exome using whole exome sequencing (WES) has the potential to discover a large number of variants implicated in many diseases including cancers [10].
The widespread use of next-generation sequencing (NGS) offers in-depth investigation into the tumor type-specific and context-driven characteristics of ITH. Next generation sequencing using WES enables researchers to create the precise genomic profile of HCC and identify protein-altering mutations per tumor in relation to clinicopathologic criteria [11]. It provides a detailed understanding of cancer pathways and the discovery of molecular mechanisms of cancer. It can throw light on the frequent somatic/genetic alterations in driver genes and the main pathways dysregulated in HCC [12, 13].
Ethnicity could contribute to global differences in the molecular profile of HCC due to the presence of various risk factors such as hepatitis B virus, hepatitis C virus, alcohol, aflatoxin exposure and metabolic syndrome [10]. Consequently, in our study we performed WES on dual HCC and surrounding non-HCC tissue samples for each HCC patient. To overview somatic mutations in the form of single nucleotide polymorphisms (SNPs) and assess their clinical relevance in HCC patients, we analyzed the type and number of SNPs in each gene and the association between them and clinicopathological information. In addition, we analyzed the pathways affected and the agreement with previous results in other populations in international databases on HCC. We analyzed the common variants between all studied patients and the presence of novel mutations aiming to generate a genomic profile for HCC Egyptian patients.
Subjects and methods
The subjects of this prospective study were Egyptian patients with HCC attending Ain Shams Centre for Organ Transplantation (ASCOT), and Hepato-pancreatico-biliary unit (HPB) Ain Shams University Hospitals, Cairo, Egypt, between October 2020 and June 2022. Our study was approved by the Faculty of Medicine Ain Shams University Research Ethics Committee (FMASU R92/2020).
Subjects
Sixteen patients underwent surgical intervention (7 patients had surgical resection and 9 patients underwent living donor liver transplantation (LDLT)). Patients were subjected to genomic profiling using whole exome sequencing.
All transplanted patients were HCC on top of HCV cirrhosis, out of 7 patients who underwent resection, 4 patients were HCV-HCC, 2 patients were HBV-HCC and 1 patient had Budd-Chiari syndrome.
Criteria for Liver transplantation selection for HCC patients (n = 9):
Age from 18–65 years.
Milan Criteria (n = 6), (single lesion greater ≥ 2 cm and ≤ 5 cm, or up to 3 lesions, ≥ 1 cm and less than or ≤ 3 cm) [14]
UCSF criteria (n = 1), (single tumor ≤ 6.5 cm or ≤ 3 tumors with the largest tumor diameter ≤ 4.5 cm and total tumor diameter ≤ 8 cm) [15]
Patients beyond UCSF are selected as case by case study after successful down staging and radiological criteria showing complete response (CR) or stable disease (ST) according to Modified RECIST (mRECIST) criteria [16] with no evidence of any macrovascular invasion or distant extra hepatic spread (n = 2).
Alpha-fetoprotein ≤ 200 ng/dl.
Criteria for HCC Eligibility for Hepatic resection (n = 7):
Methods
Whole exome sequencing was performed using Ion Torrent (Ion Chef and Ion Proton) for sequencing of HCC and non-HCC samples from 16 Egyptian HCC patients. After explanation of the process, receiving consent and signatures from participant patients, 2 tissue samples were obtained from each HCC patients, from both HCC liver tissue and surrounding non-HCC tissue. Four patients’ samples were prepared as fresh frozen paraffin embedded (FFPE) by the pathology department of Ain Shams Specialized Hospital Liver Tissue (LT) (LT2, LT6, LT7, LT8). Samples from patients LT1,3,4,5 were excluded either due to death of patient or technical problems. As a result of the observed low quality of DNA extracted from FFPE samples, as evidenced by gel electrophoresis, the following 12 patients’ samples were taken as fresh tissue samples intraoperative by surgeon on a stabilizing solution for DNA named RNALater (RNAlater™ Stabilization Solution, ThermoFisher) according to manufacturer instructions. Samples were processed within a max of three days preserved at 2–4 °C.
DNA Extraction
FFPE samples: samples were fixed in 4–10% formalin as quickly as possible after surgical removal for a fixation time of 14–24 h (longer fixation times lead to more severe DNA fragmentation, resulting in poor performance in downstream assays). As recommended by the manufacturer, samples were thoroughly dehydrated prior to embedding (residual formalin can inhibit the digestion of proteinase K). Stained sections were examined by the pathologist to ensure more than 20% cancer cells in HCC samples. DNA extraction from FFPE was done using QiaAmp DNA FFPE kit (Cat. No. / ID: 56,404, Qiagen). Starting material for DNA purification consisted of freshly cut sections of FFPE tissue, each with a thickness of up to 10 μm. Up to 8 sections, each with a thickness of up to 10 μm and a surface area of up to 250 mm2, can be combined in one preparation.
Fresh samples: For fresh tissue, samples were cut to a maximum thickness of 0.5 cm and submerged in 5 volumes of RNAlater as recommended by manufacturer (RNAlater™ Stabilization Solution, ThermoFisher). Samples were stored at 2–4 °C for a max of 3 days until processed. Fresh tissue samples were extracted using QiaAmp DNA Mini Kit (Qiagen). 25 mg of liver tissue were used as a starting material.
After extraction: Using Qubit dsDNA HS assay (ThermoFisher Scientific) DNA was quantitated and purity tested using NanoDrop Microvolume Spectrophotometer (ThermoFisher Scientific) (Supplement Table 1). Purity (the ratio of absorbance at 260 and 280 nm is used to assess DNA purity) was accepted for all samples (Ratio ~ 1.8) [19], with a mean concentration of 60.7 ng/uL (14.8–127.6), FFPE samples mean concentration 58.4 ng/uL (21–120), fresh samples 60 ng/uL (14.8–127.6). Sample LT15 was excluded from WES because of exceptionally low concentration and unaccepted purity value of DNA in addition to LT9, 12 due to a failure in the sequencing step.
Whole Exome Sequencing
The breakdown of steps in whole exome sequencing is shown in Table 1. Library preparation was executed using Ion AmpliSeq Exome RDY Library Preparation kit (ThermoFisher Scientific). The amount of DNA required was 50–100 ng in a volume not exceeding 56 uL. We amplified target regions from 50 − 100 ng of genomic DNA (gDNA) in the IonAmpliSeq™ Exome RDY plates using the 5X Ion AmpliSeq™ HiFi Mix. Target amplification reactions were combined and then amplicons partially digested with FuPa Reagent. Ligation of barcode adapters with Switch Solution and DNA Ligase followed, which then was purified. We normalized the libraries after quantification using qPCR Ion Library Taqman Quantitation Kit on Via 7 Real Time PCR machine (ThermoFisher) and diluted to 100 pM. Template preparation and amplification was implemented using emulsion PCR on Ion sphere particles (ISP) and we combined two barcoded exome libraries on a single Ion 540™ Chip using Ion PI™ Hi-Q™ Chef Kit on Ion Chef system. Finally, sequencing was performed on Ion Proton™ system using semiconductor technology based on detection of hydrogen ions released during the DNA synthesis reaction by a highly sensitive pH meter—a microchip sensor.
Table 1.
A breakdown outline for the steps used for sequencing HCC tissue samples
| No | Step | Kit | Manufacture | Instrument |
|---|---|---|---|---|
| 1 | DNA Extraction | QIAamp DNA mini Kit | Qiagen | –- |
| 2 | Measure concentration of DNA (QC Step) | Qubit dsDNA HS assay | ThermoFisher Scientific | Qubit 3.0 |
| 3 | Amplify the targets | Ion AmpliSeq™ Exome RDY Kit / Ion Xpress™ Barcode Adapters | ThermoFisher Scientific | 9700 Thermal Cycler |
| 4 | Partially digest amplicons | |||
| 5 | Ligate barcoded adapters to the amplicons | |||
| 6 | Purify the unamplified library | Agencourt™ AMPure™ XP Reagent | Beckman Coulter | DynaMag™-2 Magnet |
| 7 | Quantify the library | Ion Library Taqman Quantitation Kit | ThermoFisher Scientific | Viia 7 Real Time PCR |
| 8 | Template preparation, loading chips and sequencing | Ion PI™ Hi-Q™ Chef Kit | Ion Torrent | Ion Chef |
| 9 | Ion PI™ Chip | Ion Proton |
Bioinformatics
Reads were initially demultiplexed and aligned to hg19 human reference sequence using the variantCaller v5.12.0.4 (Torrent server software—Thermo Fisher Scientific) which ran as embedded instance within Ion Torrent S5 sequencer. The resulting alignment BAMs were further processed using Ion Reporter™ Software 5.18 pipeline (Thermo Fisher Scientific), which incorporated variant calling and annotate variants based on the most updated databases such as ClinVar, DrugBank, GO, etc. The existence of potentially significant variants was further reassessed through manual inspection of aligned reads in IGV 2.4 software.
Available clinical significance annotation was assessed in real-time from Human Gene Mutation Database Professional (https://portal.biobase-international.com/hgmd/pro/), ClinVar (https://www.ncbi.nlm.nih.gov/ClinVar/) and dbSNP.
(https://www.ncbi.nlm.nih.gov/snp). The predictions for SIFT, PolyPhen and PhyloP tools were retrieved from the IonReporter result files (tab-separated files). Frequency data was provided by Ensembl/VEP release 108.0; additionally, gnomAD v3.1 database was queried for allele frequencies of individual variants (http://gnomad.broadinstitute.org).
Variant annotation
The VCF files were uploaded on the main Galaxy public server on https://usegalaxy.org. SnpEff eff (Galaxy Version 4.3 + T.galaxy2) was used to annotate and predict the effect of these variants [20]. The SnpEff 4.3 hg19 genome data was downloaded using SnpEff download and applied as the genome source for SnpEff eff. The default settings were selected for upstream/downstream length of 5000 base, the set size for splice sites which was set to 2 bases as well as for spliceRegion Settings. For the annotation options, the following were selected: Add loss of function (LOF) and nonsense mediated decay (NMD) tags. The chromosomal position was based on the input type and “-chr” was prepended to the chromosome name. SnpSift Extract Fields was then used on the VCF output files from SnpEff eff to extract specific fields and organize them in a tabular file [20].
Data processing
MUTALISK was aided in determination of mutational signature in HCC samples [21]. A web-based version of OpenCRAVAT (https://run.opencravat.org) was used for performing genomic variant interpretation including variant impact, annotation, and scoring [22]. CancerSpecific High-throughput Annotation of Somatic Mutations (CHASM) was used as well [23]. CHASMplus discriminates somatic missense mutations as either cancer drivers or passengers. Predictions can be done in either a cancer type-specific manner or by a model considering multiple cancer types together. TARGET is a database of genes that, when somatically altered in cancer, are directly linked to a clinical action. TARGET genes may be predictive of response or resistance to a therapy, prognostic, and/or diagnostic [24]. Oncogenes and tumor suppressor genes were delineated as described by Vogelstein et al., 2013 [25]. Franklin was used to analyze variants according to ACMG classification for somatic variants [26]. David Annotation database was used for assessment of enrichment driver pathways in our HCC samples [27].
Results
Liver tissue (HCC and non-HCC) and corresponding blood samples were collected from 16 HCC patients including 15 males (94%) and 1 female (6%), with a median age of 59 years (IQR 51–60 years). Of these, 13 patients had HCV and 2 patients had HBV (LT9, LT13), while 1 patient suffered from Budd-Chiari syndrome (LT11). According to Milan staging, 6 patients followed Milan Criteria (66.7%), 1 patient followed UCSF criteria (11.1%), and 2 patients were beyond both (22.2%). Regarding BCLC clinical staging system, 7 cases were stage A (43.7%), 6 cases were stage B (37.5%), and 3 cases were stage D (18.8%). Among the sixteen enrolled patients, tissue biopsy was taken from HCC and non-HCC liver tissue from 9 patients during liver transplantation, while 7 were sampled during hepatic resection.
Variant types
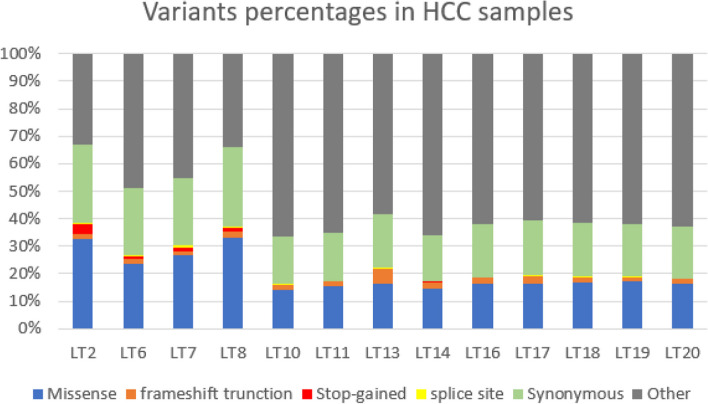
Using whole exome sequencing, the exons and surrounding noncoding genomic regions of protein-coding genes were captured in pairs of tumor and non-cancerous liver tissues. Genomic regions of variants varied from exon to non-exon variants (Supplement Table 2). Among pathogenic variants (Missense (27.9%), frameshift (1.6%), stop-gained (1.5%) and splice-site (0.4%)), missense was the highest percentage as shown in Fig. 1 and in Supplement Table 3.
Fig. 1.
Percentage of different variant sequence ontologies in HCC samples
Mutational signatures
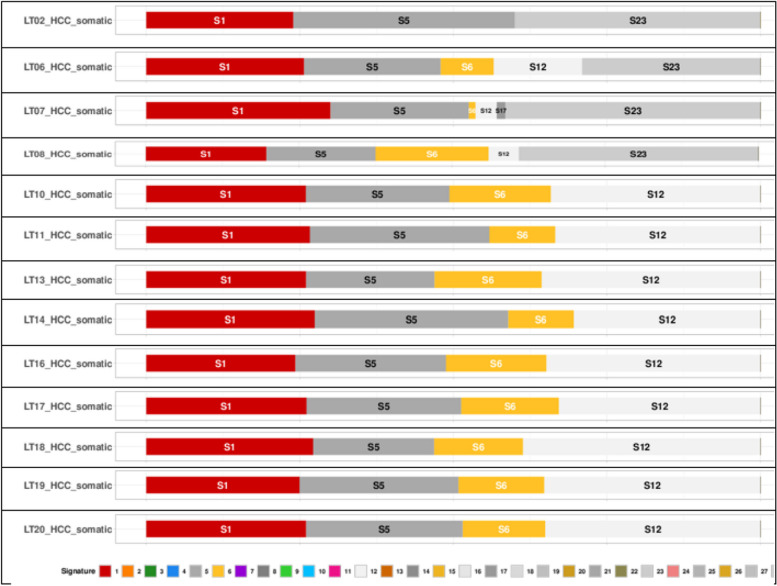
We analyzed HCC samples via Mutalisk against 30 mutational signatures COSMIC v2 [21] (Fig. 2; Supplement Table 4, Supplement Table 1). Analysis showed predominance of single base substitutions S1, S5, and S23 in FFPE samples (LT2, LT6, LT7, LT8) and S1, S5, S6, and S12 in fresh tissue samples (LT10, LT11, LT13, LT14, LT16, LT17, LT18, LT19, LT20). Signature 1 is the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. The number of S1 mutations correlates with age of cancer diagnosis. Signature 5 is found in all cancer types and most cancer samples. The etiology of S5 is unknown. Signature 6 is found in many cancer types and is most common in colorectal and uterine cancers. In most other cancer types, S6 is found in less than 3% of examined samples. S6 is associated with defective DNA mismatch repair and is found in microsatellite unstable tumors, S6 is associated with high numbers of small (< 3 bp) insertions and deletions at mono/polynucleotide repeats. Signatures 12 and 23 are found in liver cancer. Their etiology remains unknown [21]. Whole results are shown in Supplement MUTALISK Mutational signatures HCC.
Fig. 2.
Mutational signatures in HCC samples showing predominance of S1, S5 and S23 in FFPE samples (LT2, LT6, LT7, LT8) and S1, S5, S6, and S12) in fresh tissue samples (LT10, LT11, LT13, LT14, LT16, LT17, LT18, LT19, LT20) [21]
Highly mutated genes (highest number of somatic variants)
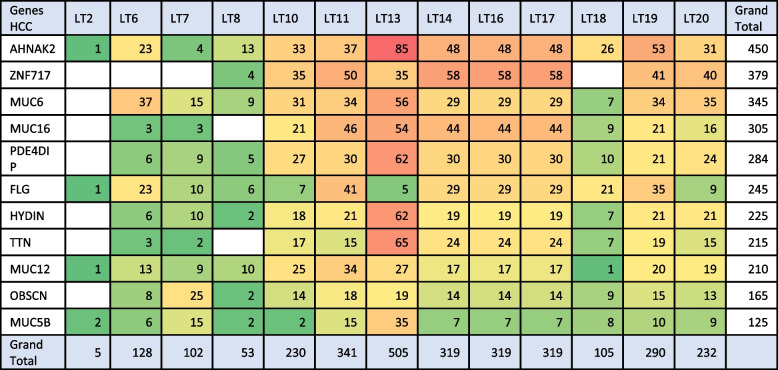
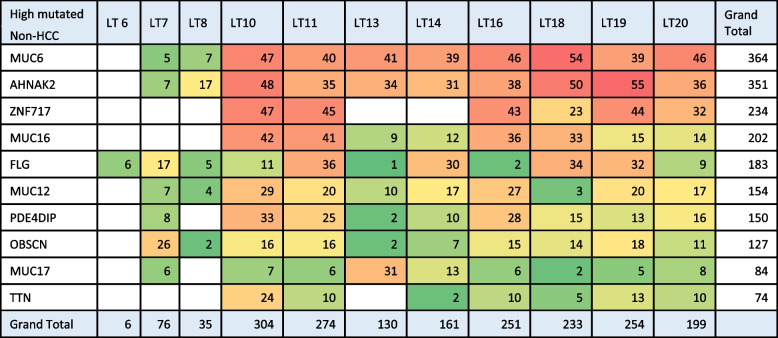
Analysis of highly mutated genes was done using OpenCRAVAT. Genes showing the highest somatic mutations in HCC and Non-HCC are shown in Tables 2, 3. Analysis of highly mutated genes in both HCC and Non-HCC revealed the presence of 10 common highly mutated genes (AHNAK2, MUC6, MUC16, TTN, ZNF17, FLG, MUC12, OBSCN, PDE4DIP, MUC5b, and HYDIN) (Supplement Tables 5, 6). Mapping of variants on the most highly mutated genes (AHNAK2, MUC16, MUC6, FLG, PDE4DIP, and HYDIN) relative to The Cancer Genome Atlas in HCC (TCGA LIHC) [28, 29] is shown in Fig. 3. Comutant OBSCN and TTN are shown in HCC and Non-HCC (Table 4).
Table 2.
Heatmap for pathogenic variants in highly mutated genes in HCC samples
Table 3.
Heatmap for pathogenic variants in highly mutated genes in Non-HCC samples
Fig. 3.
Variants in highly mutated genes HCC from our study (Above the bar), and the variants from TCGA HCC (Below the bar) are shown as a lollipop diagram, Protein domains are shown as brown bars. Variants in multiple samples are taller. Color Sequence ontology of variant
Table 4.
Commutant OBSCN and TTN HCC and Non-HCC samples
| OBSCN TTN | LT6 | LT7 | LT8 | LT10 | LT11 | LT13 | LT14 | LT16 | LT17 | LT18 | LT19 | LT20 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCC | OBSCN | 8 | 25 | 2 | 14 | 18 | 5 | 14 | 17 | 5 | 9 | 16 | 14 | 147 |
| TTN | 3 | 2 | 19 | 15 | 28 | 25 | 22 | 5 | 7 | 20 | 15 | 161 | ||
| Non-HCC | OBSCN | 26 | 2 | 16 | 16 | 2 | 7 | 15 | 2 | 14 | 18 | 11 | 129 | |
| TTN | 24 | 10 | 2 | 10 | 3 | 5 | 13 | 10 | 77 | |||||
The Cancer Genome Atlas (TCGA) significantly mutated genes
The Cancer Genome Atlas (TCGA) network performed a large-scale multi-platform analysis of HCC, including evaluation of somatic mutations in 363 patients. Whole exome sequencing on 363 HCC cases revealed that 12,136 genes had non-silent mutations, and 26 significantly mutated genes (SMGs) were determined using the MutSigCV algorithm [28, 29]. The list includes the 26 significantly mutated HCC genes identified across 10 genome sequencing studies in addition to TCGA and is shown in Supplement file TCGA HCC.
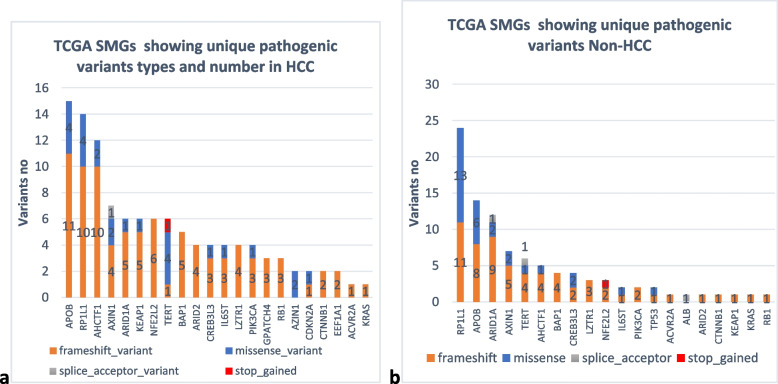
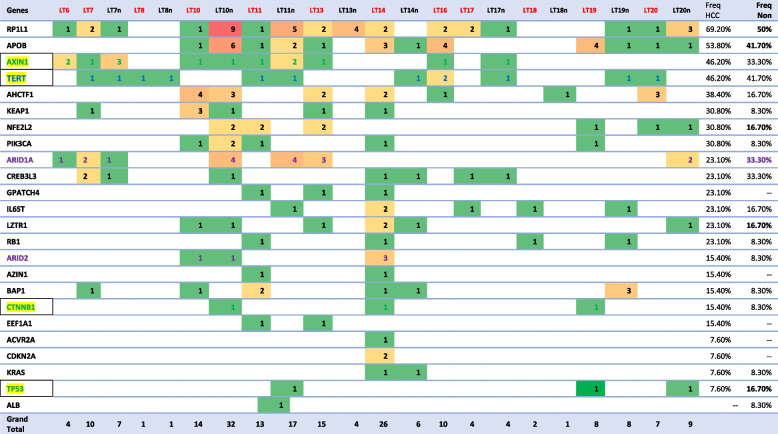
We studied the somatic variants in HCC and Non-HCC tissues regarding the 26 significantly mutated genes to determine the type and number of mutations in patients’ liver tissue. Data analysis revealed the presence of 87 unique variants in HCC and 94 in non-HCC tissues, most were frameshift truncation as demonstrated in Fig. 4. Genes APOB and RP1L1 showed the highest number of mutations in both HCC and Non-HCC tissues (Fig. 5). Frameshift mutations comprised 77% in HCC and 66% in Non-HCC tissues, while missense were 21% and 31%. respectively. Analysis of unique variants in individual samples revealed the presence of Tier 1, Tier 2 variants in SMGs in HCC and Non-HCC (TP53, PIK3CA, CDKN2A, and BAP1) among variants with CScape score for driver point mutation exceeding 0.5 [30] are listed in Table 5. CScape Coding predicts the oncogenic status (disease-driver or neutral) of somatic point mutations (missense in our list) specifically in the coding region of the cancer genome values above 0.5 are predicted to be deleterious, while those below 0.5 are predicted to be neutral or benign [30]. Only BAP1 had 2 variants with CScape exceeding 0.5 in Non-HCC TCGA SMGs. Table 6 demonstrates the distribution of different variants in TCGA SMGS in our HCC and Non-HCC patients. A list showing the frequency of TCGA SMGs and their number among HCC and Non-HCC samples is shown in Table 6.
Fig. 4.
Unique pathogenic variants involving TCGA SMGs in both (a) HCC and (b) Non-HCC
Fig. 5.
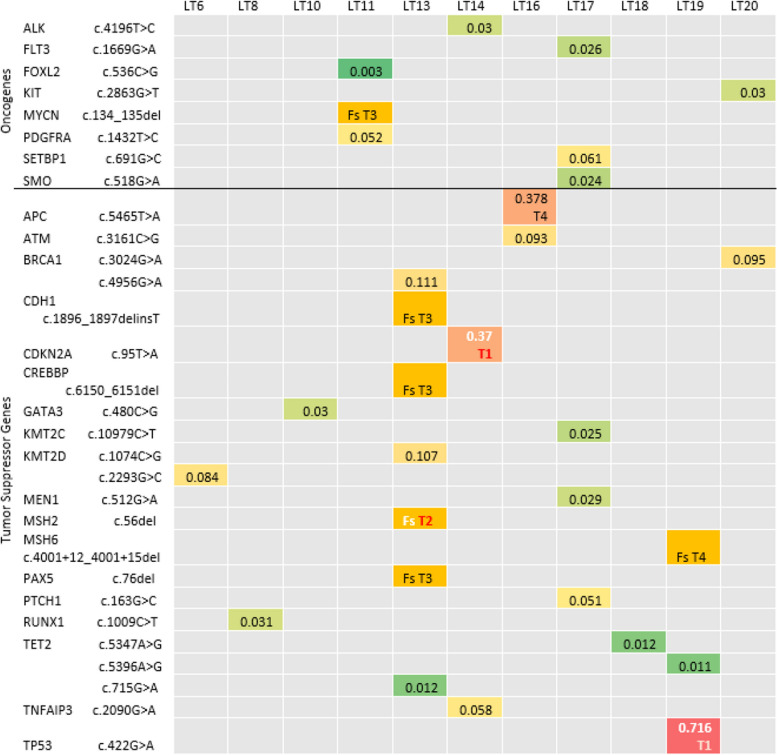
Unique pathogenic variants in Oncogenes, tumor suppressor genes (TSGs) HCC with Phred score > 20, Fs = frameshift-truncation, Sc = Splice site, T1 = Tier1, T2 = Tier 2, T3 = Tier 3, T4 = Tier4. A higher CHASM + LIHC score (liver hepatocellular carcinoma) in colored areas indicates a higher possibility of being a driver mutation in HCC
Table 5.
Unique pathogenic variants with (CScape score > 0.5) unique pathogenic variants in SMGs TCGA HCC and Non-HCC
| Chrom | Pos | Variant_note | Phred | Gene | Sequence_ontology | cDNA_change | Protein_change | Samples | CScape score |
|---|---|---|---|---|---|---|---|---|---|
| chr17 | 7,578,508 | Tier1 | 101.81 | TP53 | missense | c.422G > A | p.Cys141Tyr | LT19_HCC | 0.9312 |
| chr5 | 1,293,625 | Tier3 | 8.8584 | TERT | stop_gained | c.1376G > A | p.Trp459Ter | LT08_HCC | 0.9082 |
| chr8 | 103,855,852 | Tier3 | 30.607 | AZIN1 | missense | c.29A > G | p.Tyr10Cys | LT14_HCC | 0.8998 |
| chr19 | 10,600,008 | Tier3 | 10.393 | KEAP1 | missense | c.1568G > A | p.Gly523Glu | LT07_HCC | 0.8945 |
| chr3 | 52,437,608 | Tier3 | 6.2349 | BAP1 | missense | c.1553G > A | p.Arg518Gln | LT07_HCC | 0.8922 |
| chr3 | 178,952,085 | Tier1 | 6.2381 | PIK3CA | missense | c.3140A > G | p.His1047Arg | LT10_HCC | 0.8043 |
| chr8 | 103,845,284 | Tier3 | 6.7846 | AZIN1 | missense | c.904G > A | p.Val302Ile | LT11_HCC | 0.7754 |
| chr2 | 21,227,996 | Tier3 | 205.82 | APOB | missense | c.11744C > T | p.Ser3915Phe | LT19_HCC | 0.7374 |
| chr1 | 27,100,124 | Tier3 | 6.2383 | ARID1A | missense | c.3920C > T | p.Pro1307Leu | LT07_HCC | 0.6783 |
| chr9 | 21,974,732 | Tier2 | 25.112 | CDKN2A | missense | c.95 T > A | p.Leu32Gln | LT14_HCC | 0.5713 |
| chr1 | 27,094,369 | Tier3 | 4.1526 | ARID1A | missense | c.3077G > A | p.Arg1026His | LT07_Non | 0.9154 |
| chr16 | 354,405 | Tier3 | 11.843 | AXIN1 | missense | c.1153C > T | p.Pro385Ser | LT07_Non | 0.879 |
| chr3 | 52,442,605 | Tier2 | 5.7804 | BAP1 | frameshift | c.140_141insG | p.Ile47MetfsTer22 | LT19_Non | 0.856 |
| chr3 | 52,442,545 | Tier2 | 5.785 | BAP1 | frameshift | c.201dup | p.Asp68Ter | LT19_Non | 0.85 |
| chr2 | 178,098,885 | Tier3 | 7.2747 | NFE2L2 | frameshift | c.160_161insA | p.Leu54HisfsTer2 | LT10_Non | 0.7653 |
| chr16 | 348,239 | Tier3 | 5.0519 | AXIN1 | frameshift | c.1267_1268insC | p.Glu423AlafsTer46 | LT10_Non | 0.6795 |
| chr19 | 4,154,960 | Tier3 | 5.8058 | CREB3L3 | missense | c.92A > G | p.Asp31Gly | LT10_Non | 0.6625 |
| chr2 | 21,239,431 | Tier3 | 8.8517 | APOB | missense | c.3212 T > C | p.Val1071Ala | LT18_Non | 0.6298 |
| chr2 | 21,229,673 | Tier3 | 7.8123 | APOB | missense | c.10067 T > C | p.Leu3356Pro | LT10_Non | 0.588 |
| chr8 | 10,465,934 | Tier3 | 16.263 | RP1L1 | missense | c.5674A > G | p.Thr1892Ala | LT18_Non | 0.5742 |
Table 6.
Heatmap showing TCGA SMGs unique pathogenic variants among HCC and Non-HCC patients samples and their frequency
Cancer genome landscape
Oncogenes and tumor suppressor genes harbored pathogenic and non-pathogenic mutations. The percentage of unique TSGs mutations were higher (63%) than unique oncogenes mutations (37%) in HCC samples. Missense (60%) were more prevalent than frameshift (37%) in both with a fewer percentage of stop gained and splice site variants, and with a higher percentage of frameshift in TSGs relative to oncogenes (Supplement Table 7). Unique pathogenic variants with a Phred score exceeding 20, missense variants were sorted according to CHASMplus LIHC (Hepatocellular carcinoma) score in HCC samples, with Tier 1 and Tier 2 variants (TP53, CDKN2A, and MSH2) according to ACMG criteria by Franklin delineated in (Fig. 5) and in Supplement Table 8 including chromosome position mapped against GRCH37, targeted treatment, Phred score and zygosity. Unique pathogenic variants with Phred score more than 20, missense variants were sorted according to CHASMplus LIHC (Hepatocellular carcinoma) score in Non-HCC samples, with Tier 1 and Tier 2 variants (KMT2D and ATM) according to ACMG criteria by Franklin delineated in Fig. 6 and in Supplement Table 9.
Fig. 6.
Unique pathogenic variants in Oncogenes, tumor suppressor genes (TSGs) Non- HCC with Phred score > 20, Fs = frameshift-truncation, Ss = Splice site, T1 = Tier1, T2 = Tier 2, T3 = Tier 3, T4 = Tier4. A higher CHASM + LIHC score (liver hepatocellular carcinoma) in colored areas (CHASMplus LIHC score) indicates a higher possibility of being a driver mutation in Non-HCC
Pathways in Hepatocellular carcinoma
To reveal the potential role of mutated genes in biological process, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation using David annotation tools [27] in HCC samples, analysis of results is listed in Supplement file Go KEGG HCC. Results for the most significant KEGG pathways are shown in Table 7. For KEGG analysis, among the significantly annotated clusters were Notch signaling, Wnt signaling, PI3K-AKT pathway, Hippo signaling, Apelin signaling, Hedgehog (Hh) signaling, and MAPK signaling, in addition to ECM-receptor interaction, focal adhesion, and calcium signaling which are crucial landmarks in a hepatocellular carcinoma pathway. Pathogenic mutations in genes in all HCC samples, among HCC pathway, are illustrated in Supplement Fig. 2.
Table 7.
Enriched Kegg pathways in combined HCC samples
| Term | Gene count | % | P-values | Individual samples p < 0.05 | Individual samples p < 0.1 | Freq % |
|---|---|---|---|---|---|---|
| ECM-receptor interaction | 50 | 0.8 | 1.2E-6 | LT6, LT7, LT10, LT13, LT14, LT16, LT19, LT20 | 61% | |
| Focal adhesion | 86 | 1.4 | 3.6E-4 | LT6, LT11, LT13, LT14, LT16, LT17 | 46% | |
| Notch signaling | 33 | 0.5 | 4.1E-4 | LT7, LT8, | LT16 | 23% |
| Calcium signaling | 96 | 1.6 | 9.3E-3 | LT7, LT8 | LT17 | 23% |
| Wnt signaling | 68 | 1.1 | 1.4E-2 | LT8 | LT11 | 15% |
| PI3K-Akt signaling | 130 | 2.2 | 1.5E-2 | LT13, LT14 | LT10, LT19, LT20 | 38.4% |
| Apelin signaling | 39 | 1.0 | 2.3E-2 | LT8, LT13 | 15% | |
| Hippo signaling | 12 | 1.7 | 1.5E-2 | LT7 | 7.5% | |
| Hedgehog signaling | 11 | 0.7 | 2.0E-2 | LT13 | 7.5% | |
| MAPK signaling | 20 | 2.2 | 3.2E-2 | LT14 | 7.5% |
Tier 1 and Tier 2 variants
We performed database search on Franklin to reassure the somatic category of variants by other studies and to assess the category according to Association for Molecular Pathology (AMP) standards [26]. Eight variants proved to be (3, Tier 1 and 5, Tier 2) with a Phred score > 20 (TP53, CDKN2A, and MSH2 in individual HCC samples, Kit variant common in 3 HCC, KMT2D (3 variants in Non-HCC), and ATM in Non-HCC, in addition to other 15 variants with Phred score < 20 (KMT2D, NOTCH1, KMT2C, PIK3CA, KIT, SMARCA4, ATM, PTEN, MSH2, and PTCH1). These variants are rare in population studies as revealed by gnomAD frequency on OpenCravat less than 0.1 (Table 8). All the previous mutations are clinically targetable with specific inhibitors. A list of 134 variants (Supplement Table 10) Tier 1 and Ter 2 low frequency variants including those before, and other variants that were low confidence.
Table 8.
Tier1 and Tier2 variants in HCC and Non-HCC samples (GRCH37) all heterozygous, population frequency < 0.1
| Variant | Phred | Position | Sequence ontology/ VAF | ACMG category | HCC LT/Non-HCC LTN (VAF) | |
|---|---|---|---|---|---|---|
| 1 |
TP53 c.422G > A |
101 |
chr17 7,578,508 |
Missense | Tier1 | LT19 (22.95%) |
| 2 |
CDKN2A c.95 T > A |
25 |
chr9 21,974,732 |
Missense | Tier1 | LT14 (7.14%) |
| 3 |
MSH2 c.56del |
37.6 |
chr2 47,630,385 |
Frameshift | Tier2 | LT13(18.18%) |
| 4 |
KMT2D c.8713_8715delinsG |
20.4 |
chr12 49,432,424 |
Frameshift | Tier2 HP | LT11N (4.76%) |
| 5 |
KMT2D c.5135_5136del |
17.3 |
chr12 4,943,803 |
Frameshift | Tier2 HP | LT11N (3.86%) |
| 6 |
KMT2D c.5135del |
27.6 |
chr12 49,438,036 |
Frameshift | Tier2 HP | LT17N (10.29%) |
| 7 |
ATM c.6348-2A > G |
23 |
chr11 108,190,679 |
Splice site | Tier1 | LT20N (4.11%) |
| 8 |
KIT c.1621A > C p.Met541Leu |
119 | chr4: 55,593,464 | Missense | Tier2 | LT11, 18,19 (22%, 43%, 52%) |
| 9 |
KMT2D c.8455C > T |
8.7 | Chr12 49,432,684 | Stop_gained | Tier2 | LT7 (3.33%) |
| 10 |
KMT2D c.3553C > T |
11.8 | Chr12 49,443,818 | Stop_gained | Tier2 | LT7 (4.29%) |
| 11 |
NOTCH1 c.1773C > A |
8.9 | Chr9 139,410,065 | Stop_gained | Tier1 | LT8N (3.23%) |
| 12 |
KMT2C c.1084_1085insC |
8.3 | chr7-151,962,222 | Frameshift | Tier2 | LT10 (3.28%) |
| 13 |
PIK3CA c.1325_1326insG |
8.9 | Chr3 178,928,048 | Frameshift | Tier1 | LT11 (3.57%) |
| 14 |
KMT2D c.9279_9280insG |
7.7 | Chr12 49,431,859 | Frameshift | Tier2 | LT11N (2.2%) |
| 15 |
KIT c.1149dup |
8.5 | Chr4 55,575,619 | Frameshift | Tier1 | LT11N (4.76%) |
| 16 |
SMARCA4 c.3392del |
17.7 |
Chr 19 11,141,415 |
Frameshift | Tier2 | LT13 (7.73%) |
| 17 |
KMT2D c.15838_15839insC |
7.7 | Chr12 49,418,675 | Frameshift | Tier2 | LT14 (1.75%) |
| 18 |
SMARCA4 c.1834_1835insG |
7 | Chr19 11,113,727 | Frameshift | Tier2 | LT14 (1.85%) |
| 19 |
NOTCH1 c.3763_3764insC |
7.4 | Chr9 139,401,305 | Frameshift | Tier1 | LT17 (2.82%) |
| 20 |
ATM c.5870_5871insC |
8 | Chr11 108,180,994 | Frameshift | Tier1 | LT18N (3.17%) |
| 21 |
PTEN c.812 T > C |
7.3 | chr10 89,720,661 | Missense | Tier2 | LT18N (2.78%) |
| 22 |
MSH2 c.577C > T |
7.3 | Chr2 47,637,443 | Stop_gained | Tier2 | LT19 (2.78%) |
| 23 |
PTCH1 c.1963C > T |
8.5 | Chr9 98,231,320 | Stop-gained | Tier1 | LT7N (3.23%) |
Discussion
Currently, three types of NGS-based analytical methods are mainly used to identify genomic mutations: (i) whole exome sequencing (WES), (ii) whole-genome sequencing (WGS), and (iii) targeted sequencing (TS). This study was done to explore the mutational landscape in Egyptian patients with HCC using whole exome sequencing on both HCC and Non-HCC liver tissue.
In our study, missense was the highest prevalence, followed by synonymous variants with no significant difference between them, with frameshift and stop gained showing much lower percentages. This agrees with other cancer studies where the most called variants are commonly synonymous, followed by non-synonymous and splice site variants; however, the less common frameshift and stop gained/stop loss variants are more likely to have deleterious effects at the protein level, which guides variant prioritization. Although assumed benign and excluded, synonymous variants are claimed to harbor pathogenic properties in cancer, particularly concerning changes in protein expression and splicing [31].
The significantly lower level of variants in FFPE samples than in fresh tissue (FT) in our study can be explained by fragmentation, which is often extensive in formalin fixed samples. Thus, FFPE tissues have a significantly lower amount of amplifiable DNA templates and need special preparation kits for library preparation [32].
Regarding mutational signature, all single base substitution signatures (S1, S5, S6, S12, S17, S23) were previously observed in liver cancer, but with variable frequencies [21]. Signatures SBS1 and SBS5 are clock-like mutational processes that increase with age progress. Signatures 12, 17, and 23 are of unknown reason. Signature SBS1 (associated with spontaneous deamination) and signature SBS5 (associated with the activity of transcription-coupled nucleotide excision repair) [33]. These mutational processes are active in both normal genomes as well as in tumor cells. Signature 6 is associated with defective DNA mismatch repair and is found in microsatellite unstable tumors. It typically constitutes not more than 3% in cancers [34], but in our patients it constituted a high percentage with a median 14.2%. The mutational signature of FFPE samples is different from that of fresh samples. In their 2019 study, Bhagwate et al. also found that the DNA quality and quantity of FFPE samples are often sub-optimal, and resulting NGS-based genetics variant detections are prone to false positives, which could be the cause of the variation in mutational signature [35].
Hepatocellular carcinoma has a higher tumor mutation burden (TMB) than the average for other solid tumors. Tumor mutation burden is defined as the abundance of somatic mutations in a tumor, in the case of high TMB, immunotherapy can significantly improve overall survival than tumors with low TMB. The higher TMB differentiates the tumor from normal tissue, and exposes it to immune cells, thus it becomes more responsive to immunotherapy [36]. This is the reason we performed analysis of variants for both HCC and Non-HCC liver tissue to clarify the prevalence and type of variants present in both cancerous and non-cancerous liver tissue that increment TMB. Analysis of highly mutated genes in both HCC and Non-HCC revealed the presence of 8 highly mutated genes in HCC (AHNAK2, MUC16, TTN, MUC6, MUC12, ZNF717, OBSCN, PDE4DIP).
Mutations in AHNAK2 are listed in pathological specimens and databases from The Cancer Genome Atlas (TCGA) [29]. AHNAK2 (AHNAK nucleoprotein 2), also known as C14orf78, is a member of the AHNAK family. AHNAK mediates a negative regulation of cell growth and acts as tumor suppressor through potentiation of TGFβ signaling. Upregulated ANHAK2 activates the PI3K/AKT signaling pathway and promotes proliferation, migration, and invasion [37].
Analysis of highly mutated genes revealed that MUC16 is highly rich in somatic variants in HCC liver tissue and is ranked third among HCC and Non-HCC regarding the number of pathogenic variants arising in the gene in our study (supplement Tables 3 and 4). MUC16, an oncogene that encodes cancer antigen 125 (CA-125), sustains normal cell function, and its role in the development of numerous cancers is explained by being employed in activation of the p53 and DNA repair pathway. The critical role of the MUC16 gene in HCC is highlighted as MUC16 gene mutations which are enriched in various cancer-related pathways, such as cell cycle and metabolic processes. In addition, MUC16 mutation significantly increases TMB and represents an independent marker with high predictive value for HCC [38, 39]. In our study, AHNAK2, ZNF717, MAP2K3, HYDIN, OBSCN, PDE4DIP are among the highly mutated genes. TTN and OBSCN were most abundant in HCC and Non-HCC respectively. Our results partially agree with Shen et al. (2020) who performed WES on liver tissues collected from 10 HCC Chinese patients. Among the 25 mutant genes in their study including a different gene profile, the highest mutation frequency was found in HYDIN, TTN, OBSCN, and AHNAK2 [40].
In our data analysis, TTN and OBSCN were most frequent in HCC. In HCC samples, 11/13 (85%) samples had a commutant OBSCN and TTN while in Non-HCC 8/12 (67%) had mutation in both genes, with the remaining percentage showing only OBSCN pathogenic variants. Same observation was found previously in colorectal cancer (CRC) patients in TCGA study, TTN and OBSCN were commonly mutated, and mutations correlated with higher TMB and favorable overall survival. Patients with the commutation of TTN and OBSCN were categorized as ‘Double‐Hit’, patients with only one gene mutation TTN or OBSCN were labelled ‘Single‐Hit’, and patients with both genes wild‐type were categorized ‘Double‐WT’. Double‐Hit group revealed low tendency to malignant events, and highest TMB, immune cells infiltration abundance, as well as immune checkpoints expression compared with the other two phenotypes. This finding may extend to HCC but needs exploration through further studies; researchers suggest ‘immune‐hot’ category of tumors to have better immunotherapeutic efficacy and as a result a more favorable prognosis [41].
The HCC mutational landscape is characterized by both pronounced intra and inter-tumoral heterogeneity with a lack of a representative actionable oncogenic driver. Gene-specific filtering was performed on our WES data in both HCC and Non-HCC for the 26 significantly mutated genes in TCGA study [42] to expand data interrogation across the entire exome, and to perform candidate gene analysis based on what we obtained from up-to-date curated databases [43]. Although the Phred score for some of these variants was low, in most variants due to homopolymers errors by ion Proton semiconductor-based sequencing detection method, they passed variant assessment and had an accepted depth of coverage [44]. HCC development is mainly driven by inactivating mutations in various tumor-suppressor genes as TP53, AXIN1, ARID1A, ARID2, and CDKN2A; mutations in oncogenes as CTNNB1, PIK3CA, KRAS, NRAS, NEF2L2, which predispose hepatocytes to accumulate additional oncogenic changes that drive cancer initiation and tumorigenesis [42]. Our study shows concordant results with genes TP53, TERT, Azin1, KEAP1, BAP1, PIK3CA, APOB, ARID1A, CDKN2A, AXIN1, NFE2L2, CREB3L3, and RP1L1 showing variants with a high probability of being driver mutations in a variable number of our HCC cases having CScape score > 0.5 [30].
Exploring The Cancer Genome Atlas Significantly Mutated Genes (TCGA SMGs), in our study the most frequently mutated genes in HCC samples were APOB (53%), RP1L1 (69%), AHCTF1 (46%), AXIN1 (46%) and AHCTF1 (38%), KEAP1 (31%), NEF2L2 (31%), and PIK3CA (31%). The most frequently mutated genes in Non- HCC samples in our study were RP1L1 (50%), APOB (41.7%), TERT (41.7%), ARID1A (33%), CREB3L3 (33%), and AXIN1 (33%), some of the most established drivers of HCC [42]. In accordance with our results, mutated genes in our study were among the most frequently mutated group of HCC driver genes in TCGA. However, in TCGA the two most frequently mutated genes (> 25% of total cases), in their study on 363 HCC cases were tumor suppressor gene TP53 and the WNT pathway oncogene CTNNB1 [30]. Genes TP53 had unique pathogenic coding variants in 7.6% of HCC samples and 16.7% of Non-HCC, while CTNNB1 in 15.4% of HCC and 8.3% of Non-HCC. Genes involved in Wnt pathway as AXIN1 showed unique pathogenic variants in 46% of our HCC samples and were mutually exclusive to the activating β-catenin mutations except in one Non-HCC sample which showed both AXIN1 and CTNNB1 mutations [45]. The AXIN1 gene, on the other hand, is a negative regulator of the Wnt/β-catenin pathway. Mutually exclusive mutations occurring in AXINS are driver mutations that drive hepatocarcinogenesis in the stage of low-grade dysplastic nodules in cirrhotic liver. In their study, TCGA reported a low percentage of both NEF2L2 (3%) and KEAP1 (5%) and highlighted their vital role in cellular antioxidant defenses [9].
Variants were observed in HCC samples in TCGA SMGs in ARID1A (23%), CREB3L3 (23%), GPATCH4 (23%), IL6ST (23%), LZTR1 (23%), RB1 (23%), ARID2 (15%), AZIN1 (15.4%), BAP1 (15.4%), CTNNB1 (15.4%), EEF1A1 (15.4%), CDKN2A (7.6%), ACVR2A (7.6%), KRAS (7.6%), and TP53 (7.6%). In agreement with our results, driver mutational events were observed in TCGA study at the level of LZTR1, EEF1A1, ARID2, AZIN1, GPATCH4, CREB3L3, and ACVR2A genes [29]. Shibata, in 2021, added that low frequency mutated driver genes may trigger specific oncogenic phenotypes such as aggressive growth, invasiveness, or metastasis [46]. Driver mutations in ARID1A and ARID2 which are involved in chromatin remodeling occur early in the stage of high-grade dysplastic nodules preceding the development of HCC. One of the key pathways in HCC included cell cycle regulatory pathways driven by mutations in RB1, CDKN2A [9]. We found no variants in ALB, Shibata mentioned the uncertain role of frequent mutations of ALB and APOB in HCC. He explained that both mutations were enriched in indels and both genes are highly expressed in hepatocytes, these indels could be caused by replication errors [47]. In agreement with his explanation, out of 15 variants in APOB in our study, 11 were indels in HCC and 8/14 variants in Non-HCC samples were frameshift caused by indels. Both ALB and APOB genes are key mediators of hepatocyte function in the secretion of albumin and VLDL. Their role requires a high portion of hepatocyte transcriptional, translational, and energy supplies. This might explain APOB mutation and suppression by the malignant hepatocyte to reserve resources for cell division requirements [46].
Analysis of variants regarding the Cancer Genome Landscape revealed the presence of Tier1 variants in TP53 and CDKN2A, both are driver genes in HCC [41], in addition to Tier 2 variant in MSH2, the DNA mismatch repair gene whose defects can contribute to cancer development. In their 2018 study, Zhu et al., highlighted the role of rs2303428 of MSH2 in HCC prognosis [48]. This is an intron variant that was observed in our study in both HCC (VAF 63%) and non-HCC (VAF 54%) samples of one patient HCV-HCC. It is mainly identified in constitutional germline dependent hereditary cancers and as a somatic mutation in some sporadic cancers. As we traced in our study exome, we filtered for pathogenic coding variants in addition to splice site variants. Further studies are needed to determine the impact on prognosis. In addition, a splice site variant Tier 1 was identified in ATM gene, and 2 Tier 2 variants frameshift in KMT2D in Non-HCC samples. These tumor suppressor genes represent driver mutations in HCC, previous studies revealed many low-frequency somatic mutations that affect multiple genes, including cell cycle control as ATM in 6% of HCC patients, and recurrent inactivating mutations of members of the chromatin remodeling gene family as KMT2D in 6% of HCC patients [27].
Data analysis of the identified KEGG pathways for somatic unique variants in HCC samples highlights mutated genes’ roles during tumorigenesis, revealing biological processes and pathways implicated in carcinogenesis.
Extracellular matrix ECM-receptor interaction (61%) and Focal adhesions (46%) are significantly enriched in our HCC samples. Focal adhesions are structural links between the extracellular matrix (ECM) and actin cytoskeleton. Integrins interact with multiple proteins at cell-ECM adhesion sites forming focal adhesions (FAs). These adhesions orchestrate significant cancer related functions including cell proliferation and survival, cell invasion and epithelial mesenchymal transition. Understanding the pathogenesis of tumor cell motility can pave the way to effective therapeutic targeting to prevent cancer progression [49]. Calcium signaling was enriched in 23% of HCC samples, this pathway is reported to be associated with liver-specific diseases such as HCC, cholestasis, hepatitis, and nonalcoholic fatty liver disease (NAFLD). Calcium signaling is coordinated by ~ 1600 genes, the ultimate role of these genes is to maintain the intracellular calcium homeostasis and normal cell function. Altered calcium signaling genes expression is implicated in cancer hallmarks, such as altered cell metabolism, sustained cell proliferation, cell death resistance, angiogenesis, invasion, and metastasis [50].
Pathways PIK3AKT and Wnt had the highest number of genes enriched (130 and 68, respectively) with PIK3AKT having a frequency 38.4% in HCC samples. Regarding PI3K-Akt, several studies documented mutations of the phosphatidylinositol 3-kinase (PI3K)/the serine-threonine protein kinase (Akt)/mammalian target of the rapamycin (mTOR) signaling pathway in HCC. The PI3K/Akt/mTOR pathway is an important signaling mechanism that regulates the cell cycle, proliferation, apoptosis, and metabolism. The pathway is often dysregulated in HCC which promotes the survival and proliferation of tumor cells [51].
Signaling pathway Notch is enriched in 23% of HCC patients. Mutation in target genes of the Notch pathway is reported in HCC and it is notable that the Notch pathway has controversial effects on HCC. This pathway crosstalks with the Wnt pathway for cancer stem cell (CSC) maintenance, the PI3K/mTOR pathways for HCC proliferation, and the VEGF pathway for angiogenesis [52].
Apelin and Wnt signaling pathways are enriched in 15% of HCC patients. Activated β-catenin activates the transcription of target genes modulating the process of carcinogenesis. These genes are involved in CSC maintenance, proliferation, and epithelial mesenchymal transition (EMT) [53]. The apelin signaling pathway is a G protein-coupled receptor (GPCR) pathway that plays a crucial role in regulating various physiological processes such as angiogenesis, and energy metabolism that are involved in carcinogenesis. The apelin signaling pathway interacts with other pathways as the Wnt/β-catenin pathway, the PI3K/Akt/mTOR pathway, and the Hippo pathway to regulate the development of HCC [54].
Signaling pathways Hippo, Hedgehog (Hh) and MAPK were revealed to be enriched in 3 HCC patients. Hippo is a classical kinase cascade that phosphorylates the Mst1/2-sav1 complex and activates the phosphorylation of the Lats1/2-mob1A/B complex for inactivating Yap and Taz. Yap/Taz is the main effector molecule, which is downstream of the Hippo pathway, and its abnormal activation is related to a variety of human cancers including HCC [55]. In adult healthy liver, Hh signaling is inactive, because mature hepatocytes hardly express Hh ligands. Hh signaling is reactivated in liver diseases, activation enhances transition of quiescent hepatic stellate cells (HSCs) to myofibroblast (MF), which regenerate the liver epithelial cells. The Hh pathway can be activated by inactivating mutations of PTCH1 [56]. The mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK-ERK) is one of the molecular signaling pathways that are critical to tumor initiation, progression, and metastasis in HCC. It is a cascade of protein kinases that transmits signals from the cell surface to the nucleus. In HCC, the MAPK/ERK signaling pathway is activated in more than 50% of human HCC cases [57].
In our study, we demonstrated the presence of different unique mutations in every patient between HCC and Non-HCC samples, and interpatient molecular differences between different studied patients. Tumor heterogeneity is a hallmark of hepatocellular carcinomas (HCCs), that poses a significant challenge to the development of effective therapeutic solutions in HCC. Inter-tumor heterogeneity that can be related to de novo independent carcinogenesis on a background of cirrhosis and/or intrahepatic metastasis. The presence of variants in Non-HCC can be attributed to either intrahepatic metastases which is reported in previous studies in 20 to 40% of patients, or de novo carcinogenesis on cirrhosis [9]. We explored gene variants in significant pathways, and we demonstrated the presence of considerable heterogeneity in different liver tissues HCC and Non-HCC in the same patient at the DNA sequence level. These patients belong to the same Geographic region but have different etiologies, however they shared most KEGG pathways, except for Hh pathway in HBV HCC. Somatic mutations in similar loci of genes that are clinically actionable even though of different sequence ontology may serve as important therapeutic targets [58].
Tier 1 and Tier 2 mutations
All variants in genes showing clinically actionable Tier1 and Tier 2 variants that represent driver mutations in HCC are involved in driver pathways for HCC which we discussed earlier PI3K/mTOR, Wnt, NOTCH, Hedgehog, and MAPK [52]. In addition, some are determined through cell cycle control, chromatin remodeling and oxidative stress to be involved in the process of carcinogenesis in HCC [28]. We extended the list in supplement files, to include variants within driver genes that are abundant on COSMIC and are targetable [24, 52]. Although frameshift variants represented only 1.6% of all variants in WES, they all lied within driver genes and genes implicated in the process of carcinogenesis [28] in our patients and represented the dominant sequence ontology among pathogenic variants. Some of them on region viewer or on Franklin were found to lie in homopolymers regions [43]. These variants in addition to a considerable number of medium VAF, high and medium confidence Tier 3 variants in every HCC patient require further assessment and consideration of their clinical significance.
Conclusion
Mutational signature was mostly found in S1, S5, S6, and S12 in HCC. Analysis of highly mutated genes revealed the presence of 10 common highly mutated genes in HCC and Non-HCC (AHNAK2, MUC6, MUC16, TTN, ZNF17, FLG, MUC12, OBSCN, PDE4DIP, MUC5b, and HYDIN). Among the 26 significantly mutated HCC genes identified by TCGA, APOB and RP1L1 showed the highest number of mutations in both HCC and Non-HCC tissues. Tier 1, Tier 2 variants in TCGA SMGs in HCC and Non-HCC (TP53, PIK3CA, CDKN2A, and BAP1). Cancer Genome Landscape analysis revealed multiple variants added (MSH2) in HCC and (KMT2D and ATM) in Non-HCC. For KEGG analysis, among the significantly annotated clusters in HCC were Notch signaling, Wnt signaling, PI3K-AKT pathway, Hippo signaling, Apelin signaling, Hedgehog (Hh) signaling, and MAPK signaling, in addition to ECM-receptor interaction, focal adhesion, and calcium signaling. Tier1 and Tier 2 variants KIT, KMT2D, NOTCH1, KMT2C, PIK3CA, KIT, SMARCA4, ATM, PTEN, MSH2, and PTCH1 low frequency variants in both HCC and Non-HCC. Further assessment and confirmation for these variants and other Tier 3 ones are recommended.
Supplementary Information
Acknowledgements
All Staff members of Ain Shams Center for Organ Transplantation (ASCOT).
Abbreviations
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- ITH
Intra-tumor heterogeneity
- NGS
Next-generation sequencing
- WES
Whole exome sequencing
- SNPs
Single nucleotide polymorphisms
- EASL
European association for study of liver diseases
- UCSF
University of California San Francisco criteria
- BCLC
Barcelona clinic for cancer staging
- CR
Complete response
- ST
Stable disease
- mRECIST
Modified Response Evaluation Criteria in Solid Tumors (RECIST) criteria
- FFPE
Fresh frozen paraffin embedded
- TCGA
The Cancer Genome Atlas
Authors’ contributions
P.H.K and M.E.M chose the idea,R.M., D.A, R.A.,K.M. did the lab workup, P.H.K. M.F., M.A. prepared the figures, P.H.K, R.M,D.A. prepared the tables, P.H.K., I.M. wrote the main manuscript,I.M.,M.S., Y.M. revised the clinical data and follow up of patients, M.E.M., M.B. did the surgical procedures, M.I. prepared the pathological specimens and read the pathology, A.A. analyzed the data files for somatic extraction, M.A.H. supervised the bioinformatics and raw data files and supervised the work, P.H.K. do the data analysis., performed data analysis to assign the driver mutations and mutational signatures and pathways S.E.K supervised and consultant the project. All authors reviewed the manuscript.
Funding
This work was supported by Science and Technology Development Fund (STDF), Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229).
Availability of data and material
The data can be available on reasonable request from the corresponding author.
Declarations
Ethics approval and consent to participate
This study was reviewed and approved by the Research Ethics Committee of the Faculty of Medicine, Ain Shams University (FMASU R92/2020). All participates have assigned written informed consent form to participate in this study. The study was performed in accordance with the 1964 Declaration of Helsinki and all subsequent revisions.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
The original version of this article has been revised: Tables 2, 3, 6 have been revised to reflect their original format.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/12/2024
A Correction to this paper has been published: 10.1186/s12920-024-02045-9
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics: GLOBOCAN estimates of incidence andmortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2.Elghazaly H, GabAllah A, Eldin NB. P-019 Clinic-pathological pattern of hepatocellular carcinoma (HCC) in Egypt. Ann Oncol. 2018;29. 10.1093/annonc/mdy151.018.
- 3.Wang H, Zhou H, Li X, Wang P, Liu G, Liu W, et al. Detection of tumor- related biomarkers in hepatocellular carcinoma patients by sequencing circulating cell-free DNA. Clin Oncol. 2019;4:1645. [Google Scholar]
- 4.Maloberti T, De Leo A, Sanza V, Gruppioni E, Altimari A, Riefolo M, Visani M, Malvi D, D’Errico A, Tallini G, Vasuri F, de Biase D. Correlation of molecular alterations with pathological features in hepatocellular carcinoma: literature review and experience of an Italian center. World J Gastroenterol. 2022;28(25):2854–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C. J Hepatol. 2018;69(2):461–511. [DOI] [PubMed] [Google Scholar]
- 6.Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol. 2022;84:69-79. 10.1016/j.semcancer.2022.03.013. [DOI] [PubMed]
- 7.Ganesamoorthy D, Robertson AJ, Chen W, Hall MB, Cao MD, Ferguson K, Coin LJ. Whole genome deep sequencing analysis of cell-free DNA in samples with low tumour content. BMC Cancer. 2022;22(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunadirek P, Chuaypen N, Jenjaroenpun P, Wongsurawat T, Pinjaroen N, Sirichindakul P, Tangkijvanich P. Cell-free DNA analysis by whole-exome sequencing for hepatocellular carcinoma: a pilot study in Thailand. Cancers. 2021;13(9):2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72(2):215–29. 10.1016/j.jhep.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Imamura T, Okamura Y, Ohshima K, et al. Overview and clinical significance of multiple mutations in individual genes in hepatocellular carcinoma. BMC Cancer. 2022;22:1046. 10.1186/s12885-022-10143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59(1):5–15. [DOI] [PubMed] [Google Scholar]
- 12.Botstein D, Risch N. (2003): Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–37. [DOI] [PubMed] [Google Scholar]
- 13.Tetreault M, Bareke E, Nadaf J, Alirezaie N, Majewski J. Whole-exome sequencing as a diagnostic tool: current challenges and future opportunities. Expert Rev Mol Diagn. 2015;15(6):749–60. [DOI] [PubMed] [Google Scholar]
- 14.Mazzaferro V. et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. New England J Med. 1996;334(11). 10.1056/nejm199603143341104. [DOI] [PubMed]
- 15.Yao FY et al. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33(6). 10.1053/jhep.2001.24563. [DOI] [PubMed]
- 16.Llovet JM, Lencioni R. mRECIST for HCC: performance and novel refinements. J Hepatol. 2020;72(2):288–306. 10.1016/j.jhep.2019.09.026. PMID: 31954493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reig M, Forner A, Rimola J, Ferrer-Fàbrega J, Burrel M, Garcia-Criado Á, Kelley RK, Galle PR, Mazzaferro V, Salem R, Sangro B, Singal AG, Vogel A, Fuster J, Ayuso C. Bruix J (2022): BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76(3):681–93. 10.1016/j.jhep.2021.11.018. Epub 2021 Nov 19 PMID: 34801630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yau T, Tang VY, Yao TJ, Fan ST, Lo CM, Poon RT. Development of Hong Kong liver cancer staging system with treatment stratification for patients with hepatocellular carcinoma. Gastroenterology. 2014;146:1691-1700.e3 PMID: 24583061. [DOI] [PubMed] [Google Scholar]
- 19.Lucena-Aguilar G, Sánchez-López AM, Barberán-Aceituno C, Carrillo-Ávila JA, López-Guerrero JA, Aguilar-Quesada R. DNA source selection for downstream applications based on DNA quality indicators analysis. Biopreserv Biobank. 2016;14(4):264–70. 10.1089/bio.2015.0064. Epub 2016 May 9. PMID: 27158753; PMCID: PMC4991598. [DOI] [PMC free article] [PubMed]
- 20.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6(2):80â “92. 10.4161/fly.19695. [DOI] [PMC free article] [PubMed]
- 21.Lee J, Lee AJ, Lee JK, Park J, Kwon Y, Park S, Chun H, Ju YS, Hong D. Mutalisk: a web-based somatic MUTation AnaLyIS toolKit for genomic, transcriptional and epigenomic signatures. Nucleic Acids Res. 2018;46(w1):W102–8. 10.1093/nar/gky406. [DOI] [PMC free article] [PubMed]
- 22.Pagel KA, et al. Integrated informatics analysis of cancer-related variants. JCO Clinical Cancer Informatics. 2020;4:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. 10.1016/j.jmoldx.2016.10.002. PMID: 27993330; PMCID: PMC5707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–21. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65(5):1031–42. 10.1016/j.jhep.2016.05.035. Epub 2016 Jun 2 PMID: 27262756. [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327–1341.e23. 10.1016/j.cell.2017.05.046. PMID: 28622513; PMCID: PMC5680778. [DOI] [PMC free article] [PubMed]
- 30.Rogers MF, Shihab H, Gaunt TR, Campbell C. CScape: a tool for predicting oncogenic single-point mutations in the cancer genome. Nature Scientific Rep. 2017. http://cscape.biocompute.org.uk/ [DOI] [PMC free article] [PubMed]
- 31.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–91. [DOI] [PubMed] [Google Scholar]
- 32.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71. 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuravleva E, O’Rourke CJ, Andersen JB. Mutational signatures and processes in hepatobiliary cancers. Nat Rev Gastroenterol Hepatol. 2022;19:367–82. 10.1038/s41575-022-00587-w. [DOI] [PubMed] [Google Scholar]
- 35.Bhagwate AV, Liu Y, Winham SJ, et al. Bioinformatics and DNA-extraction strategies to reliably detect genetic variants from FFPE breast tissue samples. BMC Genomics. 2019;20:689. 10.1186/s12864-019-6056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie C, Wu H, Pan T, Zheng X, Yang X, Zhang G, Lian Y, Lin J, Peng L. A novel panel based on immune infiltration and tumor mutational burden for prognostic prediction in hepatocellular carcinoma. Aging (Albany NY). 2021;10;13(6):8563–8587. 10.18632/aging.202670. Epub 2021 Mar 10. PMID: 33714200; PMCID: PMC8034943. [DOI] [PMC free article] [PubMed]
- 37.Wang DW, Zheng HZ, Cha N, Zhang XJ, Zheng M, Chen MM, Tian LX. Down-regulation of AHNAK2 inhibits cell proliferation, migration and invasion through inactivating the MAPK pathway in lung adenocarcinoma. Technol Cancer Res Treat. 2020;19:1533033820957006. 10.1177/1533033820957006. PMID: 33000678; PMCID: PMC7533926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B, Dong Z, Lu Y, Ma J, Ma Z, Wang H. Prognostic value of MUC16 mutation and its correlation with immunity in hepatocellular carcinoma patients. Evid Based Complement Alternat Med. 2022;3478861. 10.1155/2022/3478861. PMID: 36034941; PMCID: PMC9410786. [DOI] [PMC free article] [PubMed] [Retracted]
- 39.Huang Y, Huang X, Zeng J, Lin J. Knockdown of MUC16 (CA125) enhances the migration and invasion of hepatocellular carcinoma cells. Front Oncol. 2021;11:667669. 10.3389/fonc.2021.667669. PMID: 34150633; PMCID: PMC8208084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen J, Qi L, Zou Z, Du J, Kong W, Zhao L, Wei J, Lin L, Ren M, Liu B. Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases. Sci Rep. 2020;10(1):4435. 10.1038/s41598-020-61298-3. PMID: 32157118; PMCID: PMC7064516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Wang L, Guo C, Liu L, Jiao D, Sun Z, Wu K, Zhao Y, Han X. TTN/OBSCN 'Double-Hit' predicts favourable prognosis, 'immune-hot' subtype and potentially better immunotherapeutic efficacy in colorectal cancer. J Cell Mol Med. 2021;25(7):3239–3251. 10.1111/jcmm.16393. Epub 2021 Feb 23. PMID: 33624434; PMCID. [DOI] [PMC free article] [PubMed]
- 42.Tsui YM, Chan LK, Ng IOL. Cancer stemness in hepatocellular carcinoma: mechanisms and translational potential. Br J Cancer. 2020;122:1428–40. 10.1038/s41416-020-0823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seaby EG, Pengelly RJ, Ennis S. Exome sequencing explained: a practical guide to its clinical application. Brief Funct Genomics. 2016;15(5):374–84. 10.1093/bfgp/elv054. [DOI] [PubMed] [Google Scholar]
- 44.Feng W, Zhao S, Xue D, et al. Improving alignment accuracy on homopolymer regions for semiconductor-based sequencing technologies. BMC Genomics. 2016;17(Suppl 7):521. 10.1186/s12864-016-2894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Javanmard D, Najafi M, Babaei MR, et al. Investigation of CTNNB1 gene mutations and expression in hepatocellular carcinoma and cirrhosis in association with hepatitis B virus infection. Infect Agents Cancer. 2020;15:37. 10.1186/s13027-020-00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata,. T. Genomic landscape of hepatocarcinogenesis. J Hum Genet. 2021;66:845–51. 10.1038/s10038-021-00928-8. [DOI] [PubMed] [Google Scholar]
- 47.Liang N, Yang T, Huang Q, et al. Mechanism of cancer stemness maintenance in human liver cancer. Cell Death Dis. 2022;13:394. 10.1038/s41419-022-04848-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X, Wang Z, Qiu X, Wang W, Bei C, Tan C, Qin L, Ren Y, Tan S. Rs2303428 of MSH2 is associated with hepatocellular carcinoma prognosis in a chinese population. DNA Cell Biol. 2018;37(7):634–641. 10.1089/dna.2018.4224. Epub 2018 Jun 6. PMID: 29874113. [DOI] [PubMed]
- 49.Geramoutsou C, Nikou S, Karavias D, Arbi M, Tavlas P, Tzelepi V, Lygerou Z, Maroulis I, Bravou V. (2022): Focal adhesion proteins in hepatocellular carcinoma: RSU1 a novel tumour suppressor with prognostic significance. Pathol Res Pract. 2022;235:153950. 10.1016/j.prp.2022.153950. Epub 2022 May 23 PMID: 35642986. [DOI] [PubMed] [Google Scholar]
- 50.Hernández-Oliveras A, Izquierdo-Torres E, Hernández-Martínez G, Zarain-Herzberg Á, Santiago-García J. Transcriptional and epigenetic landscape of Ca2+-signaling genes in hepatocellular carcinoma. J Cell Commun Signal. 2021;15(3):433–445. 10.1007/s12079-020-00597-w. Epub 2021 Jan 4. PMID: 33398721; PMCID: PMC8222487. [DOI] [PMC free article] [PubMed]
- 51.Sun EJ, Wankell M, Palamuthusingam P, McFarlane C, Hebbard L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines. 2021;9:1639. 10.3390/biomedicines9111639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farzaneh Z, Vosough M, Agarwal T, et al. Critical signaling pathways governing hepatocellular carcinoma behavior; small molecule-based approaches. Cancer Cell Int. 2021;21:208. 10.1186/s12935-021-01924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165. 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Wang X, Yang Y. Hepatic Hippo signaling inhibits development of hepatocellular carcinoma. Clin Mol Hepatol. 2020;26(4):742–50. 10.3350/cmh.2020.0178. PMID: 32981290; PMCID: PMC7641559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi H, Zou Y, Zhong W, et al. Complex roles of Hippo-YAP/TAZ signaling in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2023;149:15311–22. 10.1007/s00432-023-05272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao L, Zhang Z, Zhang P, Yu M, Yang T. Role of canonical Hedgehog signaling pathway in liver. Int J Biol Sci. 2018;14(12):1636. 10.7150/ijbs.28089. PMID: 30416378; PMCID: PMC6216024. [DOI] [PMC free article] [PubMed]
- 57.Moon H, Ro SW. MAPK/ERK signaling pathway in hepatocellular carcinoma. Cancers (Basel). 2021;13(12):3026. 10.3390/cancers13123026. PMID: 34204242; PMCID: PMC8234271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholas TJ, Cormier MJ, Huang X, Qiao Y, Marth GT, Quinlan AR. OncoGEMINI: software for investigating tumor variants from multiple biopsies with integrated cancer annotations. Genome Med. 2021;13(1):46. 10.1186/s13073-021-00854-6. PMID: 33771218; PMCID: PMC7995589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data can be available on reasonable request from the corresponding author.