Abstract
The 4 202 353 bp genome of the alkaliphilic bacterium Bacillus halodurans C-125 contains 4066 predicted protein coding sequences (CDSs), 2141 (52.7%) of which have functional assignments, 1182 (29%) of which are conserved CDSs with unknown function and 743 (18.3%) of which have no match to any protein database. Among the total CDSs, 8.8% match sequences of proteins found only in Bacillus subtilis and 66.7% are widely conserved in comparison with the proteins of various organisms, including B.subtilis. The B.halodurans genome contains 112 transposase genes, indicating that transposases have played an important evolutionary role in horizontal gene transfer and also in internal genetic rearrangement in the genome. Strain C-125 lacks some of the necessary genes for competence, such as comS, srfA and rapC, supporting the fact that competence has not been demonstrated experimentally in C-125. There is no paralog of tupA, encoding teichuronopeptide, which contributes to alkaliphily, in the C-125 genome and an ortholog of tupA cannot be found in the B.subtilis genome. Out of 11 σ factors which belong to the extracytoplasmic function family, 10 are unique to B.halodurans, suggesting that they may have a role in the special mechanism of adaptation to an alkaline environment.
INTRODUCTION
Generally, alkaliphilic Bacillus strains cannot grow or grow poorly under neutral pH conditions, but grow well at pH >9.5. Since 1969 we have isolated a great number of alkaliphilic Bacillus strains from various environments and have purified many alkaline enzymes (1). Over the past two decades our studies have focused on the enzymology, physiology and molecular genetics of alkaliphilic microorganisms to elucidate their mechanisms of adaptation to alkaline environments. Industrial applications of these microbes have been investigated and some enzymes, such as proteases, amylases, cellulases and xylanases, have been commercialized. It is well recognized that these commercial enzymes have brought great advantages to industry (1). Thus, it is clear that alkaliphilic Bacillus strains are quite important and interesting not only academically but also industrially.
An alkaliphilic bacterium, strain C-125 (JCM9153), isolated in 1977, was identified as a member of the genus Bacillus and reported as a β-galactosidase (2) and xylanase producer (3). It is the most thoroughly characterized strain, physiologically, biochemically and genetically, among those in our collection of alkaliphilic Bacillus isolates (1). Recently, this strain was re-identified as Bacillus halodurans based on the results of 16S rDNA sequence and DNA–DNA hybridization analyses (4).
Analysis of the entire genome of Bacillus subtilis, which is taxonomically related to alkaliphilic B.halodurans strain C-125 (Fig. 1), except for the alkaliphilic phenotype, has been completed (5). Knowledge of the complete nucleotide sequence of the B.subtilis genome will definitely facilitate identification of common functions in bacilli and specific functions in alkaliphilic Bacillus strains. We have determined the complete genomic sequence of alkaliphilic B.halodurans C-125 and its genome is compared with that of B.subtilis in this study. Alkaliphilic B.halodurans is the second Bacillus species whose whole genomic sequence has been completely defined.
Figure 1.
General view of the alkaliphilic B.halodurans C-125 chromosome. Arrows indicate the direction of transcription. Genes are colored according to their classification into nine broad categories (green, protein conserved in comparison with other organisms, including B.subtilis; striped green, unknown; red, protein conserved in comparison with organisms other than B.subtilis; striped red, protein conserved in comparison with organisms other than members of Bacillus; striped yellow, protein conserved in comparison with other members of Bacillus, including B.subtilis; striped blue, protein conserved in comparison only with B.subtilis; striped pink, protein related to alkaliphily in C-125; purple, protein conserved in comparison with members of Bacillus other than B.subtilis; see Table S1). Ribosomal RNA genes are colored purple and shown as thin arrows. tRNA genes are marked as vertical bars on the line.
MATERIALS AND METHODS
Preparation of the whole genome shotgun library
A 20 µg aliquot of chromosomal DNA was sonicated for 5–25 s with a Bioruptor UCD-200TM (Tosho Denki Co., Japan). The sonicated DNA fragments were blunt-ended using a DNA blunting kit (Takara Shuzo, Kyoto, Japan) and fractionated by 1% agarose gel electrophoresis. DNA fragments 1–2 kb in length were excised from the gel and eluted by the freeze–squeeze method (6). The DNA recovered was ligated to the SmaI site of pUC18, which had been previously treated with BAP, and introduced into competent XL1-Blue cells by the standard method (7). Transformants with a frequency of 5–6 × 105/µg DNA were cultivated in LB liquid medium at 37°C and 1 µl of culture broth was used for template DNA. The insert in the plasmid, amplified using a standard PCR method, was used for sequencing.
Sequencing and assembly of the whole genome shotgun library
The genome of alkaliphilic B.halodurans C-125 was basically sequenced by the whole genome random sequencing method as described (8–10). The DNA fragment inserted into pUC18 was amplified by PCR using M13-20 and reverse primers. PCR fragments, treated with exonuclease I and shrimp alkaline phosphatase (Amersham, OH, USA) to eliminate excess primers in the PCR reaction mixture, were used for sequencing analysis as template DNA. Sequencing was performed with an ABI Prism 377 DNA sequencer using a Taq Dye Terminator Cycle Sequencing Kit (Perkin Elmer, CT, USA). DNA sequences determined by means of the ABI sequencer were assembled into contigs using Phrap (http://bozeman.mbt. washington.edu/phrap.docs/phrap.html ) with default parameters and without quality scores. At a statistical coverage of 7.1-fold, the assembly using Phrap yielded 656 contigs. Sequences were obtained from both ends of 2000 randomly chosen clones from a λ library (11). These sequences were then assembled with consensus sequences derived from the contigs of random phase sequences using Phrap. Gaps between contigs were closed by shotgun sequencing of λ clones which bridged the contigs of random phase sequences. The final gaps were closed by direct sequencing of the products amplified by long accurate PCR with a LA PCR Kit v.2 (Takara Shuzo).
Annotation and clustering
The predicted protein coding regions were initially defined by searching for open reading frames (ORFs) longer than 100 codons using the Genome Gambler program (10). Coding potential analysis of the entire genome was performed with the GeneHacker Plus programs using hidden Markov models (12) trained with a set of B.halodurans ORFs longer than 300 nt. This program evaluates quality of the Shine–Dalgano sequence (SD) and codon usage for a series of two amino acids. The SD sequence was complementary to one found at the 3′-end of 16S rRNA. The SD sequence (UCUUUCCUCCACUAG…) of alkaliphilic B.halodurans C-125 (13) is the same as that of B.subtilis. Searches of the protein databases for amino acid similarities were performed using BLAST2 sequence analysis tools (14) with subsequent comparison of protein coding sequences (CDSs) showing significant homology (>10–5 significance) performed using the Lipman–Pearson algorithm (15). Significant similarity was defined as at least 30% identity observed over 60% of the CDS, although those CDSs showing <30% identity over >60% of the protein were also included. A search for paralogous gene families, such as σ factors, ATPases, antiporters and ATP-binding cassette (ABC) transporters, in the B.halodurans genome was performed with stepwise BLAST2P, identifying pairwise matches above P < 10–5–10–80 over 50% of the query search length, and subsequently by single linkage clustering of these matches into multigene families (16).
RESULTS AND DISCUSSION
Genome analysis
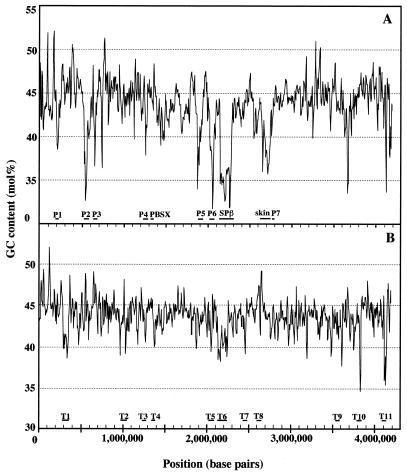
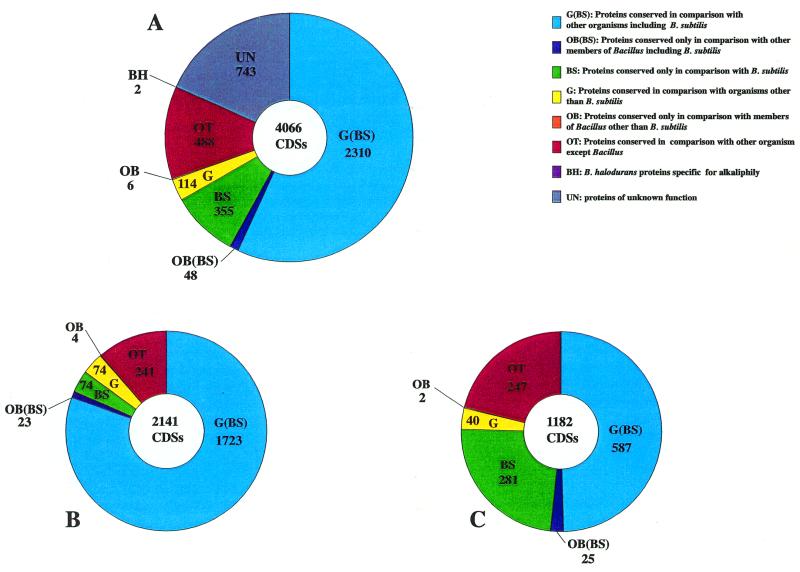
The genome of B.halodurans is a single circular chromosome (17) consisting of 4 202 353 bp (Fig. 1) with an average G+C content of 43.7% (Table 1). The G+C content of DNA in the coding regions and non-coding regions is 44.4 and 39.8%, respectively. On the basis of analysis of the G+C ratio and G–C skew (G–C/G+C), we estimated that the site of termination of replication (terC) is nearly 2.2–2.3 Mb (193°) from the origin, but we could not find the gene encoding the replication termination protein (rtp) in the genome of B.halodurans. Several A+T-rich and G+C-rich islands are likely to reveal the signature of transposons or other inserted elements (Fig. 2). We identified 4066 CDSs (Fig. 1 and Supplementary Table S1 available at NAR Online), on average 877 nt in size, using the coding region analysis program GeneHacker Plus (12) and the Genome Gambler system (10). We have not annotated CDSs that largely or entirely overlap existing genes. It was found that the termination codon in BH1054, annotated as a transposase, disappeared due to a frameshift and as a result this CDS was combined with the adjacent CDS, presumably coding for an ABC transporter/ATP-binding protein. We identify it as a gene coding for a transposase in this case. Coding sequences cover 85% of the chromosome. We found that 78% of the genes started with ATG, 10% with TTG and 12% with GTG, as compared with 87, 13 and 9%, respectively, in the case of B.subtilis (Table 1). The average size of the predicted proteins in B.halodurans is 32.841 kDa, ranging from 1.188 to 199.106 kDa. Predicted protein sequences were compared with sequences in a non-redundant protein database and biological roles were assigned to 2141 (52.7%) of them. In this database search 1182 predicted coding sequences (29.1%) were identified as conserved proteins of unknown function in comparison with proteins from other organisms, including B.subtilis, and for 743 (18.3%) there was no database match (Table 1). Among all of the CDSs found in the B.halodurans genome, 2310 (56.8%) were widely conserved in other organisms, including B.subtilis, and 355 (8.7%) of the CDSs matched the sequences of proteins found only in B.subtilis (Fig. 3). The ratio of proteins conserved in various organisms, including B.subtilis, among functionally assigned CDSs (2141) and among the CDSs (1182) matched with hypothetical proteins from other organisms was 80.5 and 49.7%, respectively, as shown in Figure 3. Of 1183 CDSs, 23.8% matched hypothetical proteins found only in the B.subtilis database, showing relatively high similarity values (Fig. 3).
Table 1. Comparison of the general features of the B.halodurans and B.subtilis genomes.
| Genome features | B.halodurans | B.subtilisa |
|---|---|---|
| Size (bp) | 4 202 353 | 4 214 810 |
| G+C content (mol%) | ||
| Total genome | 43.7 | 43.5 |
| Coding region | 44.4 | 44.3 |
| Non-coding region | 39.8 | 39.3 |
| Open reading frames | ||
| Percent of genome (coding) | 85 | 87 |
| Predicted no. | 4066 | 4100 |
| Conserved with function assigned | 2144 | 2379 |
| Conserved with unknown function | 1182 | 668 |
| Non-conserved | 743 | 1053 |
| Percent AUG initiation codons | 78 | 78 |
| Percent GUG initiation codons | 12 | 9 |
| Percent UUG initiation codons | 10 | 13 |
| Insertion elements | ||
| PBSX prophage-related protein | 2 | 27 |
| Transposase and related protein | 112 | 10 |
| RNA elements | ||
| Stable RNA (percent of genome) | 1.02 | 1.27 |
| 16S, 23S and 5S rRNA | 8 | 10 |
| tRNA | 78 | 86 |
aKunst et al. (5).
Figure 2.
Comparison of G+C profile between the genomes of B.halodurans and B.subtilis. Distribution of A+T-rich islands along the chromosome, in sliding windows of 10 000 nt, with a step of 5000 nt. Known phage (PBSX, SPβ and skin) are indicated by their names and prophage-like elements in the B.subtilis genome are numbered from P1 to P7. The A+T-rich or G+C-rich regions containing transposases in the B.halodurans genome are also indicated from T1 to T11. (A) B.subtilis; (B) B.halodurans.
Figure 3.
Summary of conserved CDSs identified in the genome of B.halodurans C-125. (A) All CDSs; (B) CDSs with a known functional role; (C) CDSs of unknown function
General features
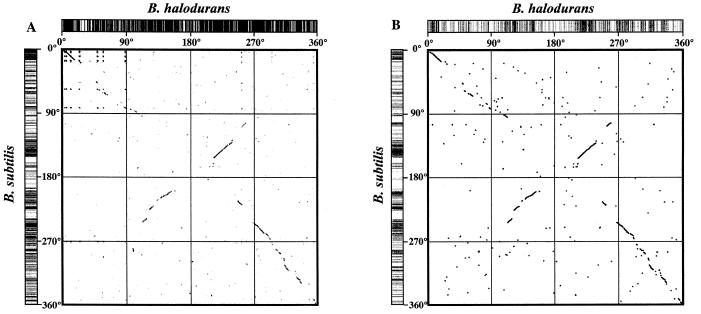
Bacillus halodurans C-125 is quite similar to B.subtilis in terms of genome size, G+C content of the genomic DNA and the physiological properties used for taxonomical identification, except for the alkaliphilic phenotype (4). Also, the phylogenetic placement of C-125 based on 16S rDNA sequence analysis indicates that this organism is more closely related to B.subtilis than to other members of the genus Bacillus. Therefore, the question arises of how does the genome structure differ between two Bacillus strains which have similar properties except for alkaliphily. As a first step to answer this question, we analyzed the genome structure both at the level of the whole genomic sequence and at the level of orthologous proteins, comparing the B.halodurans and B.subtilis genomes continuously from the replication origin region (oriC). The dots in Figure 4A were plotted when more than 20 bases in the B.halodurans nucleotide sequence continuously matched those of B.subtilis in a sliding window 100 nt wide, with a step of 50 nt. Figure 4B shows the distribution of orthologous proteins, comparing B.halodurans and B.subtilis, and the dot patterns in these figures resemble each other. About 1500 genes, some of which constitute operons, mainly categorized as genes associated with the following functions, are well conserved in the region common to B.halodurans and B.subtilis: mobility and chemotaxis, protein secretion, cell division, the main glycolytic pathways, the TCA cycle, metabolism of nucleotides and nucleic acids, metabolism of coenzymes and prosthetic groups, DNA replication, RNA modification, ribosomal proteins, aminoacyl-tRNA synthetases, protein folding, etc. On the other hand, the region around 112–153° in the B.halodurans genome corresponds to the region around 212–240° in the B.subtilis genome, as suggested in a previous paper (17).
Figure 4.
Structural analysis of the B.halodurans and B.subtilis genomes. (A) Distribution of regions having a nucleotide sequence common to the two Bacillus spp. The dots were plotted when more than 20 bases of the nucleotide sequence matched continuously with that of B.subtilis in a sliding window 100 nt wide with a step of 50 nt. The columns appearing as a bar code clearly show the regions shared between the B.halodurans and B.subtilis genomes. (B) Distribution of orthologs between the two Bacillus spp. The dots were plotted at the positions where the gene for an orthologous protein exists when comparing the two genomes. The columns appearing as a bar code clearly show the positions of the genes for orthologous proteins in B.halodurans and B.subtilis.
Transposable elements
One hundred and twelve CDSs in the B.halodurans genome showed significant similarity to the transposases or recombinases from various species, such as Anabena sp. Rhodobacter capsulatus, Lactococcus lactis, Enterococcus faecium, Clostridium beijerinckii, Staphylococcus aureus and Yersinia pseudotuberculosis, indicating that these have played an important evolutionary role in horizontal gene transfer and also in internal rearrangement of the genome. These CDSs were categorized into 27 groups by similarity pattern and the genes are widely spread throughout the genome (Fig. 1 and Table S1). As shown in Figure 2, at least 11 A+T-rich and G+C-rich islands containing tranposases (T1–T11) occur in the B.halodurans genome. This is one of the notable features of this genome, as B.subtilis has only 10 transposons and transposon-related proteins. The G+C content of transposases varies from 37.4 to 49.2% and codon usage in transposases, especially for termination, is obviously different from other indigenous genes in B.halodurans. In other bacterial genomes it has been reported that Synecosystis sp. PC6803 (18), Escherichia coli MG1655 (19), Mycobacterium tuberculosis (20), Deinicoccus radiodurans (21) and Lactococcus lactis (22) contain many transposase genes, as well as B.halodurans C-125.
On the other hand, we could not find any prophage which seemed to be active, although several phage-related proteins were identified in the B.halodurans genome. The B.halodurans genome contains no intact prophage, such as Spβ, PBSX or skin, found in the B.subtilis genome (5), as shown in Figure 2. We confirmed that B.halodurans has the gene for σK as the complete form, which is divided into two parts, spoIVCB (N-terminal) and spoIIIC (C-terminal) by a prophage (skin element) in the B.subtilis genome.
Origin of replication
There are 14 CDSs in the oriC region of the chromosome of B.halodurans. The organization of the CDSs in the region is basically similar to those of other bacteria. The region from gidB to gyrA (BH4060–BH4066 and BH1–BH7), especially, was found to be the same as in B.subtilis. On the other hand, it was found that there are 10 CDSs (BH8–BH18), including three CDSs previously identified in the 13.3 kb of the oriC region (23), between gyrB and the rrnA operon, corresponding to the rrnO operon in the B.subtilis genome (Fig. 1 and Table S1), although there is no CDS between gyrA and rrnO in B.subtilis. Of these 10 CDSs, only one (BH8) was found to have a homolog in another organism, interestingly, not in the genus Bacillus; the others were unique to the B.halodurans genome (Table S1).
Transcription and translation
Genes encoding the three subunits (α, β and β′) of the core RNA polymerase have been identified in B.subtilis along with the genes for 19 σ factors (24). σ factors belonging to the σ70 family (σA, σB, σD, σE, σF, σG, σH and σK) required for sporulation and σL are well conserved between B.halodurans and B.subtilis. Of 11 σ factors identified in B.halodurans, belonging to the extracytoplasmic function (ECF) family, σW is also found in B.subtilis, but the other 10 (BH620, BH672, BH1615, BH2026, BH3117, BH3216, BH3223, BH3362, BH3632 and BH3882) are unique to B.halodurans. These unique σ factors may have a role in the special physiological mechanisms by which B.halodurans is able to live in an alkaline environment, because it is well known that ECF σ factors are present in a wide variety of bacteria and they serve to control the uptake or secretion of specific molecules or ions and to control responses to a variety of extracellular stress signals (25).
Seventy-nine tRNA species, organized into 11 clusters involving 71 genes plus eight single genes, were identified (Fig. 1 and Table S1). Of the 11 clusters, six were organized in association with rRNA operons. Eight rRNA operons are present in the C-125 genome and their organization is the same as that in B.subtilis (tRNA–16S–23S–5S, 16S–tRNA–23S–5S and 16S–23S–5S–tRNA). With respect to tRNA synthetases, the C-125 genome lacks the glutaminyl-tRNA synthetase gene (glnS), one of two threonyl-tRNA synthetase genes (thrZ) and one of two tyrosyl-tRNA synthetase genes (tyrS). The B.subtilis genome has all of these tRNA synthetase genes except for the glutaminyl-tRNA gene. It is likely that glutamyl-tRNA synthetase aminoacylates tRNAGln with glutamate followed by transamidation by Glu-tRNA amidotransferase (26) in both Bacillus species.
Competence and sporulation
Out of 20 genes related to competence in B.subtilis, 13 (cinA, comC, comEA, comEB, comEC, comER, comFA, comFC, comGA, comGB, comGC, comGD and mecA), mainly expressed in the late stage of competence, were identified in the B.halodurans genome, but we could not find any of the genes expressed in the early stage of competence. Among six genes whose products are known to serve as components of the DNA transport machinery, only three (comGB, comGC and comGD), but not the others well conserved in B.subtilis, were identified in B.halodurans C-125. Actually, competence has not been experimentally demonstrated in C-125 although we attempted to use standard (27) and modified methods, changing such conditions as pH, temperature and medium, for transformation. It has become clear that this is due to lack of some of the necessary genes, especially those expressed in the early stages, such as comS, srfA and rapC. Only 68 genes related to sporulation were identified in the C-125 genome, in contrast to 138 genes found in the B.subtilis genome. Although the minimum set of genes for sporulation was well conserved, as in the case of B.subtilis, the C-125 genome lacks some genes encoding key regulatory proteins (the response regulator for aspartate phosphatase and the phosphatase regulator) and the spore coat protein for sporulation conserved in the B.subtilis genome. The rap (rapA–rapK) and phr (phrA, phrC, phrE–phrG, phrI and phrK) genes especially were not found in the C-125 genome, suggesting that C-125 may have another type(s) of regulatory gene(s) for control of sporulation in a manner the same as or different from that in B.subtilis, as we have observed sporulation in B.halodurans.
Cell walls
The peptidoglycan of alkaliphilic B.halodurans C-125 appears to be similar to that of neutrophilic B.subtilis. However, the cell wall components in C-125 are characterized by an excess of hexosamines and amino acids compared to that of B.subtilis. Glucosamine, muramic acid, d- and l-alanine, d-glutamic acid, meso-diaminopimelic acid and acetic acid were found in cell wall hydrolysates (1). Although some variation was found in the amide content of the peptidoglycan isolated from alkaliphilic B.halodurans C-125, the pattern of variation was similar to that known to occur in B.subtilis. All genes related to peptidoglycan biosynthesis, such as mraY, murC–murG, cwlA, ddlA and glnA, confirmed to be present in the B.subtilis genome were also conserved in the C-125 genome (1). A bacitracin resistance gene found in the B.subtilis genome is duplicated in the C-125 genome (BH464 and BH1521). On the other hand, although the tagH and tagG genes were identified in B.halodurans C-125 (Fig. 1 and Table S1), 13 other genes for teichoic acid biosynthesis found in B.subtilis (dltA–dltE, ggaA, ggaB, tagA–tagC, tagE, tagF and tagO) are missing from the B.halodurans genome. Bacillus halodurans also lacks six genes (tuaB–tuaF and tuaH) for teichuronic acid biosynthesis, all except tuaA and tuaG, in comparison with those of B.subtilis. In addition to peptidoglycan, the cell wall of alkaliphilic B.halodurans is known to contain certain acidic polymers, such as galacturonic acid, glutamic acid, aspartic acid and phosphoric acid. A teichuronopeptide (TUP) is present as a major structural component of the cell wall of C-125, which is a co-polymer of polyglutamic acid and polyglucuronic acid. Thus, the negative charges on acidic non-peptidoglycan components may give the cell surface the ability to absorb sodium and hydronium ions and to repel hydroxide ions and, as a consequence, may contribute to allowing the cells to grow in alkaline environments. A mutant defective in TUP synthesis grows slowly at alkaline pH. The upper limit of pH for growth of the mutant is 10.4, whereas that of the parental C-125 strain is 10.8. The tupA gene encoding TUP has been cloned from the C-125 chromosomal DNA (28). In this study, it has become clear that B.halodurans C-125 has no paralog of tupA in the genome and an ortholog of tupA cannot be found in the B.subtilis genome.
Membrane transport and energy generation
Bacillus halodurans C-125 requires Na+ for growth under alkaline conditions. The presence of sodium ions in the surrounding environment has been proved to be essential for effective solute transport through the cytoplasmic membrane of C-125 cells. According to the chemi-osmotic theory, a proton-motive force is generated across the cytoplasmic membrane by the electron transport chain or by extrusion of H+ derived from ATP metabolism through the action of ATPase. We identified four types of ATPases (preprotein translocase subunit, class III heat shock ATP-dependent protease, heavy metal transporting ATPase and cation transporting ATPase). These ATPases are well conserved between B.halodurans and B.subtilis.
Through a series of analyses such as a BLAST2 search, clustering analysis by the single linkage method examining all CDSs identified in the B.halodurans C-125 and B.subtilis genomes (8166 CDSs) and multiple alignment (16), 18 CDSs were grouped into the category of antiporter- and transporter-related protein genes in the C-125 genome. In this analysis it was found that five CDSs are candidates for Na+/H+ antiporter genes (BH1316, BH1319, BH2844, BH2964 and BH3946). However, we could not find any gene encoding antibiotic resistance proteins in the C-125 genome, whereas the B.subtilis genome has nine different ones. Eleven genes for multidrug resistance proteins were identified in the C-125 genome, six fewer than in B.subtilis. A non-alkaliphilic mutant strain (mutant 38154) derived from B.halodurans C-125 which is useful as a host for cloning genes related to alkaliphily has been isolated and characterized (29). A 3.7 kb DNA fragment (pALK fragment) from the parent strain restored growth of mutant 38154 under alkaline pH conditions. This fragment was found to contain CDS BH1319, which is one of the Na+/H+ antiporter genes in B.halodurans. The transformant was able to maintain an intracellular pH lower than the external pH and the cells expressed an electrogenic Na+/H+ antiporter driven only by Δψ (membrane potential, interior negative) (1,29). Bacillus subtilis has an ortholog (mprA) of BH1319 and it has been reported that a mprA-deficient mutant of B.subtilis showed a sodium-sensitive phenotype (30). On the other hand, a mutant of strain C-125 with a mutation in BH1317 adjacent to BH1319 has been isolated and it showed an alkali-sensitive phenotype, although whether the Na+/H+ antiporter encoded by BH1317 is active in this mutant has not been confirmed experimentally yet. In addition, it has been reported that BH2819, the function of which is unknown and which is unique to the C-125 genome, is also related to the alkaliphilic phenotype (31).
Bacillus halodurans C-125 has a respiratory electron transport chain and the basic gene set for it is conserved as compared with B.subtilis, but the gene for cytochrome bd oxidase (BH3775 and BH3776) is duplicated in the C-125 genome. It is also clear that two genes for bo3-type cytochrome c oxidase (BH739 and BH740) not seen in B.subtilis are present in the C-125 genome. The C-125 genome has a F1F0-ATP synthase operon (Fig. 1 and Table S1). The gene order in this operon (ɛ subunit–β subunit–γ subunit–α subunit–δ subunit–subunit b–subunit c–subunit a) is identical to that seen in B.subtilis. In addition to the F1F0-ATP synthase operon, the operon for a Na+-transporting ATP synthase and the operon for a flagellar-specific ATP synthase are also conserved between B.halodurans and B.subtilis.
ABC transporters
Members of the superfamily of ABC transport systems couple the hydrolysis of ATP to the translocation of solutes across a biological membrane (32). ABC transporter genes are the most frequent class of protein coding genes found in the B.halodurans genome, as in the case of B.subtilis. They must be extremely important in Gram-positive bacteria such as Bacillus, because these bacteria have an envelope consisting of a single membrane. ABC transporters allow such bacteria to escape the toxic action of many compounds. Through the series of analyses described above, 75 genes coding for ABC transporter/ATP-binding proteins were identified in the B.halodurans genome. In this analysis 67 CDSs were grouped into the category of ATP-binding protein genes, although 71 ATP-binding protein genes have been identified in the B.subtilis genome (5). We found that B.halodurans has eight more oligopeptide ATP-binding proteins, but four fewer amino acid ATP-binding proteins, as compared with B.subtilis. We could not find any other substantial difference between B.halodurans and B.subtilis in terms of the other ATP-binding proteins, although it should be noted that the specificity of some of these proteins is not known. The genes for oligopeptide ATP-binding proteins (BH27, BH28, BH570, BH571, BH1799, BH1800, BH2077, BH2078, BH3639, BH3640, BH3645, BH3646, AppD and AppF) are distributed throughout the C-125 genome. We speculate that these may contribute to survival under highly alkaline conditions, although there is no direct evidence to support this. On the other hand, 43 CDSs were identified as ABC transporter/permeases in the B.halodurans genome. Surprisingly, B.halodurans has only one amino acid permease, in contrast to the 12 present in the B.subtilis genome. In addition, it is clear that B.halodurans lacks the sodium permease gene present in B.subtilis, whereas B.subtilis lacks the nickel permease gene present in B.halodurans.
CONCLUSION
Alkaliphilic B.halodurans is the second Bacillus species whose whole genomic sequence has been completely defined. The genomic sequence of B.halodurans offers a wealth of basic information regarding gene conservation and diversity in Bacillus spp. and systematic information that would be difficult, if not impossible, to obtain by any other approach. A more complete understanding of the biochemistry of this organism derived from genome analysis will provide the foundation for clarification of the mechanisms of adaptation to extreme environments, especially to a highly alkaline environment, as a first step. A new database specifically established for the B.halodurans sequence, ExtremoBase, will be accessible through the World Wide Web server at http://www.jamstec.go.jp/jamstec-e/bio/DEEPSTAR/FResearch.html . The sequence has been deposited in DDBJ/EMBL/GenBank with the accession nos AP001507–AP001520.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to A. Ohyama, K. Doga, and T. Kozuki of Mitsui Knowledge Inc. Ltd for computing assistance. We thank Dr T. Yada of RIKEN Genomic Science Centre for his help with operating the GeneHacker Plus program. Thanks are also due to Dr T. Sakiyama for his technical assistance.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +81 468 67 3895; Fax: +81 468 66 6364; Email: takamih@jamstec.go.jp AP001507–AP001520
REFERENCES
- 1.Horikoshi K. (1999) Microbiol. Rev., 63, 735–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikura Y. and Horikoshi,K. (1979) Agric. Biol. Chem., 43, 85–88. [Google Scholar]
- 3.Honda H., Kudo,T., Ikura,Y. and Horikoshi,K. (1985) Can. J. Microbiol., 31, 538–542. [Google Scholar]
- 4.Takami H. and Horikoshi,K. (1999) Biosci. Biotechnol. Biochem., 63, 943–945. [DOI] [PubMed] [Google Scholar]
- 5.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 6.Thuring R.W., Sanders,J.M.P. and Borst,P. (1975) Anal. Biochem., 66, 213–220. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook J., Frisch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J., Dougherty,B.A., Merrick,J.M. et al. (1995) Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 9.Takami H. and Horikoshi,K. (2000) Extremophiles, 4, 99–108. [DOI] [PubMed] [Google Scholar]
- 10.Sakiyama T., Takami,H., Ogasawara,N., Kuhara,S., Doga,K., Ohyama,A. and Horikoshi,K. (2000) Biosci. Biotechnol. Biochem., 64, 670–673. [DOI] [PubMed] [Google Scholar]
- 11.Takami H., Nakasone,K., Ogasawara,N., Hirama,C., Nakamura,Y., Masui,N., Fuji,F., Takaki,Y., Inoue,A. and Horikoshi,K. (1999) Extremophiles, 3, 29–34. [DOI] [PubMed] [Google Scholar]
- 12.Yada T. and Hirosawa,M. (1996) In Proceedings of the Fourth International Conference on Intelligent Systems and Molecular Biology. AAAI Press, St Louis, MO, pp. 252–260.
- 13.Takami H., Inoue,A., Fuji,F. and Horikoshi,K. (1997) FEMS Microbiol Lett., 152, 279–285 [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson W.R. and Lipman,D.J. (1988) Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogiwara A. (1998) Genome Informatics, 1998, 310–311. [Google Scholar]
- 17.Takami H., Nakasone,K., Hirama,C., Takaki,Y., Masui,N., Fuji,F., Nakamura,Y. and Inoue,A. (1999) Extremophiles, 3, 21–28. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko T., Sato,S., Kotani,H., Tanaka,A., Asamizu,E., Nakamura,Y., Miyajima,N., Hirosawa,M., Sugiura,M., Sasamoto,S. et al. (1996) DNA Res., 3, 109–136. [DOI] [PubMed] [Google Scholar]
- 19.Blattner F.R., Plunkett,G.III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 20.Cole S.T., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S.V., Eiglmeier,K., Gas,S., Barry,C.E.III et al. (1998) Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 21.White O., Eisen,J.A., Heidelberg,J.F., Hickey,E.K., Peterson,J.D., Dodson,R.J., Haft,D.H., Gwinn,M.L., Nelson,W.C., Richardson,D.L. et al. (1999) Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolotin A., Mauger,S., Malarme,K., Ehrlich,S.D. and Sorokin,A. (1999) Antonie Van Leeuwenhoek, 76, 27–76. [PubMed] [Google Scholar]
- 23.Takami H., Masui,N., Nakasone,K. and Horikoshi,K. (1999) Biosci. Biotechnol. Biochem., 63, 1134–1137. [DOI] [PubMed] [Google Scholar]
- 24.Haldenwang W.G. (1995) Microbiol. Rev., 59, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonetto M.A., Brown,K.L., Rudd,K.E. and Buttner,M.J. (1994) Proc. Natl Acad. Sci. USA, 91, 7573–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freist W., Gauss,D.H., Ibba,M. and Soll,D. (1997) Biol. Chem., 378, 1103–1117. [PubMed] [Google Scholar]
- 27.Dubnau D. and Davidoff-Abelson,R. (1971) J. Mol. Biol., 56, 209–221 [DOI] [PubMed] [Google Scholar]
- 28.Aono R., Ito,M. and Machida,T. (1999) J. Bacteriol., 181, 6600–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudo T., Hino,M., Kitada,M. and Horikoshi,K. (1990) J. Bacteriol., 172, 7282–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosono S., Ohhashi,Y., Kawamura,F., Kitada,M. and Kudo,T. (2000) J. Bacteriol., 182, 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider E. and Hunke,S. (1998) FEMS Microbiol. Rev., 22, 1–20. [DOI] [PubMed] [Google Scholar]
- 32.Aono R., Hashimoto,M., Hayakawa,A., Nakamura,S. and Horikoshi,K. (1992) Biosci. Biotechnol. Biochem., 56, 842–844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.