Abstract
DNA ligases are enzymes required for the repair, replication and recombination of DNA. DNA ligases catalyse the formation of phosphodiester bonds at single-strand breaks in double-stranded DNA. Despite their occurrence in all organisms, DNA ligases show a wide diversity of amino acid sequences, molecular sizes and properties. The enzymes fall into two groups based on their cofactor specificity, those requiring NAD+ for activity and those requiring ATP. The eukaryotic, viral and archael bacteria encoded enzymes all require ATP. NAD+-requiring DNA ligases have only been found in prokaryotic organisms. Recently, the crystal structures of a number of DNA ligases have been reported. It is the purpose of this review to summarise the current knowledge of the structure and catalytic mechanism of DNA ligases.
INTRODUCTION
Polynucleotide ligases are ubiquitous cell proteins that are required for a number of important cellular processes, including the replication, repair and recombination of DNA. DNA ligases catalyse the formation of phosphodiester bonds at single-strand breaks between adjacent 3′-hydroxyl and 5′-phosphate termini in double-stranded DNA (for reviews see 1–3). DNA ligases have also found widespread use as a tool for in vitro DNA manipulation and cloning techniques. DNA ligases can be divided into two broad classes, those requiring NAD+ as cofactor and those requiring ATP. The eukaryotic, viral and archaebacteria encoded enzymes all require ATP. NAD+-requiring DNA ligases are found exclusively in eubacteria. The ATP-dependent ligases range in size from 30 to >100 kDa but the NAD+-dependent enzymes are highly homologous and are monomeric proteins of 70–80 kDa. The sequence similarity between the two classes was until recently thought to be limited to a conserved KxDG sequence motif. This conserved motif (I), one of six co-linear sequence motifs (I–VI) known to be at the active site of the nucleotidyl transferase superfamily of enzymes (4), including all ATP-dependent DNA ligases (5,6), RNA ligases (7) and tRNA ligases (8) as well as the eukaryotic mRNA ‘capping’ enzymes (9,10). However, a new iterative sequence search method (11) showed that five of the six motifs are also present in the NAD+-dependent ligases.
All DNA ligases catalyse the synthesis of phosphodiester bonds in a very similar manner, by esterification of a 5′-phosphoryl to a 3′ hydroxyl group (1,2). The reaction mechanism can be split into three distinct catalytic events (Fig. 1). The first involves activation of the ligase through the formation of a covalent protein–AMP intermediate. The nucleotide has been shown to be linked to the enzyme through a phosphoramidate bond (Fig. 1) to the ɛ amino group of a conserved active site lysine (5,12). In the second step of the reaction, the AMP moiety is transferred from the ligase to the 5′-phosphate group at the single-strand break site. Finally, DNA ligase catalyses the DNA ligation step with loss of free AMP. In spite of these similarities between the two classes of enzymes, the manner by which the eubacterial and ‘eukaryotic’ proteins become activated is rather different (Fig. 2). For eukaryotic ligases, the enzyme–AMP complex is formed after reaction of the enzyme and ATP with the release of free pyrophosphate. The bacterial ligases become adenylated in an unusual reaction which involves cleavage of NAD+ and release of nicotinamide mononucleotide (1) (Fig. 1).
Figure 1.
Mechanisms of DNA ligation. This figure shows the partial reactions catalysed by ATP- and NAD+-dependent DNA ligases. The structure of the phosphoramidate intermediate common to both reactions is also illustrated.
Figure 2.
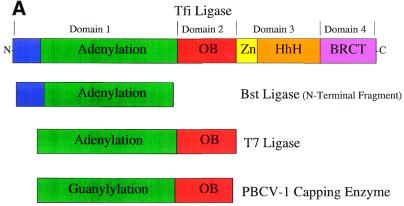
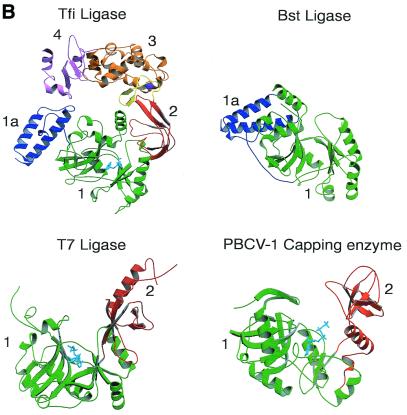
Domain structure of DNA ligases. (A) Schematic representation of the domain architecture of the known DNA ligase structures. The domains are colour coded: subdomain 1a, blue; subdomain 1b (adenylation), green; domain 2 (oligo-binding), red; subdomain 3a (zinc finger), yellow; subdomain 3b (helix–hairpin–helix), orange; domain 4 (BRCA1 C-terminus), pink. (B) A ribbon diagram representation of the structures of the NAD+-dependent ligases encoded by T.filiformis (Tfi) and B.stearothermophilus (Bst) (N-terminal domain only), the ATP-dependent DNA ligase of bacteriophage T7 (T7) and the capping enzyme encoded by Chlorella virus PBCV-1. The conserved structural domains have been numbered and coloured as in (A) to allow for ease of identification in the different structures.
The discovery of DNA ligases over 30 years ago was followed shortly afterwards by elucidation of the catalytic mechanism (1,2). However, it is only in recent years that we have glimpsed the molecular architecture of a DNA ligase, the ATP-dependent ligase of bacteriophage T7 (13). This report was closely followed by the structure of the related RNA capping enzyme from PBCV-1 (14). Determination of these crystal structures has given us valuable molecular insights into the enzyme mechanism of nucleotidyl transferases. More recently, elucidation of the structures of the adenylation domain of Bacillus stearothermophilus (Bst) ligase (15) and the full-length Thermus filiformis (Tfi) ligase (16) has greatly advanced our understanding of the multidomain NAD+-dependent ligases. Here we review the current knowledge of the structures of DNA ligases and discuss the important structural and mechanistic implications arising from these recent advances.
Modular architecture of DNA ligases
The elucidation of crystal structures of the ATP-dependent ligase from bacteriophage T7 (13), the N-terminal fragment of Bst ligase (15) and intact Tfi ligase (16), both NAD+-dependent ligases, has revealed that DNA ligases have a highly modular architecture consisting of a unique arrangement of two or more discrete domains (Fig. 2). Currently five classes of motifs [nucleotide-binding domain, oligomer-binding (OB) fold, zinc finger, helix–hairpin–helix (HhH) motif and BRCA1 C-terminus (BRCT) domain] have been detected at both the sequence and more recently at the structural level (Fig. 2). The structure and role of these motifs in the ligase mechanism are discussed.
Adenylation domain
The structure of bacteriophage T7 ligase (13) revealed that the enzyme consists of two domains (Fig. 2), a larger N-terminal domain (green), domain 1, and a small C-terminal domain (red), domain 2 (Fig. 2). Domain 1 (residues 1–240) consists of three mainly antiparallel β-sheets that are flanked by six α-helices. This domain contains the ATP-binding site, which is situated in a pocket beneath one of the β-sheets (Fig. 2). Doherty and Wigley (17) demonstrated that domain 1 itself has intrinsic adenylation activity and therefore this domain has been named the ‘adenylation domain’. A similar fold was subsequently found in the PBCV-1 capping enzyme (14) and the N-terminal domains of the Bst (15) and Tfi ligases (16) (Fig. 2), as well as in a number of other enzymes which bind ATP and GTP (18). Domain 1 of the NAD+-dependent ligases (15,16) contains a subdomain (blue), 1a (residues 1–80) (Fig. 2), which is mainly α-helical; this structure is not present in the T7 ligase.
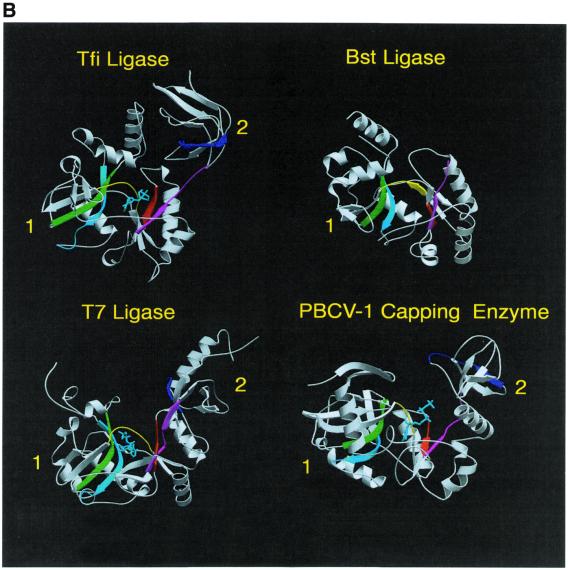
Many of the residues that line the ATP-binding pocket of the adenylation domain of the T7, Bst and Tfi ligases belong to five (motifs I–V) of the six sequence elements (Fig. 3A) conserved among covalent nucleotidyl transferases (4). These motifs contain residues known to be essential for catalysis (4). Examination of the positions of these motifs within the DNA ligase structures indicates that they cluster around the ATP-binding site and they also form the sides of the groove between domains 1 and 2 (Fig. 3B). Many of the residues within these motifs make specific conserved interactions with the ATP molecule (13,16). A similar nucleotide binding mode is conserved in non-covalent and covalent complexes of Chlorella virus mRNA guanylyl transferase (capping enzyme) (Fig. 3B) (14).
Figure 3.
(A) Sequence motifs conserved in DNA ligases and RNA capping enzymes. Conserved sequence elements define a superfamily of covalent nucleotidyl transferases. Six sequence elements, designated motifs I, III, IIIa, IV, V and VI, are conserved in NAD+- and ATP-dependent DNA ligases and guanylyl transferases as shown. The alignment includes the NAD+-dependent ligases (Lig) encoded by T.filiformis (Tfi), B.stearothermophilus (Bst), E.coli (Eco) and T.thermophilus (Tth). Below these are aligned sequences for the ATP-dependent DNA ligases (Lig) of bacteriophage T7 (BT7), vaccinia virus (Vac), Saccharomyces cerevisiae (Sce), Schizosaccharomyces pombe (Spo) and human ligases I (Hu1), 3 (Hu3) and 4 (Hu4). The alignment also contains the amino acid sequences for capping enzymes (CE) encoded by Chlorella virus PBCV-1 (ChV), S.cerevisiae, S.pombe, Candida albicans (Cal), African swine fever virus (ASF), vaccinia virus and Caenorhabditis elegans (Cel). The numbers of amino acid residues separating the motifs are indicated. The active site lysine is shown in red. (B) The location of the conserved sequence motifs as described in (A) are indicated by the corresponding colours in the ribbon diagrams of the crystal structures of the Tfi, Bst and T7 DNA ligases and Chlorella virus RNA capping enzyme. The bound nucleotides are shown in cyan.
OB fold domain
In all DNA ligases adenylation domain 1 is connected to a conserved domain 2 (Fig. 2, in red). The T7 and Tfi DNA ligase structures (13,16) revealed that this domain has an OB fold, a derivative of a Greek key motif also found in the structures of many proteins that bind to single-stranded and double-stranded (ds)DNA and RNA (for a review see 19). This fold is found in a diverse range of protein families, including the bacterial ribosomal proteins S1 (20) and S17 (21), the subunits of replication protein A (22), the telomere end-binding protein (23), bacterial cold shock proteins CspA and CspB (24), translation initiation factor (IF) 5A (25), translation IF1 (26), SSB protein (27), RuvA DNA recombination protein (28,29), staphylococcal nuclease (30) and several tRNA synthetases (30). A number of co-crystal structures of these domains bound to DNA and RNA have established that the OB fold mediates polynucleotide recognition (19). Many of these OB fold proteins bind their ligands on the surface of the β-barrel (19,30). Biochemical studies (17) have shown that the OB domain of T7 ligase binds dsDNA and also dramatically enhances the adenylation activity of domain 1. In the T7 ligase structure (13) this domain is distant from domain 1 (Fig. 2B), therefore, domain 2 must undergo a profound conformational change if it is to stimulate the adenylation activity of domain 1. A direct physical interaction between these domains has been demonstrated by gel filtration (17). Two crystal structures of PBCV-1 mRNA capping enzyme (14) have provided conclusive evidence for such a conformational change during the guanylylation reaction in the capping enzymes. This involves a 13 Å movement of the C-terminal OB domain 2 towards domain 1. This reaction is equivalent to the adenylation reaction catalysed by DNA ligases (4). During this conformational change by PBCV-1 capping enzyme, conserved residues in motifs V and VI, located in the OB domain, are positioned in the active site and form specific interactions with the nucleotide (14). Motif VI encompasses the last strand (in blue) of the ‘OB fold’ domain 2 (Fig. 2B). Two residues (R295 and K298) from the corresponding motif in the capping enzyme bind and position the triphosphate tail of GTP for a direct in-line attack by the active site lysine (14). It is likely that this motif also plays a similar role in the adenylation reaction of ligases (Fig. 4) and this is supported by mutagenesis studies on the PBCV-1 DNA ligase (31). Site-directed mutagenesis studies on human DNA ligase III have implicated this motif in the interaction with nicked DNA (32).
Figure 4.
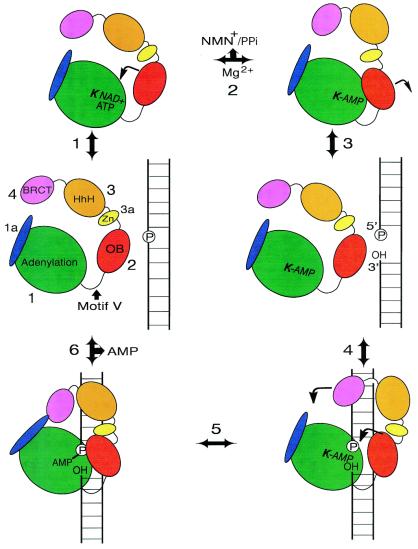
Proposed mechanism of nick recognition and ligation by DNA ligases. In this schematic diagram we propose the following ligase reaction pathway. (1) NAD+ or ATP binds in the nucleotide-binding pocket, followed by closure of domain 2. (2) AMP is transferred to the active site lysine (bold) in a transadenylation reaction involving magnesium, and NMN+ or PPi is released. (3) Domain 2 opens up and the enzyme is ready for nick binding. (4) The adenylated enzyme binds to phosphorylated nicked DNA. Domain 2 closes in on the nick site. In the larger ligases, such as Tfi ligase, additional domains may wrap around the DNA. (5) AMP is transferred to the 5′-phosphate of the nick. (6) The ligase catalyses in-line attack of the 3′-OH on the adenylated 5′-phosphate of the nick, forming a phosphodiester bond and sealing the break, with the concomitant release of AMP. The enzyme opens up and falls off the dsDNA ready to undergo another catalytic cycle.
Zinc finger motif
Four cysteine residues are conserved in the C-terminal region of NAD+-dependent ligases and they have been implicated in zinc binding and interaction with DNA. Atomic emission spectroscopy confirmed that Tfi ligase binds zinc ions (16). In the Tfi ligase structure a zinc ion is tetrahedrally liganded by the four conserved cysteine residues (Cys406, Cys409, Cys422 and Cys427). This single zinc finger forms a subdomain (3a) of the larger domain 3 of Tfi ligase (Fig. 2B, in yellow). It is expected that all the NAD+-dependent ligases have a similar zinc finger, since the four cysteine residues that coordinate the zinc ion are strictly conserved among eubacterial DNA ligases. Zinc fingers and related motifs can act as DNA recognition modules, often recognising specific DNA sequences (33). The overall fold of the Tfi ligase zinc finger is similar to other Cys4 type zinc fingers, including the first of the two zinc fingers in the DNA-binding domain of steroid/nuclear hormone receptors (33–35). In all these cases the phosphate backbone of DNA interacts with residues on the β-hairpin and the α-helix of the first zinc finger, with the latter helix sitting in the major groove of dsDNA.
It is interesting to note that human DNA ligase III possesses a Cys-Cys/His-Cys type zinc finger motif that is homologous to the two zinc fingers present in human poly(ADP-ribose) polymerase (32,36). The second of these zinc finger motifs is involved in specific recognition of dsDNA nicks (37). It has been demonstrated that the human DNA ligase III zinc finger forms a specific complex with nicked duplex DNA (32). Analogously, the Tfi ligase zinc finger motif (subdomain 3a) may also mediate interactions with nicked DNA (Fig. 4). This suggestion is consistent with the results of mutagenesis of the zinc-coordinating cysteines of Thermus thermophilus and Escherichia coli ligases (38,39). These mutations did not affect the adenylation or deadenylation activity but reduced or abolished DNA binding and ligation. Zinc fingers can also play a structural role in proteins. It has been proposed (16) that the zinc finger in Tfi ligase may also act as a structural support for subdomain 3b and domain 4 (Fig. 4).
HhH motif domain
Doherty et al. (40) predicted the presence of four copies of a conserved HhH motif in the C-terminal region of NAD+-dependent ligases. This motif consists of two helices, of conserved length, connected by a type II β-turn. HhH motifs are present in a large number of DNA repair enzymes (40,41), including endonuclease III (41), AlkA (42), MutY (43), DNA polymerase β (44) and RuvA (28,29). The HhH motif has been implicated in non-sequence-specific DNA binding (40,41). The recent determination of the NAD+-dependent Tfi ligase structure (16) confirmed that four HhH motifs are present and provided a unique example in which the four clustered HhH motifs form a single compact structure (subdomain 2b) (Fig. 2B, in orange). The hairpins are located in a linear chain at the bottom of this subdomain (16) (Fig. 2B). This surface is rich in positively charged residues and it has been suggested that this forms one of the two DNA-binding sites in Tfi and other NAD+-dependent ligases (Fig. 4).
BRCT domain
The final structural motif currently found in both ATP- and NAD+-dependent DNA ligases is a member of the BRCT domain superfamily (45,46). BRCT domains are present in NAD+-dependent ligases and eukaryotic ligases III and IV. The structure of Tfi ligase (16) is the first case in which a BRCT domain has been seen as part of a multidomain protein. The Tfi ligase BRCT domain consists of a four-stranded parallel β-sheet flanked by three α-helices (Fig. 2B, in pink). This fold is similar to that of the C-terminal BRCT domain of the XRCC1 protein (47). XRCC1 is a multidomain protein involved in the repair of single-strand breaks in DNA. The BRCT domain of XRCC1 forms a specific complex with the C-terminal BRCT domain of mammalian DNA ligase III (48). It has also been reported that the DNA repair ligase, ligase IV, interacts with XRCC4, a protein implicated in DNA repair processes, via a tandem repeat of the BRCT motif (49,50). These interactions appear to be constitutive and it is likely that these DNA ligase complexes are recruited to sites of DNA damage by protein–protein interactions that directly involve the BRCT motifs of these proteins. It has been suggested that BRCT domains may also act as signal transducers that transmit the signals from DNA damage detectors to other components of the DNA repair machinery via specific protein–protein interactions (45).
The Tfi ligase crystal structure revealed that the BRCT domain is very mobile in the open conformation but its mobility is restricted in the closed conformation, where the BRCT domain comes into close proximity to the conserved segment around the N-terminus of helix B of domain 1a (Fig. 2B) (16). The orientation of subdomain 1a relative to subdomain 1b in the N-terminal fragment of Bst ligase is significantly different from that of Tfi ligase (15). Subdomain 1a of Bst ligase is rotated by ∼90° around Pro68, which is also conserved in Tfi ligase. Lee et al. (16) suggest that if subdomain 1a of Tfi ligase adopts the same orientation as Bst ligase then its interaction with the BRCT domain is more extensive (Fig. 4). One possible role of the BRCT domain may be to form a contact with helix B in domain 1a, forming a toroidal conformation (Fig. 4). The BRCT domain may therefore act as a gate which regulates DNA binding and release (Fig. 4). It is possible that Tfi ligase operates as a sliding clamp, like PCNA, moving along the DNA until it encounters a nick, however, this remains to be demonstrated.
Nick recognition by DNA ligases
What are the key determinants for nick recognition by DNA ligases? A DNA nick typically consists of 5′-phosphorylated and 3′-OH termini on opposites sides of a break in the phosphodiester backbone. Shuman and colleagues have shown that the 5′-phosphate is absolutely required for nick recognition by ATP-dependent ligases but the 3′-OH is dispensable for DNA binding (51–54). Formation of nick-specific complexes is reduced significantly if the nicks lack a 5′-phosphate (53–55). Ligases also discriminate between nicks and gaps. The introduction of a ≥1 nt gap in a nick site effectively abolishes DNA binding (51,52). The DNA nick binding sites of the minimal sized ATP-dependent ligases from T7 and Chlorella virus have been physically mapped using different DNA footprinting approaches (54,55). These footprinting studies revealed that the enzymes bind asymmetrically to nicks, extending 7–12 nt on the 5′-phosphate side of the nick and 3–8 nt on the 3′-OH side.
There are currently no structures of a DNA ligase in complex with nicked DNA in the protein database. Molecular modelling studies (55), using the crystal structure of T7 ligase, suggest that dsDNA binds predominantly in the positively charged interdomain cleft (Fig. 5) with domain 2 acting as a movable ‘thumb’ which can open, rotate or close in response to ligand association/dissociation (Figs 4 and 5). DNA binding experiments with recombinant domains 1 and 2 of T7 ligase (17) showed that the larger N-terminal domain 1 has a much higher affinity for DNA than domain 2. DNA can be docked closely into the positively charged cleft region, which extends between the two domains of the protein (Fig. 5). This charged groove is lined by a number of conserved motifs and residues which have a strong positive potential. The most favourable DNA-binding site is asymmetrical with respect to the nick site, consistent with the DNA footprinting studies. This model is supported by a number of key observations. Odell and Shuman (54) have shown that the interdomain linker region (motif V) of PBCV-1 ligase is less accessible to proteolysis when the enzyme is bound to nicked DNA. Photocrosslinking and mutagenesis studies (55) have implicated two conserved lysine residues of motif V in DNA nick recognition. Doherty and Wigley (17) reported that domains 1 and 2 of T7 ligase can independently and non-specifically bind to DNA and they suggested that nick sensing is a product of these distinct DNA-binding activities. This suggests that domain 1 together with domain 2 make up the minimal unit for all the ATP-dependent ligases and, probably, NAD+-dependent bacterial DNA ligases. It is likely that these two domains are responsible for the inherent nick sensing and ligation activities of these enzymes. The additional domains, found in the larger ligases, may enhance DNA binding or target the enzymes to regions of DNA damage or replication, but they are not essential for ligase activity. This is supported by complementation experiments which showed that the smaller PBCV-1 ligase can function in lieu of the more complex ligases I and IV in yeast (39).
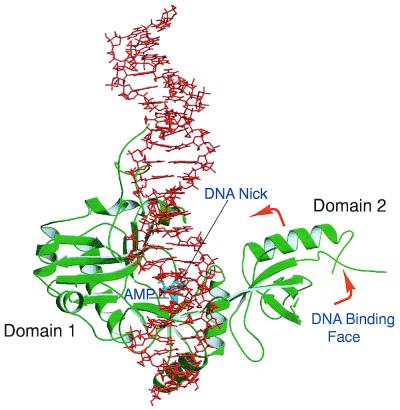
Figure 5.
A view of the model T7 ligase–DNA complex. The DNA–protein complex was achieved by placing the DNA nick site adjacent to the active site followed by energy minimisation and molecular dynamics protocols as detailed by Doherty and Dafforn (55). After the enzyme is adenylated (AMP shown in cyan) domain 2 rotates around, exposing the DNA-binding face to the active site and allowing DNA to bind.
Nick recognition requires ligases to be adenylated at the active site lysine (51). A structural basis for this requirement can now be proposed based on the T7 and Tfi ligase structures. As discussed above, the OB fold (domain 2) has been shown to bind to dsDNA. In the non-adenylated structure of the T7 ligase–ATP complex the proposed DNA-binding face of the OB domain is rotated away from the active site cleft (Fig. 2B). In contrast, in the covalently bound AMP structure of the NAD+-dependent Tfi ligase the equivalent OB domain is facing towards the active site cleft (Fig. 2B). Why is this domain found in such different conformations in the two structures? In the non-adenylated T7 ligase structure it is likely that the OB domain is orientated to prevent DNA binding in the active site cleft until the conserved active site lysine has become adenylated. In contrast, the OB domain in the adenylated Tfi ligase structure is rotated around, positioning the DNA-binding surface towards the active site awaiting nick binding. The OB fold appears to play a dual role in the ligation mechanism. Residues on one face of this domain position the β and γ phosphate tail of ATP away from the incoming nucleophile, enhancing the adenylation reaction. Adenylation of the active site lysine acts as a conformational switch, facilitating rotation of the DNA-binding surface (β-barrel face) of the OB fold towards the active site cleft (Fig. 4). In this way only adenylated ligase can bind to nicked DNA. This mechanism prevents the formation of non-productive ligase–DNA complexes.
The larger ligases appear to have additional DNA-binding motifs which contribute to the DNA-binding surface. This is most obvious in the Tfi ligase structure, which has two additional C-terminal domains. Domain 3 consists of a zinc finger subdomain (3a) and four HhH motifs (3b). Lee et al. (16) proposed that this domain forms a second DNA-binding site, named the ‘non-catalytic’ DNA-binding site as it is distant from the active site. This proposal is supported by the result of a limited proteolysis study on the homologous Bst ligase (56). Timson and Wigley (56) have shown that the DNA-binding activity of a C-terminal fragment of Bst ligase is comparable to full-length enzyme. Mackey et al. (32) identified two functionally distinct regions within mammalian ligase III that interact with nicked DNA. Currently no structural information is available on the mammalian ligases (I–IV) but it is obvious at the sequence level that these enzymes, in common with Tfi ligase, have a catalytic core (domains 1 and 2) with additional domains, such as BRCT and zinc finger domains, forming distinct N- and C-terminal extensions.
A conserved catalytic mechanism for DNA ligases and nucleotidyl transferases
As we discussed above, DNA ligases are members of the nucleotidyl transferase family of enzymes and proceed through a covalent AMP–enzyme intermediate in which the AMP is attached to the enzyme via a lysine residue (1,2). This lysine residue is part of the conserved motif I (5), one of six co-linear sequence motifs, also found in capping enzymes, RNA ligases and tRNA ligases, with a similar spacing between them (Fig. 3A) (4). Shuman and Schwer (4) proposed that all of these enzymes share a common nucleotidyl transfer mechanism and are likely to have a similar structure. This has since been confirmed by biochemical analysis of mutant enzymes (7,12,53,57–59) and, more recently, by the structures of T7 DNA ligase (13), Chlorella virus RNA capping enzyme (14), the N-terminal domain 1 of Bst ligase (15) and Tfi ligase (16). Examination of the positions of the conserved sequence motifs (Fig. 3A) within these DNA ligase and RNA capping structures (13,14) reveals that they are clustered around the NTP-binding site and they form the sides of the groove between domains 1 and 2 (Fig. 3B). The crystal structures of the T7 and Tfi ligases (in bold) reveal a conserved role for many of the residues in these motifs. Motif 1 contains the active site lysine (K34, K116) which forms the covalent AMP adduct. Motif III contains a glutamate residue (E93, E114) which forms hydrogen bonds with the ribose of the ATP, while the tyrosine (Y149, Y221) in motif IIIa is stacked against the adenine ring and the essential lysine in motif V (K238, K312) contacts the α-phosphate group. Examination of the conservation of sequence between the DNA ligases and the capping enzymes, coupled with the crystal structures, provides clues about how nucleotide specificity is achieved. There are two important interactions between the 6-amino group of the adenine ring and ligases, one via the main chain carbonyl of I33/H115 and the other via the side chain of E32/E114. This latter residue is usually a Glu, though occasionally an Asp or Gln, in ATP-dependent ligases (6). In contrast, this residue is highly variable in the capping enzymes but is never a Glu, Asp or Gln (4). In T7 ligase Lys222 and Glu32 form an ion pair at the base of the ATP-binding pocket. A similar ion pair is also present in the Tfi NAD-dependent DNA ligase (Lys288 and Glu114) (13). Currently there is no structural information on the binding specificity for the base at the nicotinamide position of NAD+ in NAD+-dependent ligases. A complete structural understanding of the nucleotide specificity of the two classes of ligases will be essential for the design and development of specific inhibitors of bacterial ligases as potential antibacterial compounds.
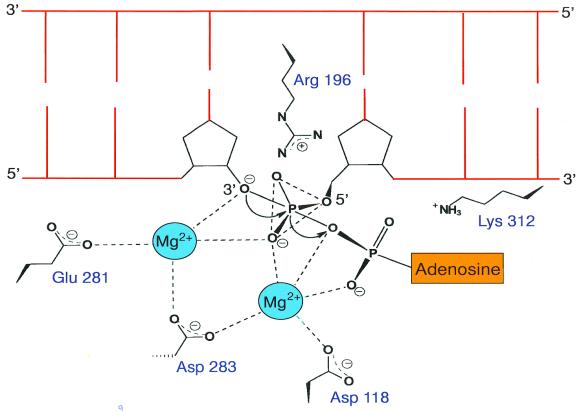
Lee et al. (16) have proposed that the final step of the ligation reaction, deadenylation of the adenylated DNA intermediate and phosphodiester bond formation, is analogous to the polymerising step catalysed by DNA polymerases. The adenylated DNA intermediate in ligation corresponds to deoxyribonucleoside 5′-triphosphate in polymerisation. They argue that DNA ligases are also likely to utilise a similar two divalent metal ion mechanism, proposed previously for DNA polymerases (60). They have identified a putative metal ion-binding site Asp118, Glu281 and Asp283 in Tfi ligase (Fig. 6) which matches the two cobalt ion-binding site in E.coli methionine aminopeptidase. These three residues form a highly negatively charged pocket near the active site lysine residue (Lys116) (Fig. 6). The strictly conserved Arg196 is likely to interact with the 5′-phosphate end of the nicked strand. A schematic model proposed for the ligase active site is shown in Figure 6. It is very likely that a similar active site architecture exists in all DNA ligases and related nucleotidyl transferases.
Figure 6.
Schematic model of the Tfi ligase active site. Residues that are likely to participate in binding divalent metal ions and the 5′-phosphate end of the nick are indicated.
The biochemical properties of a chimeric polypeptide ACE (ATP-dependent capping enzyme), consisting of the N-terminal domain 1 of T7 DNA ligase fused to the C-terminal OB domain 2 of the PBCV-1 capping enzyme, have recently been reported (61). The ACE protein can become adenylated at the active site lysine in common with DNA ligases. However, ACE is deficient in DNA ligase activity, but remarkably this chimeric enzyme can ‘cap’ RNA molecules by transferring AMP specifically to diphosphate terminated 5′-ends of RNA. The OB domain of ACE confers a novel binding specificity for the 5′-end of mRNA rather than for nicked RNA. The OB fold appears to determine the polynucleotide specificity of these enzymes. This report supports the idea that nucleotidyl transferases have a conserved catalytic mechanism.
CONCLUSIONS
DNA ligases are ideal candidates for structural studies as all of the intermediates in the ligation pathway are stable and readily purifiable. We currently have a number of structural ‘snapshots’ of NAD+- and ATP-dependent ligases and the closely related capping enzymes in different catalytic conformations. These studies have shed considerable light on the mode of action of these nucleotidyl transferases and suggest that this family of enzymes share a conserved catalytic mechanism. Despite having considerably different amino acid sequences, both the NAD+- and ATP-dependent DNA ligases appear to have a common core architecture responsible for nick recognition and sealing. The crystal structures have confirmed and clarified the catalytic roles of the six conserved sequence motifs and the structural basis for nucleotide specificity. The virally encoded ligase enzymes, such as T7 and Chlorella viruses, have essentially the same catalytic properties as the larger bacterial and eukaryotic ligases. The larger ligases have additional domains that are likely to enhance certain properties of these enzymes, such as DNA binding, nick recognition and targeting of the enzymes to sites of DNA damage, replication and recombination, but they do not appear to be directly involved in catalysis. One big question remains to be answered, what is the structural basis for DNA nick recognition? As discussed above, DNA ligases have exquisite specificity for nicked DNA and stable ligase–DNA complexes can be readily produced. The challenge now is to trap and crystallise the various ligase–DNA intermediates and build up a complete picture of the ligase catalytic pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Louise Serpell for assistance with the figures and for critically reading this manuscript. A.J.D. is a Royal Society University Research Fellow. S.W.S. is supported by the BK21 program of the Ministry of Education and the Center for Molecular Catalysis (KOSEF).
REFERENCES
- 1.Lehman I.R. (1974) Science, 186, 790. [DOI] [PubMed] [Google Scholar]
- 2.Engler M.J. and Richardson,C.C. (1982) In Boyer,P.D. (ed.), The Enzymes. Academic Press, New York, NY, Vol. XV, pp. 3–29.
- 3.Lindahl T. and Barnes,D.E. (1992) Annu. Rev. Biochem., 61, 251–281. [DOI] [PubMed] [Google Scholar]
- 4.Shuman S. and Schwer,B. (1995) Mol. Microbiol., 17, 405–410. [DOI] [PubMed] [Google Scholar]
- 5.Tomkinson A.E., Totty,N.F., Ginsburg,M. and Lindahl,T. (1991) Proc. Natl Acad. Sci. USA, 88, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kletzin A. (1992) Nucleic Acids Res., 20, 5389–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heaphy S. Singh,M. and Gait,M.J. (1987) Biochemistry, 18, 2980. [Google Scholar]
- 8.Baymiller J., Jennings,S., Kienzle,B., Gorman,J.A., Kelly,R. and McCullough,J.E. (1994) Gene, 142, 129–134. [DOI] [PubMed] [Google Scholar]
- 9.Fresco L.D. and Buratowski,S. (1994) Proc. Natl Acad. Sci. USA, 91, 6624–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuman S., Liu,Y. and Schwer,B. (1994) Proc. Natl Acad. Sci. USA, 91, 12046–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L. and Koonin,E.V. (1999) J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 12.Kodama K., Barnes,D.E. and Lindahl,T. (1991) Nucleic Acids Res., 19, 6093–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanya H.S., Doherty,A.J. Ashford,S.R. and Wigley,D.B. (1996) Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]
- 14.Håkansson K., Doherty,A.J., Shuman,S. and Wigley,D.B. (1997) Cell, 89, 545–553. [DOI] [PubMed] [Google Scholar]
- 15.Singleton M.R., Håkansson,K., Timson,D.J. and Wigley,D.B. (1999) Structure, 7, 35–42. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.Y., Chang,C., Song,H.K., Moon,J., Yang,J.K., Kim,H.K., Kwon,S.T. and Suh,S.W. (2000) EMBO J., 19, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty A.J. and Wigley,D.B. (1999) J. Mol. Biol., 285, 63–71. [DOI] [PubMed] [Google Scholar]
- 18.Murzin A.G. (1996) Curr. Opin. Struct. Biol., 6, 386–394. [DOI] [PubMed] [Google Scholar]
- 19.Suck D. (1997) Nat. Struct. Biol., 4, 161–165. [DOI] [PubMed] [Google Scholar]
- 20.Bycroft M., Hubbard,T.J.P., Proctor,M., Freund,S.M.V. and Murzin,A.G. (1997) Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- 21.Jaishree T.N., Ramakrishnan,V. and White,S.W. (1996) Biochemistry, 35, 2845–2853. [DOI] [PubMed] [Google Scholar]
- 22.Bochkarev A., Bochkareva,E., Frappier,L. and Edwards,A.M. (1999) EMBO J., 18, 4498–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Cell, 9, 963–974. [DOI] [PubMed] [Google Scholar]
- 24.Schindler T., Perl,D., Graumann,P., Sieber,V., Marahiel,M.A. and Schmid,F.X. (1998) Proteins, 30, 401–406. [DOI] [PubMed] [Google Scholar]
- 25.Peat T.S., Newman,J., Waldo,G.S., Berendzen,J. and Terwilliger,T.C. (1998) Structure, 6, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 26.Sette M., van Tilborg,P., Spurio,R., Kaptein,R., Paci,M., Gualerzi,C.O. and Boelens,R. (1997) EMBO J., 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghunathan S., Ricard,C.S., Lohman,T.M. and Waksman,G. (1997) Proc. Natl Acad. Sci. USA, 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafferty J.B., Sedelnikova,S.E., Hargreaves,D., Artymiuk,P.J., Baker,P.J., Sharples,G.J., Mahdi,A.A., Lloyd,R.G. and Rice,D.W. (1996) Science, 274, 415–421. [DOI] [PubMed] [Google Scholar]
- 29.Roe S.M., Barlow,T., Brown,T., Oram,M., Keeley,A., Tsaneva,I.R. and Pearl,L.H. (1998) Mol. Cell, 2, 361–372. [DOI] [PubMed] [Google Scholar]
- 30.Murzin A.G. (1993) EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriskanda V. and Shuman,S. (1998) Nucleic Acids Res., 26, 4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackey Z.B., Niedergang,C., Murcia,J.M., Leppard,J., Au,K., Chen,J., de Murcia,G. and Tomkinson,A.E. (1999) J. Biol. Chem., 274, 21679–21687. [DOI] [PubMed] [Google Scholar]
- 33.Klug A. and Schwabe,J.W.R. (1995) FASEB J., 9, 597–604. [PubMed] [Google Scholar]
- 34.Schmiedeskamp M. and Klevit,R.E. (1994) Curr. Opin. Struct. Biol., 4, 28–35. [Google Scholar]
- 35.Mackay J.P. and Crossley,M. (1998) Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- 36.Taylor R.M., Whitehouse,J., Cappelli,E., Frosina,G. and Caldecott,K.W. (1998) Nucleic Acids Res., 26, 4804–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gradwohl G., Menissier de Murcia,J.M., Molinete,M., Simonin,F., Koken,M., Hoeijmakers,J.H. and de Murcia,G. (1990) Proc. Natl Acad. Sci. USA, 87, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo J. and Barany,F. (1996) Nucleic Acids Res., 24, 3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sriskanda V., Schwer,B., Ho,C.K. and Shuman,S. (1999) Nucleic Acids Res., 27, 3953–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labahn J., Scharer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- 43.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) Nat. Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 44.Mullen G.P. and Wilson,S.H. (1997) Biochemistry, 36, 4713–4717. [DOI] [PubMed] [Google Scholar]
- 45.Bork P., Hofmann,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- 46.Callebaut I. and Mornon,J.P. (1997) FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X., Morera,S., Bates,P.A., Whitehead,P.C., Coffer,A.I., Hainbucher,K., Nash,R.A., Sternberg,M.J., Lindahl,T. and Freemont,P.S. (1998) EMBO J., 17, 6404–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 49.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 50.Critchlow S.E., Bowater,R.P. and Jackson,S.P. (1997) Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 51.Shuman S. (1995). Biochemistry, 34, 16138–16147. [DOI] [PubMed] [Google Scholar]
- 52.Ho C.K., Vanetten,J.L. and Shuman,S. (1996) J. Virol., 70, 6658–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sriskanda V. and Shuman,S. (1998) Nucleic Acids Res., 26, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odell M. and Shuman,S. (1999) J. Biol. Chem., 274, 14032–14039. [DOI] [PubMed] [Google Scholar]
- 55.Doherty A.J. and Dafforn,T.R. (2000) J. Mol. Biol., 296, 41–54. [Google Scholar]
- 56.Timson D.J. and Wigley,D.B. (1999) J. Mol. Biol., 285, 73–83. [DOI] [PubMed] [Google Scholar]
- 57.Cong P.J. and Shuman,S. (1995) Mol. Cell. Biol., 15, 6222–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shuman S. and Ru,X.M. (1995) Virology, 211, 73–83. [DOI] [PubMed] [Google Scholar]
- 59.Wang S.P., Deng,L., Ho,C.K. and Shuman,S. (1997) Proc. Natl Acad. Sci. USA, 94, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steitz T.A., Smerdon,S.J., Jäger,J. and Joyce,C.M. (1994) Science, 266, 2022–2025. [DOI] [PubMed] [Google Scholar]
- 61.Doherty A.J. (1999) Nucleic Acids Res., 27, 3253–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]