Abstract
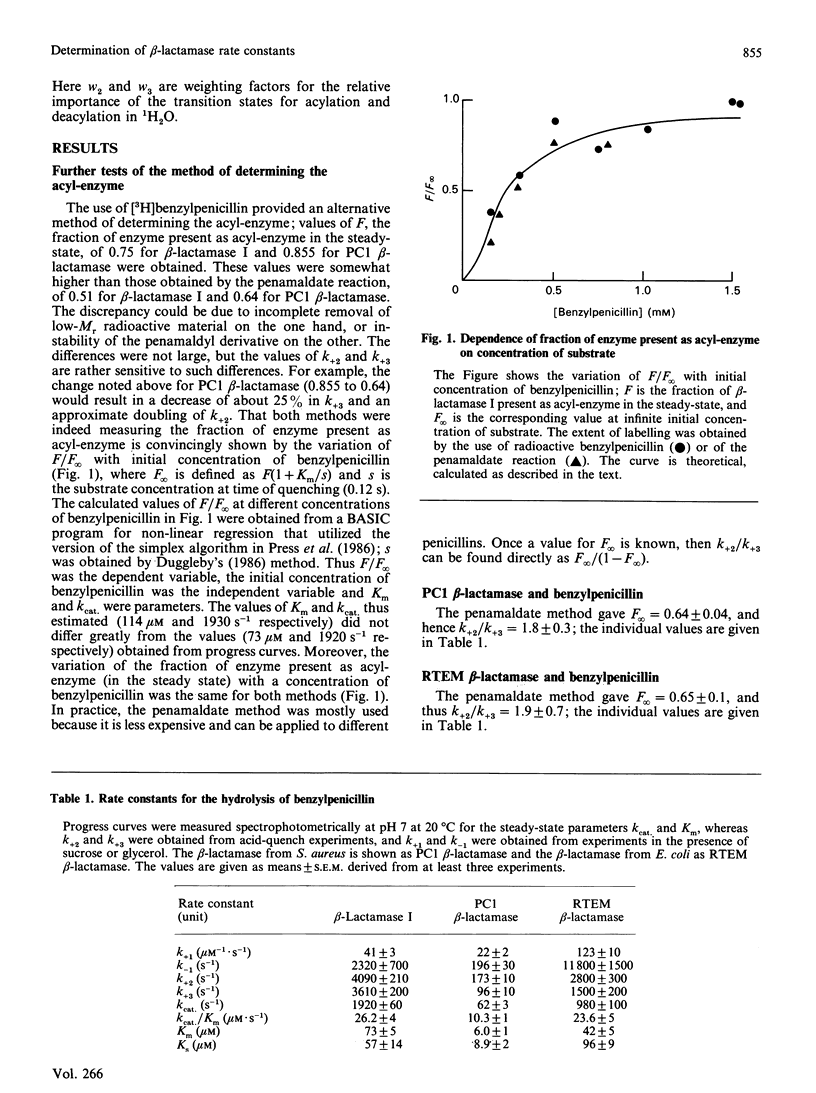
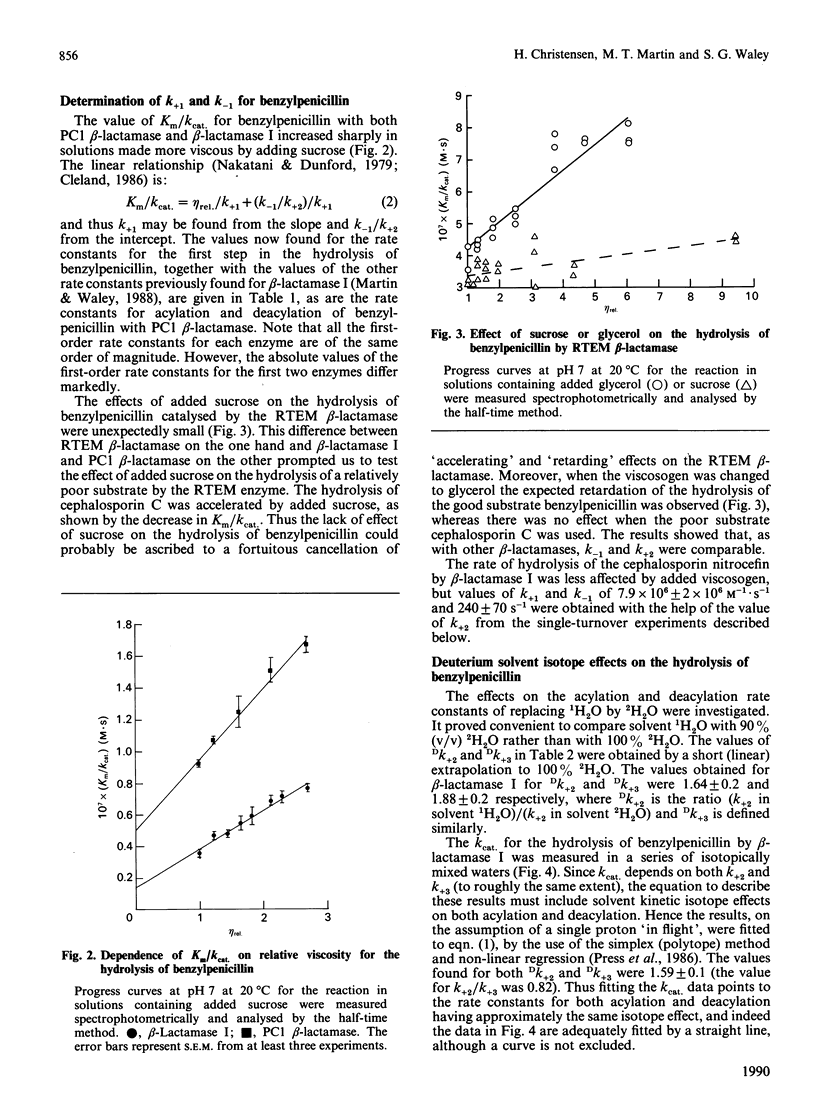
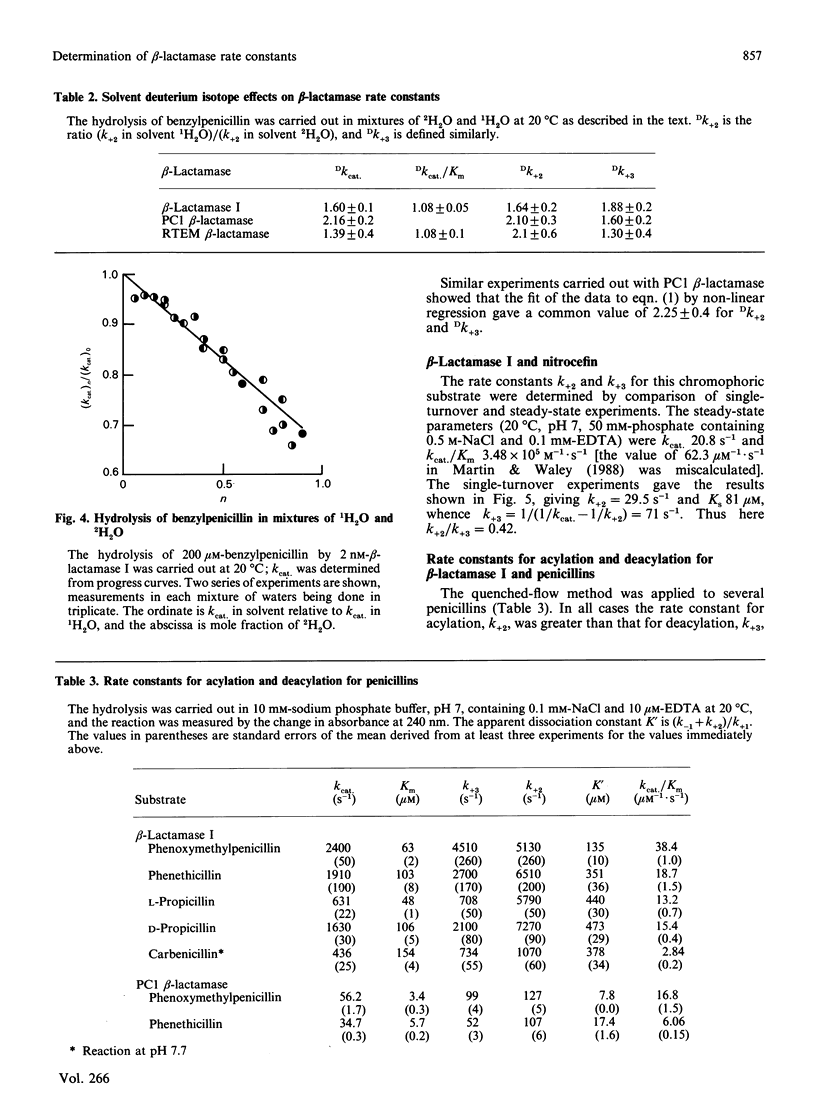
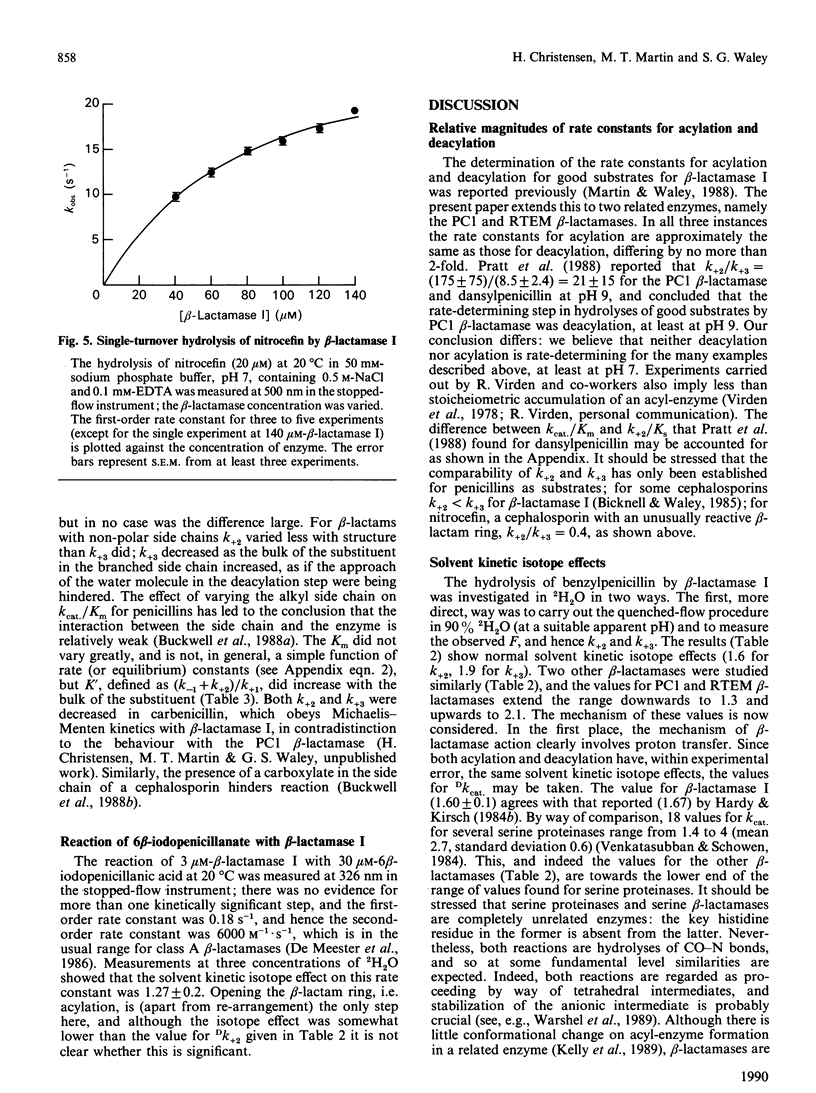
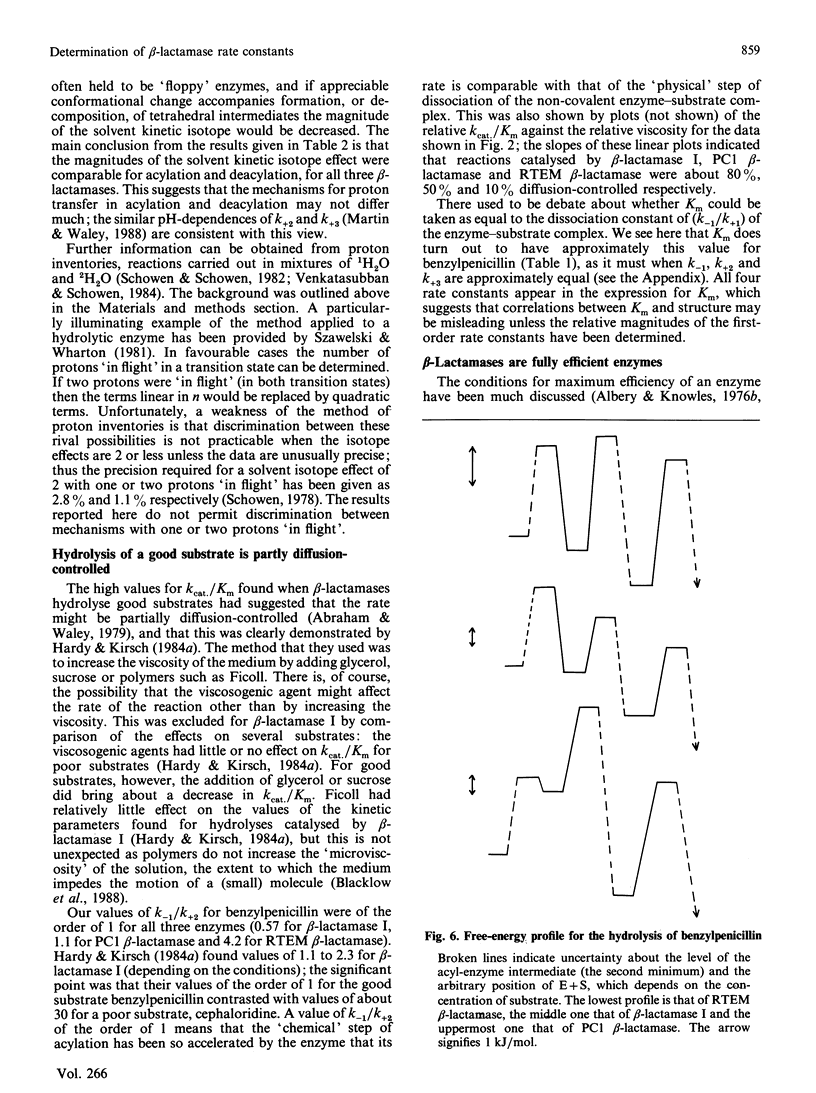
The rate constants for both acylation and deacylation of beta-lactamase PC1 from Staphylococcus aureus and the RTEM beta-lactamase from Escherichia coli were determined by the acid-quench method [Martin & Waley (1988) Biochem. J. 254, 923-925] with several good substrates, and, for a wider range of substrates, of beta-lactamase I from Bacillus cereus. The values of the acylation and deacylation rate constants for benzylpenicillin were approximately the same (i.e. differing by no more than 2-fold) for each enzyme. The variation of kcat./Km for benzylpenicillin with the viscosity of the medium was used to obtain values for all four rate constants in the acyl-enzyme mechanism for all three enzymes. The reaction is partly diffusion-controlled, and the rate constant for the dissociation of the enzyme-substrate complex has approximately the same value as the rate constants for acylation and deacylation. Thus all three first-order rate constants have comparable values. Here there is no single rate-determining step for beta-lactamase action. This is taken to be a sign of a fully efficient enzyme.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albery W. J., Knowles J. R. Efficiency and evolution of enzyme catalysis. Angew Chem Int Ed Engl. 1977 May;16(5):285–293. doi: 10.1002/anie.197702851. [DOI] [PubMed] [Google Scholar]
- Albery W. J., Knowles J. R. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976 Dec 14;15(25):5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- Albery W. J., Knowles J. R. Free-energy profile of the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1976 Dec 14;15(25):5627–5631. doi: 10.1021/bi00670a031. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Baldwin G. S., Edwards G. F., Kiener P. A., Tully M. J., Waley S. G., Abraham E. P. Production of a variant of beta-lactamase II with selectively decreased cephalosporinase activity by a mutant of Bacillus cereus 569/H/9. Biochem J. 1980 Oct 1;191(1):111–116. doi: 10.1042/bj1910111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R., Waley S. G. Cryoenzymology of Bacillus cereus beta-lactamase II. Biochemistry. 1985 Nov 19;24(24):6876–6887. doi: 10.1021/bi00345a021. [DOI] [PubMed] [Google Scholar]
- Blacklow S. C., Raines R. T., Lim W. A., Zamore P. D., Knowles J. R. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988 Feb 23;27(4):1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- Campbell J. I., Scahill S., Gibson T., Ambler R. P. The phototrophic bacterium Rhodopseudomonas capsulata sp108 encodes an indigenous class A beta-lactamase. Biochem J. 1989 Jun 15;260(3):803–812. doi: 10.1042/bj2600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrey E. A., Pain R. H. Conformation of a stable intermediate on the folding pathway of Staphylococcus aureus penicillinase. Biochim Biophys Acta. 1978 Mar 28;533(1):12–22. doi: 10.1016/0005-2795(78)90542-1. [DOI] [PubMed] [Google Scholar]
- Davies R. B., Abraham E. P. Separation, purification and properties of beta-lactamase I and beta-lactamase II from Bacillus cereus 569/H/9. Biochem J. 1974 Oct;143(1):115–127. doi: 10.1042/bj1430115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Frère J. M., Waley S. G., Cartwright S. J., Virden R., Lindberg F. 6-beta-Iodopenicillanate as a probe for the classification of beta-lactamases. Biochem J. 1986 Nov 1;239(3):575–580. doi: 10.1042/bj2390575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzou P., Petsko G. A. Proteins at work: "stop-action" pictures at subzero temperatures. Adv Protein Chem. 1984;36:245–361. [PubMed] [Google Scholar]
- Duggleby R. G. Progress-curve analysis in enzyme kinetics. Numerical solution of integrated rate equations. Biochem J. 1986 Apr 15;235(2):613–615. doi: 10.1042/bj2350613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D., Benner S. A. Free energy differences between enzyme bound states. J Theor Biol. 1987 Aug 21;127(4):491–506. doi: 10.1016/s0022-5193(87)80145-5. [DOI] [PubMed] [Google Scholar]
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Hardy L. W., Kirsch J. F. Diffusion-limited component of reactions catalyzed by Bacillus cereus beta-lactamase I. Biochemistry. 1984 Mar;23(6):1275–1282. doi: 10.1021/bi00301a040. [DOI] [PubMed] [Google Scholar]
- Hardy L. W., Kirsch J. F. pH dependence and solvent deuterium oxide kinetic isotope effects on Bacillus cereus beta-lactamase I catalyzed reactions. Biochemistry. 1984 Mar 13;23(6):1282–1287. doi: 10.1021/bi00301a041. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Knox J. R., Zhao H., Frère J. M., Ghaysen J. M. Crystallographic mapping of beta-lactams bound to a D-alanyl-D-alanine peptidase target enzyme. J Mol Biol. 1989 Sep 20;209(2):281–295. doi: 10.1016/0022-2836(89)90277-5. [DOI] [PubMed] [Google Scholar]
- Madgwick P. J., Waley S. G. beta-lactamase I from Bacillus cereus. Structure and site-directed mutagenesis. Biochem J. 1987 Dec 15;248(3):657–662. doi: 10.1042/bj2480657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. T., Waley S. G. Kinetic characterization of the acyl-enzyme mechanism for beta-lactamase I. Biochem J. 1988 Sep 15;254(3):923–925. doi: 10.1042/bj2540923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchinson C., Pain R. H. Effects of sulphate and urea on the stability and reversible unfolding of beta-lactamase from Staphylococcus aureus. Implications for the folding pathway of beta-lactamase. J Mol Biol. 1985 Jul 20;184(2):331–342. doi: 10.1016/0022-2836(85)90384-5. [DOI] [PubMed] [Google Scholar]
- Persaud K. C., Pain R. H., Virden R. Reversible deactivation of beta-lactamase by quinacillin. Extent of the conformational change in the isolated transitory complex. Biochem J. 1986 Aug 1;237(3):723–730. doi: 10.1042/bj2370723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt R. F., McConnell T. S., Murphy S. J. Accumulation of acyl-enzyme intermediates during turnover of penicillins by the class A beta-lactamase of Staphylococcus aureus PC1. Biochem J. 1988 Sep 15;254(3):919–922. doi: 10.1042/bj2540919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samraoui B., Sutton B. J., Todd R. J., Artymiuk P. J., Waley S. G., Phillips D. C. Tertiary structural similarity between a class A beta-lactamase and a penicillin-sensitive D-alanyl carboxypeptidase-transpeptidase. 1986 Mar 27-Apr 2Nature. 320(6060):378–380. doi: 10.1038/320378a0. [DOI] [PubMed] [Google Scholar]
- Schowen K. B., Schowen R. L. Solvent isotope effects of enzyme systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- Schultz S. C., Richards J. H. Site-saturation studies of beta-lactamase: production and characterization of mutant beta-lactamases with all possible amino acid substitutions at residue 71. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1588–1592. doi: 10.1073/pnas.83.6.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., DeGrado W. F., Thomas B. J., Petteway S. R., Jr Purification and properties of thiol beta-lactamase. A mutant of pBR322 beta-lactamase in which the active site serine has been replaced with cysteine. J Biol Chem. 1984 Apr 25;259(8):5327–5332. [PubMed] [Google Scholar]
- Szawelski R. J., Wharton C. W. Kinetic solvent isotope effects on the deacylation of specific acyl-papains. Proton inventory studies on the papain-catalysed hydrolyses of specific ester substrates: analysis of possible transition state structures. Biochem J. 1981 Dec 1;199(3):681–692. doi: 10.1042/bj1990681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubban K. S., Schowen R. L. The proton inventory technique. CRC Crit Rev Biochem. 1984;17(1):1–44. doi: 10.3109/10409238409110268. [DOI] [PubMed] [Google Scholar]
- Virden R., Bristow A. F., Pain R. H. Reversible inhibition of penicillinase by quinacillin: evaluation of mechanisms involving two conformational states of the enzyme. Biochem Biophys Res Commun. 1978 Jun 14;82(3):951–956. doi: 10.1016/0006-291x(78)90875-6. [DOI] [PubMed] [Google Scholar]
- Waley S. G. Beta-lactamases: a major cause of antibiotic resistance. Sci Prog. 1988;72(288 Pt 4):579–597. [PubMed] [Google Scholar]
- Warshel A., Naray-Szabo G., Sussman F., Hwang J. K. How do serine proteases really work? Biochemistry. 1989 May 2;28(9):3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- Wharton C. W., Szawelski R. J. Half-time analysis of the integrated Michaelis equation. Simulation and use of the half-time plot and its direct linear variant in the analysis of some alpha-chymotrypsin, papain- and fumarase-catalysed reactions. Biochem J. 1982 May 1;203(2):351–360. doi: 10.1042/bj2030351. [DOI] [PMC free article] [PubMed] [Google Scholar]


