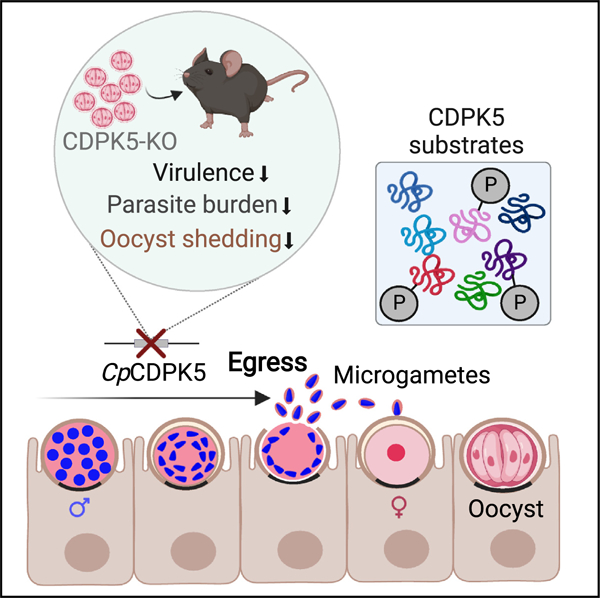
SUMMARY
The protozoan parasite Cryptosporidium is a leading cause of diarrhea in young children. The parasite’s life cycle involves a coordinated and timely progression from asexual to sexual stages, leading to the formation of the transmissible oocyst. Underlying molecular signaling mechanisms orchestrating sexual development are not known. Here, we describe the function of a signaling kinase in Cryptosporidium male gametogenesis. We reveal the expression of Cryptosporidium parvum calcium-dependent protein kinase 5 (CDPK5) during male gamete development and its important role in the egress of mature gametes. Genetic ablation of this kinase results in viable parasites, indicating that this gene is dispensable for parasite survival. Interestingly, cdpk5 deletion decreases parasite virulence and impacts oocyst shedding in immunocompromised mice. Using phosphoproteomics, we identify possible CDPK5 substrates and biological processes regulated by this kinase. Collectively, these findings illuminate parasite cell biology by revealing a mechanism controlling male gamete production and a potential target to block disease transmission.
Graphical Abstract
In brief
Nava et al. demonstrate that the Cryptosporidium calcium-dependent protein kinase 5 (CDPK5) functions during microgamont development and is important for the egress of male gametes. Phosphoproteomics reveals putative substrates of CDPK5. Kinase deletion reduces parasite virulence, disease severity, and oocyst shedding and thus identifies a potential target for blocking disease transmission.
INTRODUCTION
Cryptosporidium spp. is a leading cause of diarrheal disease and mortality among children living in resource-limited settings.1–4 Epidemiological studies have found an association of multiple episodes of Cryptosporidium infection with malnutrition, growth defects, and developmental abnormalities in young children.5,6 Besides being a pediatric pathogen, it is a major threat to the lives of immunocompromised individuals such as those infected with HIV/AIDS and organ transplant recipients. Both C. hominis and C. parvum infect humans, but C. parvum also naturally infects young ruminant animals such as calves and goat kids, resulting in neonatal diarrhea, poor growth, and mortality.7 Even in countries with good sanitation practices and adequate water treatment facilities, Cryptosporidium is a major cause of outbreaks associated with oocyst contamination of recreational water or farm facilities.8 These infectious oocysts are difficult to eliminate due to their resistance to standard disinfection procedures. Regarding treatment options, there is only one FDA-licensed drug, nitazoxanide, that is approved for treating human cryptosporidiosis. However, nitazoxanide shows poor clinical efficacy in malnourished children and the immunocompromised population.9,10 Besides the unavailability of an effective drug, there is no vaccine available to prevent infection. Thus, the tremendous public health burden posed by cryptosporidiosis can only be curbed by developing effective drugs and vaccines.4 Advancements in treatment and vaccine development against Cryptosporidium require a comprehensive understanding of fundamental parasite biology such as the molecular processes that underlie the development of asexual and sexual life cycle stages. Targeting sexual stages and oocyst development could provide a route toward breaking the within-host re-infection cycle as well as blocking disease transmission.
Cryptosporidium exhibits an extremely timed progression of developmental events throughout its life cycle (Figure S1). The cycle begins with the ingestion of an infectious oocyst that excysts to release four haploid sporozoites. These sporozoites infect intestinal epithelial cells and replicate to release merozoites. After three rounds of asexual merogony (36 h), the parasite switches to the sexual developmental cycle ~40 h post-infection. Type I meronts (8-merozoite stage) directly develop into the male gamont (microgamont) and female gamont (macrogamont).11,12 The microgametes (also called “male gametes”) fertilize the macrogamont, resulting in a diploid zygote that undergoes meiosis and sporogony to produce an infectious oocyst containing four haploid sporozoites.13 These oocysts excyst to cause re-infection within the same host and are also shed in the feces for transmission of infection to another host. There is a critical gap in our knowledge regarding the molecular processes and underlying signaling machinery that control the development of these stages for a coordinated progression and completion of the parasite’s life cycle.
With respect to the Cryptosporidium sexual developmental cycle, we lack an understanding of the microgametogenesis process that results in the production of male gametes that fertilize a macrogamont. The dearth of molecular markers and antibodies makes it hard to identify and track different stages during male gametogenesis. The only known male-specific markers to date for Cryptosporidium are HAP2 and AP2-M. HAP2 is a membrane fusion protein that has been shown to localize to a single pole on the mature male gamete,11 and AP2-M is a transcription factor that localizes to early males.14 Ultrastructural studies in Cryptosporidium suggest that during the male gametogenesis process, sixteen non-flagellated wedge-shaped microgametes bud and remain within the microgamont, and these differentiated microgametes have characteristic microtubules that extend longitudinally along their spindle-shaped nuclei.15,16 How these gametes are released from the microgamont and reach the macrogamont for fertilization is not understood.
In the related parasite, Plasmodium, the well-characterized microgametogenesis (also called male gametogenesis) process involves gametocyte activation and exflagellation, characterized by distinct cell rounding events and the release of eight motile male gametes.17,18 However, the cell biological events during microgametogenesis that dictate the formation and, ultimately, release (egress) of male gametes remain unknown in Cryptosporidium.
Work done on Plasmodium spp. and Toxoplasma gondii has delineated the critical role of secondary messengers (such as calcium, cyclic nucleotides) and the activation of plant-like calcium-dependent protein kinases (CDPKs) in controlling parasite processes like motility, invasion and egress of asexual stages, and sexual stage development.19–24 Although the Cryptosporidium genome encodes for these conserved CDPKs, it is likely that their functions can be markedly different from Toxoplasma and Plasmodium. This is due to the adaptation of Cryptosporidium to the intestinal environment and its greater evolutionary relatedness to eugregarines, a group of apicomplexan parasites with an extracellular lifestyle inhabiting the gut of terrestrial and marine invertebrates.25,26 Thus far, the essential role of only Cryptosporidium CDPK1 in asexual proliferation has been described.27 However, the signaling mechanism that controls microgametogenesis and specifically how these male gametes escape from the microgamont have not been characterized for Cryptosporidium.
In this study, we address this knowledge gap by using a combination of genetically modified parasites, cellular biology, phosphoproteomics, and animal infection experiments to dissect the process of male gametogenesis in Cryptosporidium. We identified the important function of the signaling kinase CDPK5 in male gamete egress, revealed its putative substrates, and demonstrated the role of this kinase in parasite virulence. Overall, our findings enhance the understanding of Cryptosporidium biology by elucidating the role of this kinase in male gametogenesis and controlling parasite burden in infected animals.
RESULTS
Cryptosporidium CDPK5 is a sexual-stage kinase
To gain an insight into the molecular signaling mechanisms controlling development of sexual stages, we sought to identify CDPKs in Cryptosporidium that show stage-specific expression. By searching the Cryptosporidium database (CryptoDB)28 (https://cryptoDB.org), we identified a “calcium/calmodulin dependent protein kinase” (gene ID: cgd2_1300) belonging to the CDPK family. This protein has the characteristic serine/threonine kinase domain and a calmodulin-like domain containing EF-hand motifs for binding calcium ions (Figure 1A). We interrogated the RT-PCR data available in the CryptoDB on parasite transcript abundance at different time points after host cell infection29 and found this gene to be highly expressed at the 48 h time point (Figure 1B). Based on this late expression profile, we speculated that this kinase may be involved in the parasite’s sexual cycle, since this time point coincides with the development of male and female gamont stages.
Figure 1. CDPK5 is expressed only in the sexual stage of C. parvum.
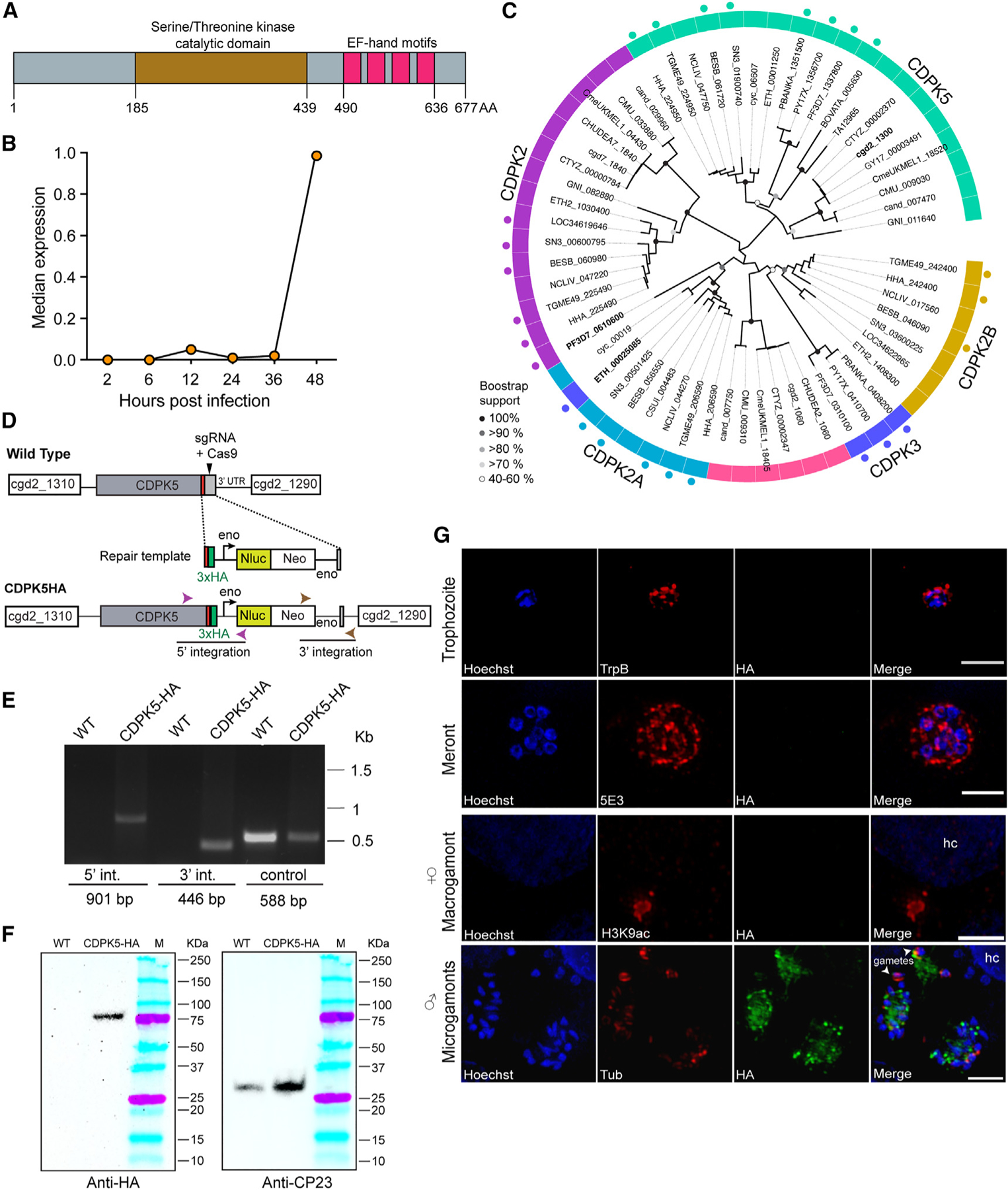
(A) Domain architecture of CDPK5 showing the serine/threonine kinase catalytic domain and four calcium-binding (EF-hand) motifs. AA, amino acids.
(B) Median transcript expression of cgd2_1300 gene over 2, 6, 12, 24, 36, and 48 h infection time points. Gene expression was normalized to C. parvum 18S rRNA. Data shown here have been replotted from Mauzy et al.29
(C) Maximum likelihood phylogenetic tree showing the relationship of Cryptosporidium CDPK5 with selected apicomplexan kinases. Tip labels indicate the gene IDs of the kinases. Dots outside the ring represent annotated CDPKs. Abbreviated species names for Cryptosporidium parvum, C. hominis, C. andersoni, C. muris, C. meleagridis, C. tyzzeri, Gregarina niphandrodes, Toxoplasma gondii, Plasmodium falciparum, P. yoelli, P. berghei, Hammondia hammondi, Neospora caninum, Eimeria tenella, Cyclospora cayetanensis, Cystoisospora suis, Babesia bovata, Besnoitia besnoiti, Sarcosystis neurona, and Theileria annulata are shown, and detailed annotations are provided in Table S1. Bootstrap support values for 1,000 replicates are indicated on the branches.
(D) Schematic for endogenous tagging of cdpk5 gene locus with 3X-HA epitope tag. The locations of the single guide RNA (sgRNA) in the 3ʹ UTR, the Cas9 cut site (arrow), and the repair template for homologous recombination are shown. Nluc, nanoluciferase; Neo, neomycin resistance marker; Eno, enolase promoter and 3ʹ UTR.
(E) PCR mapping of the correct integration events in the CDPK5-HA transgenic parasite. Fecal genomic DNA extracted from wild-type (WT) and CDPK5-HA transgenic parasites were used for PCRs. The locations of primers for checking 5ʹ and 3ʹ integrations are indicated in (A), and their sequences are provided in Table S2.
(F) Western blot detection of 80 kDa C. parvum CDPK5 protein in CDPK5-HA strain using anti-HA antibody. WT, wild type C. parvum. CP23 antibody was used to detect the presence of protein in both WT and transgenic strains.
(G) Immunofluorescence assay and super-resolution microscopy of CDPK5-HA transgenic parasites showing localization only in the male (microgamont stage). Asexual stages (trophozoite and 8-nuclei meront) were visualized after 24 h of HCT-8 infection using tryptophan B (TrpB) and 5E3 antibodies. Female and male sexual stages at 48 h post-infection were detected using H3K9ac and α-tubulin (tub) antibodies, respectively, and are shown in red. HA, anti-HA antibody (green). Parasite nuclei were detected with Hoechst stain (blue). hc, host cell. Maximum intensity projection images are shown. Scale bars, 2 μm. Representative data from three independent experiments are shown.
To further investigate the evolution of this C. parvum CDPK and its relationship to other related apicomplexan kinases, we conducted a maximum likelihood phylogenetic analysis (Figure 1C; Table S1). For this analysis, we used a protein BLAST search to identify parasite homologs with an e-value cutoff of >1e−10. These were further filtered using the OrthoMCL database (https://orthomcl.org) to include only core sequences, and any peripheral hits (such as CDPK1, CDPK4, CDPK6, and CDPK7) were removed. Our analysis revealed that this Cryptosporidium kinase is related to CDPK5 from Plasmodium falciparum (PF3D7_1337880) and Toxoplasma gondii (TGME49_ 224950) as well as to other annotated apicomplexan CDPK5 (aquamarine color ring in Figure 1B, annotated CDPK5 in other apicomplexan parasites are shown as dots outside this ring). As expected, Cryptosporidium CDPK5 shows evolutionary relatedness to the kinase (GNI_011640) from an “early evolving” apicomplexan parasite, Gregarina niphandrodes, that colonizes the gut of the yellow mealworm beetle (Figure 1C). Functional studies done in Plasmodium spp. have demonstrated that CDPK5 is required for parasite survival, its expression in asexual replicative stages and its key function in the egress of blood-stage schizonts and liver-stage merozoites, but its role in the sexual cycle has not been tested.21,24,30 Other kinases in Plasmodium, namely CDPK1, CDPK2, and CDPK4, have been shown to play critical roles in male gamete exflagellation during its sexual stage cycle.19,20,31 On the contrary, we and others have reported that the Cryptosporidium CDPK1 is expressed only during the asexual cycle (in trophozoite and 8-nuclei meront stages) and plays an essential role in asexual proliferation.12,27 Thus, the evolutionary relatedness of Cryptosporidium CDPK5 to gregarines, along with other contributing factors such as the parasite’s adaptation to the intestinal environment and a single-host life cycle, likely contributed toward shaping a different cellular function for this kinase. To evaluate sequence variability in CDPK5 across Cryptosporidium species, we analyzed publicly available whole-genome sequences for C. parvum (n = 158) and C. hominis (n = 96). We found no changes in sequence within C. parvum or C. hominis isolates, thus indicating that this kinase is highly conserved in Cryptosporidium (Figure S2).
To assess the expression and localization of CDPK5 in Cryptosporidium, we introduced a hemagglutinin (3X-HA) epitope tag at the C terminus of this kinase using CRISPR-Cas9 genome editing and interferon γ (IFN-γ) knockout (KO) mouse model system to generate a stable transgenic parasite strain (CDPK5-HA) (Figure 1D). The correct 5ʹ and 3ʹ integration for the tagged cdpk5 gene locus was confirmed using PCR (Figure 1E). Next, we infected HCT-8 cells with CDPK5-HA transgenic oocysts purified from mice feces and harvested cells after 48 h for western blot. We detected the expected 80 kDa protein using an antibody against the HA tag (Figure 1F). To assess the expression and localization of CDPK5, infected cultures were fixed at 24 or 48 h for immunofluorescence assays and super-resolution microscopy. We did not detect expression of CDPK5 during asexual stages using C. parvum tryptophan B (TrpB) and 5E3 antibodies that stain trophozoites and the apical ends of merozoites, respectively,32,33 thus validating the gene expression data (Figure 1G). To further evaluate the expression of this kinase during sexual stages that are predominant at 48 h, we used antibodies that have been reported to mark male (microgamonts) and female (macrogamont) stages.11,27 Since the female gamont nucleus does not stain well with DNA stains such as Hoechst, we used an acetylated histone antibody (H3K9Ac) that has been reported to react with accumulated histones in its nucleus.11,27 Using the anti-HA antibody, we could not detect any expression of CDPK5 in the female gamont (Figure 1G). To visualize the male stages, we used nuclear staining and an α-tubulin antibody that has been reported to stain the microtubules surrounding the male gametes.11 Interestingly, we detected a high signal of CDPK5 in microgamonts. Thus, these results clearly demonstrate that CDPK5 is expressed only during the male developmental stage of the parasite’s life cycle.
CDPK5 localization and expression changes during male gametogenesis
We next sought to investigate the localization and expression of CDPK5 during the process of male gametogenesis that leads to the development of mature male gametes from the microgamont. To assess this, we analyzed the developmental stages during this process using immunofluorescence assay followed by super-resolution structured illumination microscopy and three-dimensional (3D) image reconstruction.
There are no defined markers available to track male gametogenesis process in Cryptosporidium, but electron microscopy and recent molecular studies have demonstrated that male gametes lack flagella and that nuclei shape changes from round to more bullet- or spindle-like in mature male gametes.11,12,15,16 Each male gamete that emerges from the microgamont contains a set of microtubules that can be stained with an α-tubulin antibody. These parallel microtubules run outside the bullet-shaped nucleus of the mature male gamete.11 Therefore, to differentiate between the different stages of development, we relied on counting the number of nuclei and changes in nuclear morphology during the process of male gametogenesis, as well as utilizing the α-tubulin antibody to visualize the newly formed gametes. We infected HCT-8 cells with CDPK5-HA parasites and fixed cells at 42 and 48 h post-infection. The 42 h time point correlates with the emergence of male gamonts in culture.11 By nuclear morphology, we observed the 8 nuclei arranged in a rosette at this early stage but found no CDPK5 signal, thus indicating that the kinase was not expressed during early stages of male gametogenesis. However, we could detect expression of CDPK5 at the 48 h time point during gamont development and the formation of male gametes. We defined these stages as “developing” (12–16 round nuclei, very weak tubulin staining), “mature” (compact 16 nuclei, intense tubulin staining), “egress ready” (male gametes with bullet-shape nuclei getting ready to escape, tubulin staining only in mature male gametes), and “egressed” (mature male gametes with bullet-shape nuclei released from the gamont). It is important to note here that only a few developing and mature male gamont stages were seen, while the egress-ready and egressed gamonts and “free” gametes were readily observed. As depicted in Figures 2 and S3, CDPK5 expression changed during male gametogenesis.
Figure 2. Localization of CDPK5 during male gametogenesis.
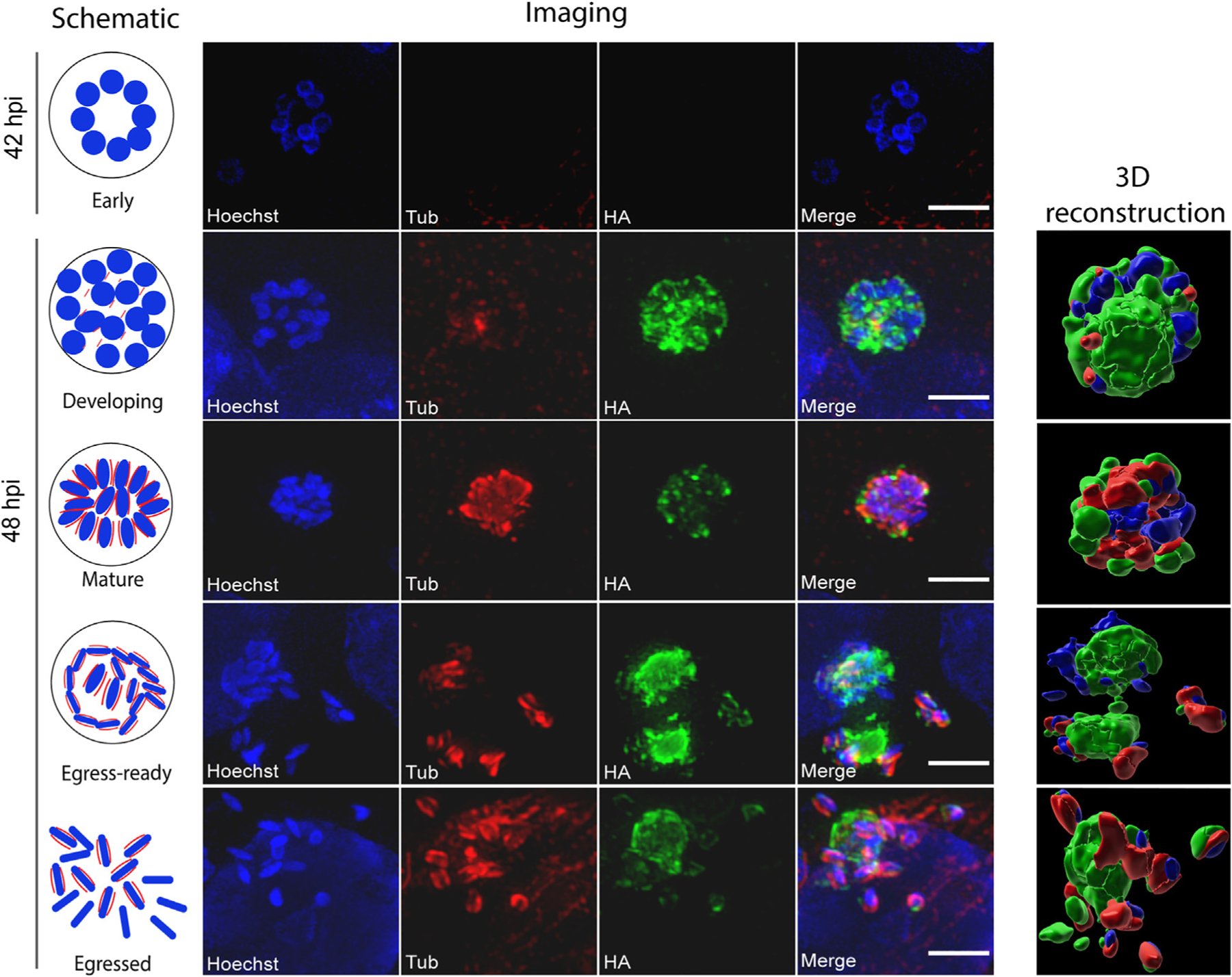
Immunofluorescence assay and super-resolution imaging of CDPK5-HA transgenic parasites after infection of HCT-8 cells. Early males were detected at 42 h, while the other stages (developing, mature, egress ready, and egressed) were imaged after the 48 h time point. Anti-HA (green) and anti-alpha-tub (tub, red) antibodies and Hoechst staining of parasite nuclei (blue) were used to follow gamont development. 3D reconstructions of images using Imaris software are shown. Scale bars, 2 μm. Representative data from three independent experiments are shown. Additional images are provided in Figure S3.
We observed a vacuolar staining for CDPK5 in the developing microgamont, while the mature gamont exhibited a vesicular or punctate staining with the kinase relocalizing to the cell periphery. Notably, there was an intense signal of CDPK5 right before egress (egress-ready stage) that seemed to dampen after the release of mature male gametes (egressed stage).
To better visualize these developmental stages, we conducted a 3D reconstruction on representative immunofluorescence images (Figure 2). The 3D image reconstruction analysis provided an enhanced understanding of CDPK5 and tubulin expression dynamics during different stages of male gametogenesis (Figure 2). Collectively, these results demonstrate that the localization and expression of CDPK5 change during male gametogenesis with an increase in signal intensity prior to the egress of mature male gametes.
Genetic ablation of CDPK5 impairs egress of mature male gametes
To understand the function of CDPK5 in Cryptosporidium, we generated a transgenic strain with the cdpk5 gene deleted. This genetic KO line (CDPK5-KO) contained a nanoluciferase reporter-neomycin resistance cassette driven by the C. parvum enolase promoter that replaced the cdpk5 gene (Figure 3A). Diagnostic PCRs using primers flanking the recombination site revealed the correct 5ʹ and 3ʹ integration events, while gene-specific primers confirmed the absence of the cdpk5 gene (Figure 3B; Table S2). Further, no cdpk5 transcript was detected at 48 h by RT-PCR, thus validating the loss of this gene in the CDPK5-KO transgenic line (Figure S4).
Figure 3. Knockout of cdpk5 leads to a defect in the egress of male gametes from the microgamont.
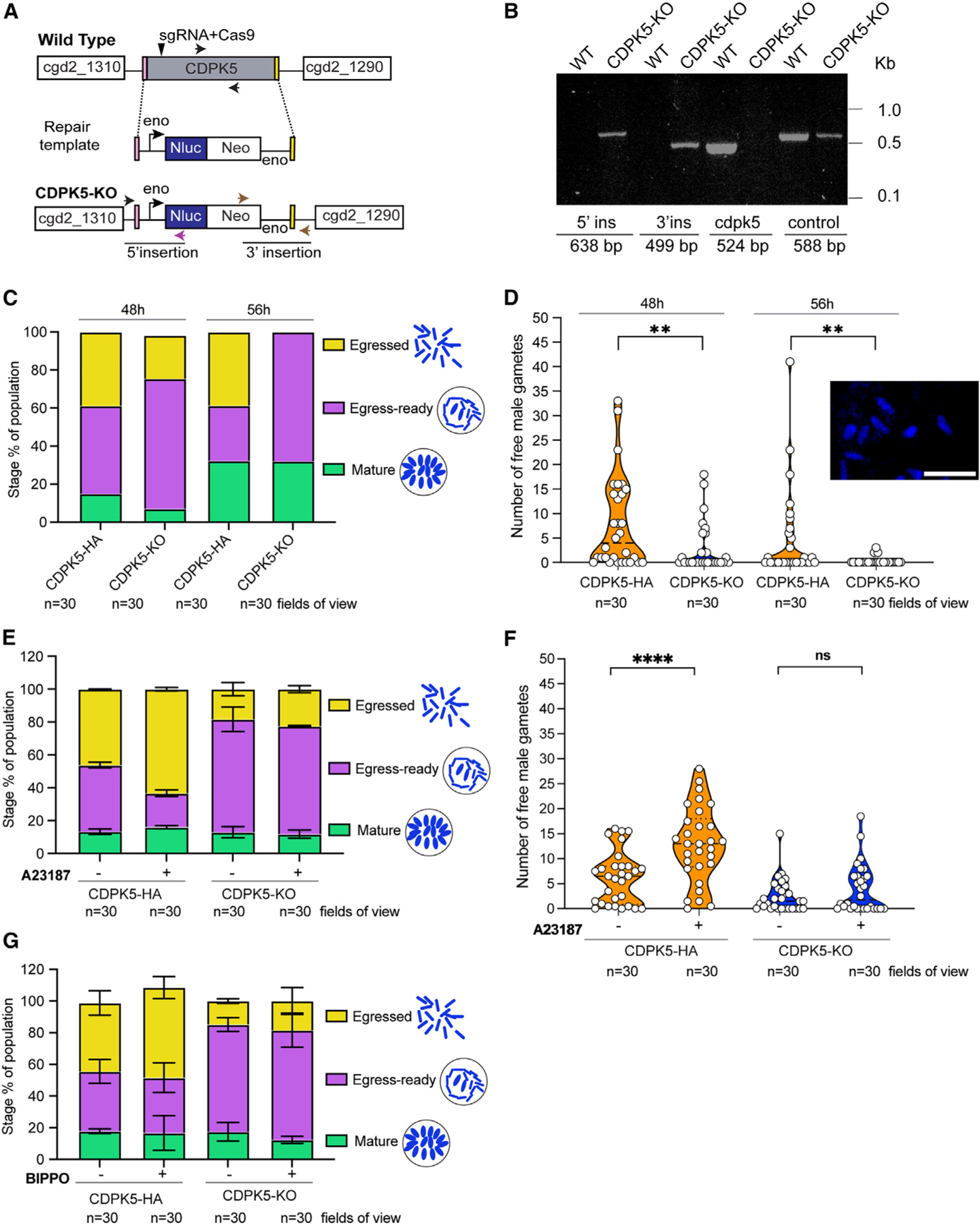
(A) Schematic showing C. parvum CDPK5-KO transgenic line. The cdpk5 gene was replaced with an Nluc-Neo cassette by homologous recombination. The locations of the sgRNA within the gene and the Cas9 cut site (arrow) are shown. Nluc, nanoluciferase; Neo, neomycin resistance marker; Eno, enolase promoter and 3ʹ UTR.
(B) PCR mapping of the CDPK5-KO transgenic to confirm the correct 5ʹ and 3ʹ integration events. Fecal genomic DNA extracted from WT and CDPK5-KO transgenic parasites were used for PCRs. The locations of primers for integrations are indicated in (A), and their sequences are provided in Table S2.
(C) Quantification of microgamont stages in CDPK5-HA and CDPK5-KO transgenics at 48 and 56 h post-infection of HCT-8 cells.
(D) Violin plot showing distribution of free male gamete counts after egress in CDPK5-HA and CDPK5-KO parasites at 48 and 56 h post-infection of HCT-8 cells. The inset shows a representative image of Hoechst-stained free male gametes. Scale bar, 2 μm. Data represented are from two independent experiments. Mann-Whitney test: two-tailed **p = 0.063 for 48 h and **p = 0.0016 for 56 h comparisons.
(E) Quantification of microgamont stages in CDPK5-HA and CDPK5-KO parasites treated with or without 4 μM A23187. Data represented are mean ± SEM (standard error of mean) from two independent experiments.
(F) Violin plot showing distribution of free male gamete counts after egress in CDPK5-HA and CDPK5-KO at 48 h post-infection of HCT-8 cells incubated for 30 min with or without A23187. Mann-Whitney test: two-tailed ****p < 0.0001; ns, not significant. Data represented are from two independent experiments.
(G) Quantification of microgamont stages in CDPK5-HA and CDPK5-KO parasites treated with or without 50 μM BIPPO. Data represented are mean ± SEM from two independent experiments.
For (C)–(G), images captured and quantified were from three technical replicates representing 30 fields of view for each strain. Representative images for (C) and (D) are provided in Figures S5 and S6.
To assess the effect on the parasite upon deletion of cdpk5, we infected HCT-8 cells with CDPK5-KO and CDPK5-HA transgenic oocysts and fixed cells for immunofluorescence assays and microscopy. Since the α-tubulin staining is observed only in fully mature microgamonts and emerging male gametes, we used nuclear staining to distinguish the various stages during male gametogenesis. In addition, we used Vicia villosa lectin (VVL) fluorescein for parasite staining, since this lectin stain has been previously reported to detect parasite stages.11,34 The VVL stain can identify most, but not all, intracellular stages; therefore, we relied on nuclear morphology to carefully visualize and quantify various stages of male gametogenesis in cells infected with CDPK5-HA and CDPK5-KO strains (Figures 3C, S5, and S6). At 48 h post-infection, we found a higher number (68.4%) of microgamonts arrested at the egress-ready gamont stage for the CDPK5-KO as compared to the CDPK5-HA (46.3%) strain. These egress-ready microgamonts were readily distinguished by their bullet-shaped nuclei and the arrangement of the mature gametes in a circular or wreath-like pattern (Figures S5 and S6). Interestingly, we observed a significantly lower percentage of egressed microgamonts (22.8%) in the CDPK5-KO strain compared to the CDPK5-HA (38.8%) strain (unpaired t test with Welch’s correction, two-tailed p = 0.0297, 30 fields of view counted for each strain), thus indicating that genetic ablation of this kinase impacted egress (Figure 3C).
Gamont development in Cryptosporidium has been reported to occur at 48 h post-infection by both fixed-cell and live-imaging experiments using reporter parasite strains.11,12 However, time-lapse microscopy studies tracking the development of microgamonts from type I meronts and male gamete egress events indicate that male gamete egress occurs at 48 h and continues up to an average time point of 56 h.12 Therefore, we decided to quantify microgamont stages at this time point (56 h) after the infection of HCT-8 cells with CDPK5-HA and CDPK5-KO parasites. Corroborating our 48 h results, we found the same percentage (68%) of microgamonts stuck in the egress-ready stage at 56 h for the CDPK5-KO strain (Figure 3C). For egressed-stage microgamonts, we detected the same percentage (38.7%) at 56 h post-infection as observed for 48 h (38.8%) in the CDPK5-HA strain. On the contrary, we did not observe any egressed gamonts in the CDPK5-KO strain at 56 h compared to the CDPK5-HA strain (unpaired t test with Welch’s correction, two-tailed p = 0.0052, 30 fields of view counted at each time point) (Figure 3C). Overall, these findings demonstrate that the block in egress for CDPK5-KO parasites is not released even at this later (56 h) time point, thus suggesting an important role of cdpk5 in male gametogenesis.
To directly assess the outcome of egress, we manually counted free male gametes released by CDPK5-HA and CDPK5-KO strains at 48 and 56 h post-infection. At both time points, there was a significant reduction in free male gametes for the CDPK5-KO compared to the CDPK5-HA strain (Figure 3D). The inset in Figure 3D shows a representative image of free male gametes that were recognized with their characteristic bullet-shaped nuclei. At the 56 h time point, we detected even fewer male gametes for the CDPK5-KO strain, likely due to the end of the egress window. Based on these results, we determined that 48 h was a more suitable time point to evaluate all stages of egress and thus used this time point for all further experiments.
Next, we wanted to investigate if we could artificially induce egress pharmacologically by increasing intracellular calcium levels in the parasite. We individually tested two different compounds, a calcium ionophore, A23187, that directly increases the calcium level and benzyl-3-isopropyl-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-one (BIPPO), which is an inhibitor of 3ʹ,5ʹ-cyclic nucleotide phosphodiesterases (PDEs). In Plasmodium and Toxoplasma, regulation of 3ʹ,5ʹ-cyclic guanosine monophosphate (cGMP) levels has been demonstrated to control the egress of asexual stages and gametogenesis.20,21,35–38 An increase in cGMP levels has been shown to activate cGMP-dependent protein kinase G (PKG), leading to calcium-dependent activation of CDPKs for the egress of asexual stages and the activation of gametogenesis in these parasites (Figure S7). The egress process can be artificially induced in these parasites by elevating cGMP levels using the fast-acting PDE inhibitor BIPPO.36 Exposure with 2 μM BIPPO in Plasmodium and 50 μM in Toxoplasma for few seconds to 30 min has been reported to induce egress.21,36,39 The Cryptosporidium genome encodes for a PKG (cgd8_750) and three PDEs (cgd6_4020, cgd3_2320, and cgd6_500), while the Plasmodium and Toxoplasma genomes encode for four and eighteen PDEs, respectively. The C. parvum pkg gene (cgd8_750) is expressed both during asexual (24 h, merogony) and sexual (48 h) stages, although transcript expression is higher at the earlier time point. PKG has been reported to play an essential role during merozoite egress,40 but its role during the sexual cycle of Cryptosporidium biology has not been elucidated.
We performed two separate experiments where HCT-8 cells were infected with CDPK5-HA and CDPK5-KO parasites for 47.5 h and then incubated with either 4 μM A23187 or a high concentration of BIPPO (50 μM) for 30 min. Cells were fixed at 48 h for immunofluorescence assays and super-resolution microscopy. The quantification of microgamont stages for CDPK5-HA parasites treated with A23187 revealed a significant reduction in egress-ready and an increase in egressed stages as compared to the no-treatment control (two-way ANOVA, Holm’s Sidak’s comparison, two-tailed p = 0.0040 for egress ready and p = 0.0112 for egressed). For the CDPK5-KO strain that lacked a functional kinase, there was no change in the percentage of egress-ready or egressed microgamonts upon treatment with A23187 (Figure 3E). We also quantified the number of free male gametes and found a significantly high number of gametes in the CDPK5-HA strain treated with A23187 as compared to the no-treatment control (Figure 3F). However, there was no significant difference in male gamete counts for the CDPK5-KO strain upon A23187 treatment with this calcium ionophore (Figure 3F). These results collectively suggest that directly increasing calcium levels by A23187 induces male gamete egress in the presence of a functional CDPK5.
Interestingly, in our BIPPO experiments, we did not observe any change in egress-ready or egressed microgamont stages in the CDPK5-HA strain upon treatment (Figure 3G). Similarly, the percentages of egress-ready and egressed gamonts for the CDPK5-KO strain also remained unchanged upon BIPPO treatment (Figure 3G). These findings demonstrate that BIPPO cannot rescue the male gamete egress block in CDPK5-KO parasites, thus suggesting that the gamete egress mechanism in Cryptosporidium is likely PKG independent.
Deletion of CDPK5 attenuates parasite virulence and increases survival of infected immunocompromised mice
To determine the role of the Cryptosporidium CDPK5 in virulence and parasite infectivity, we performed multiple animal infection experiments. We infected IFN-γ KO mice (n = 4 mice per group) orally with 5,000 oocysts of CDPK5-HA or CDPK5-KO transgenic parasites and followed individual mouse body weight and survival post-infection. We found that mice infected with CDPK5-HA oocysts exhibited a sharp decline in body weight, while mice infected with CDPK5-KO oocysts steadily maintained their weight (Figure 4A). As depicted in the survival curve analysis, all mice infected with CDPK5-HA oocysts succumbed to infection earlier (days 13–15 post-infection) compared to the CDPK5-KO-infected mice that survived longer, thus suggesting that ablation of CDPK5 reduces parasite virulence (Figure 4B). Two mice from the CDPK5-KO cage that were lost at an earlier time point (days 9 and 12) died of an unrelated cause (polycystic kidney disease) but not due to Cryptosporidium infection. Since both these transgenic strains express the Nluc reporter, we also measured an increase in fecal luminescence as a proxy for infection in these infected animals. Mice infected with CDPK5-HA oocysts showed high infection that peaked at days 12–14 (Figure 4C), while CDPK5-KO-infected mice showed an overall lower infection that was sustained over the 4 week period (Figure 4D). It is worth noting here that the CDPK5-KO-infected mice looked healthy with no signs of infection until day 32, and after this time, mice were euthanized following our approved animal protocol guidelines. Oocysts purified from the feces of CDPK5-KO mice were used for infecting naive IFN-γ KO mice to purify oocysts for downstream experiments. We observed similar infection dynamics during multiple infection passages, thus indicating that the oocysts produced were not defective in infection (Figure S8). Overall, our results suggest that CDPK5 plays an important role in infectivity and virulence.
Figure 4. Ablation of CDPK5 reduces virulence and infection, parasite intestinal burden, and oocyst shedding in infected immunocompromised mice.
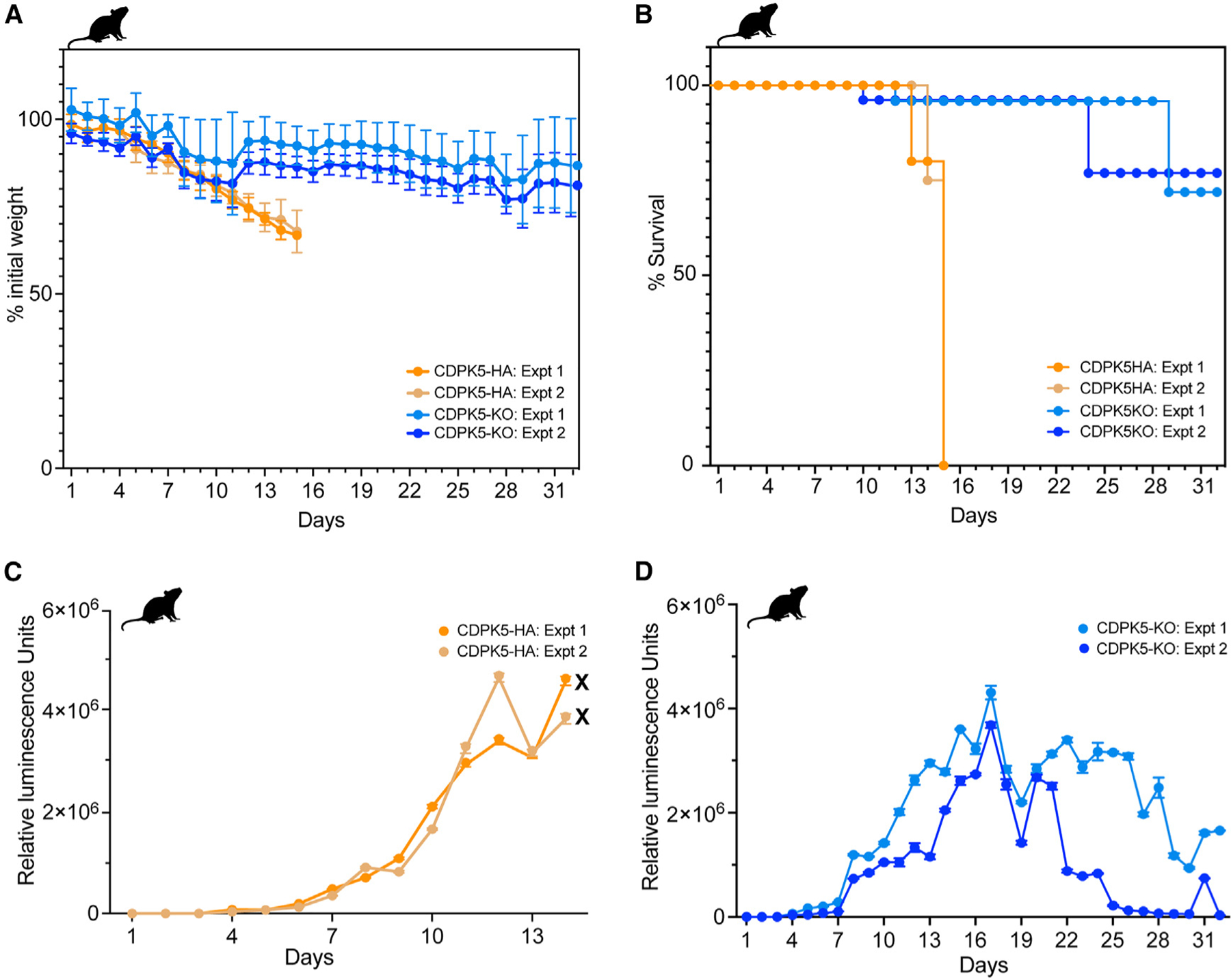
(A) Measurement of weight of IFN-γ KO mice orally infected with CDPK5-HA or CDPK5-KO oocysts. Data (mean ± SD) from two representative experiments with n = 4 mice per group are shown. Weight measurements could be obtained only up to day 14 for the CDPK5-HA group, since all mice succumbed to infection after this time point.
(B) Survival curve analysis for IFN-γ KO mice infected with CDPK5-HA or CDPK5-KO. Data from two independent experiments with n = 4 mice per group are shown. By day 14, all CDPK5-HA-infected mice died of infection (indicated by the symbol “X” on the graph), while the CDPK5-KO mice showed increased survival for a longer time.
(C) Fecal luminescence measurements of mice infected with CDPK5-HA oocysts.
(D) Fecal luminescence measurements of mice infected with CDPK5-KO oocysts.
For (C) and (D), data (mean ± SD) for three technical replicates from two independent experiments with n = 4 mice per group are shown.
CDPK5 deletion impacts intestinal parasite burden, disease severity, and oocyst production in immunocompromised mice
Given that deletion of cdpk5 resulted in a decrease in male gamete production in vitro and reduced infection in vivo, we next wanted to assess if the loss of this kinase impacted oocyst shedding and disease burden in infected animals. We infected mice orally with 5,000 oocysts of CDPK5-HA or CDPK5-KO and collected feces every 3 days (completion of one parasite life cycle) starting at day 5 post-infection. We used these fecal samples to measure oocyst shedding by quantitative real-time PCR using C. parvum 18S rRNA primers (Table S2). Since none of the CDPK5-HA-infected mice survived after day 14 post-infection, we could only collect and quantify oocyst shedding until this day. As shown in Figure 5A, mice infected with CDPK5-HA showed significantly higher oocyst shedding compared to the CDPK5-KO-infected mice (area under the curve, unpaired t test, p < 0.0001). These results suggests that the deletion of cdpk5 decreased oocyst shedding in immunocompromised mice.
Figure 5. Deletion of CDPK5 impacts parasite intestinal burden and oocyst shedding in infected immunocompromised mice.
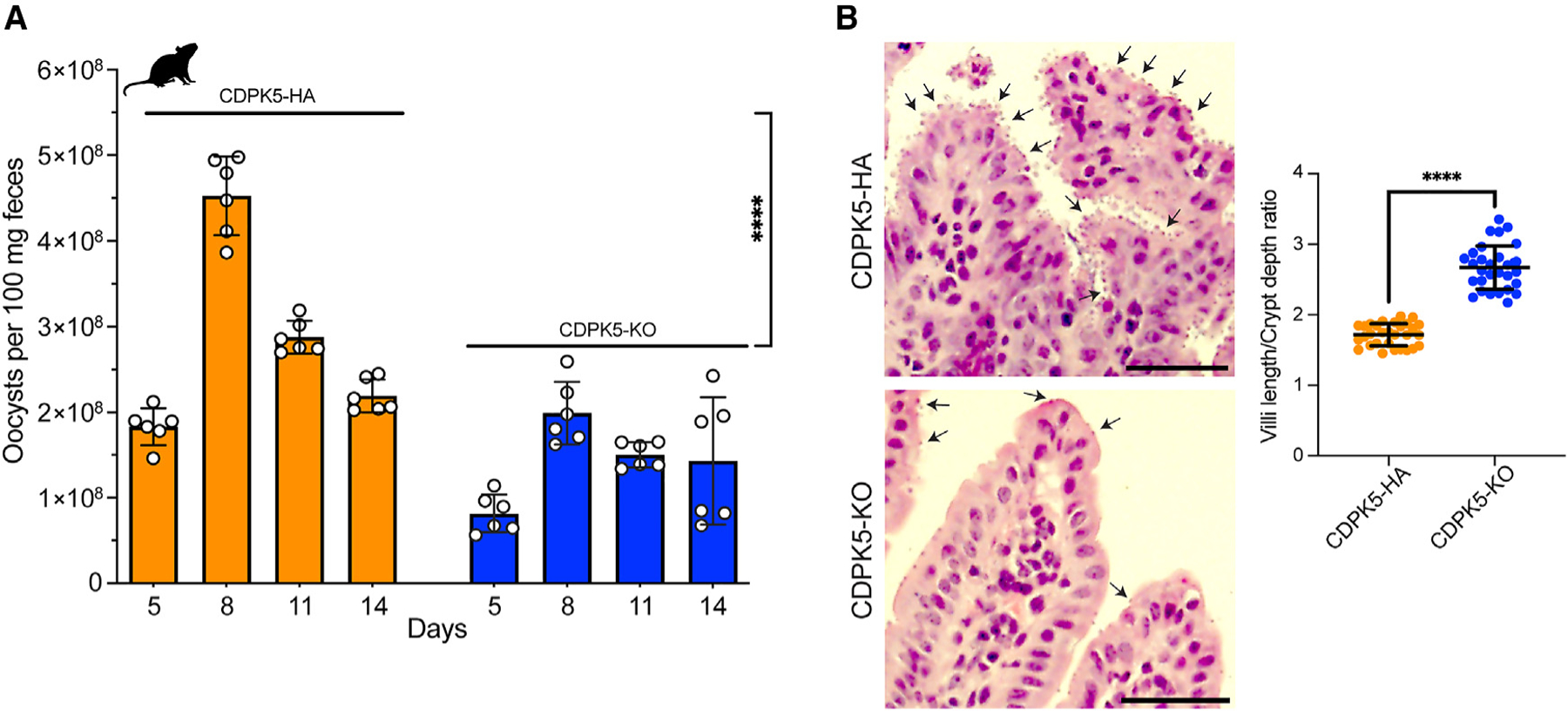
(A) Quantification of oocyst shedding per 100 mg feces in mice infected with CDPK5-HA and CDPK5-KO transgenics. IFN-γ KO mice (n = 4 per group) were infected with 5,000 oocysts from each TG strain. Oocyst quantification was performed only until day 14 post-infection, since none of the CDPK5-HA-infected mice survived after this time point. Data shown are mean ± SD of six technical replicates from two independent experiments. Unpaired t test: two-tailed ****p < 0.0001.
(B) Histology showing increased villi blunting and crypt hyperplasia in small intestines of mice infected with CDPK5-HA compared to CDPK5-KO. Representative arrows indicate C. parvum infection. Scale bars, 50 μm. The graph depicts measurements of the villus length and crypt depth ratio from IFN-γ KO mice (n = 3 per group) infected with the CDPK5-HA or CDPK5-KO strain. For each group, 30 measurements of villi length and crypt depth from n = 3 mice are shown as mean ± SD. Mann-Whitney test: two-tailed ****p < 0.0001.
Next, we wanted to compare the intestinal parasite burden upon infection of mice with the CDPK5-HA and CDPK5-KO strains. For this, we infected another set of IFN-γ KO mice (n = 3 per cage) with these transgenic oocysts, euthanized them at the peak of infection (day 13), and resected the small intestine for histopathological examination. Hematoxylin and eosin staining on the distal part of the small intestine tissues from CDPK5-HA-infected mice revealed a very heavy infection, with many parasites seen on the villi surface (Figure 5B). Contrarily, only a few parasites could be detected adhering to the villi in the CDPK5-KO intestinal sections (Figures 5B and S9). We further quantified intestinal damage by examining villi blunting and crypt hyperplasia, which are hallmarks of C. parvum infection. There was acute intestinal damage in sections from CDPK5-HA-infected mice, while the CDPK5-KO sections exhibited a healthy intestinal architecture (Figures 5B and S9). Upon measuring the villi length and crypt depth, we found a significant decrease in this ratio for the CDPK5-HA- compared to the CDPK5-KO-infected mice (Mann-Whitney test, two-tailed p < 0.0001), thus demonstrating that the high parasite burden resulted in these gross intestinal abnormalities. Overall, our findings suggest that the genetic deletion of CDPK5 leads to a significant reduction in intestinal parasite burden and oocyst production.
Phosphoproteomics reveals putative CDPK5 substrates and biological processes downregulated upon kinase deletion
To identify the potential substrates of C. parvum CDPK5, we conducted phosphoproteomics on HCT-8 cultures infected with CDPK5-HA and CDPK5-KO oocysts for 48 and 56 h (Figure 6A). In our global proteome analysis, we detected a total of 457 C. parvum proteins, with 384 proteins present in both runs. After phosphoprotein enrichment using iron immobilized metal ion affinity chromatography (Fe-IMAC) and mass spectrometry analysis, there were 296 phosphosites mapping to 113 parasite phosphoproteins. For further analysis, we filtered to only include proteins that were present in both time points and runs, and this resulted in 69 C. parvum phosphoproteins with 230 phosphosites (Table S3).
Figure 6. Identification of putative substrates of CDPK5 using phosphoproteomics.
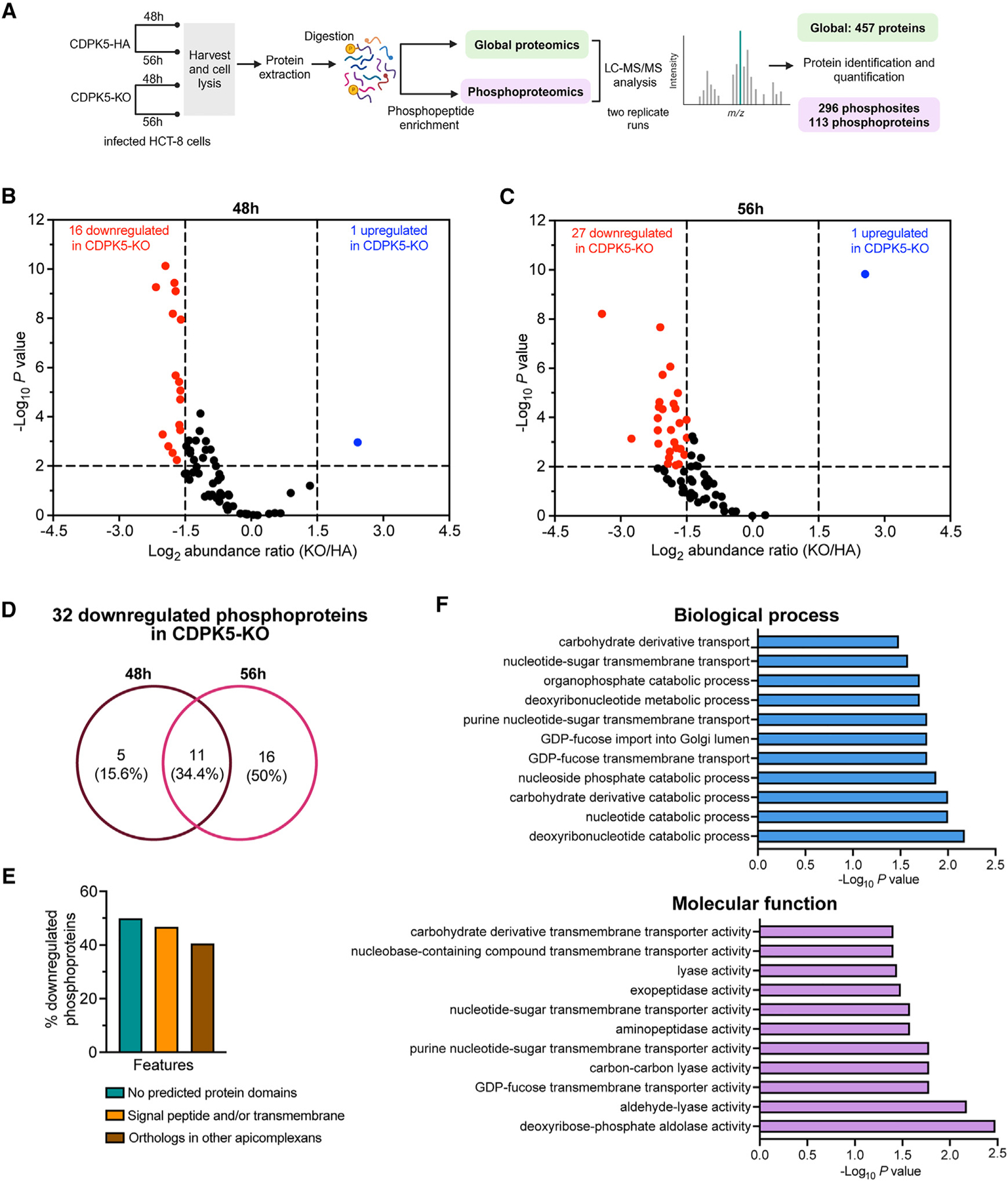
(A) Schematic illustrating the workflow for proteomic analysis of CDPK5-HA and CDPK5-KO at two infection time points.
(B) Volcano plot showing abundance ratio (CDPK5-KO/CDPK5-HA) of phosphoproteins and adjusted p value at 48 h.
(C) Volcano plot showing abundance ratio (CDPK5-KO/CDPK5HA) of phosphoproteins and adjusted p value at 56 h.
For (B) and (C), red circles represent significantly downregulated phosphoproteins, and blue circles represent significantly upregulated phosphoproteins. A cutoff of log2 fold change of < −1.5 or >1.5 was used, and p < 0.05 indicates significantly downregulated or upregulated phosphoproteins in the CDPK5-KO strain. A list of downregulated and upregulated phosphoproteins is provided in Table S4. For (B) and (C), data from two technical replicate runs for each strain and time point are shown.
(D) Venn diagram showing distribution of the 32 significantly downregulated phosphoproteins in the CDPK5-KO at 48 and 56 h and common phosphoproteins at both time points. A list of proteins is provided in Table S5.
(E) Bar graph showing the percentage of the 32 downregulated phosphoproteins lacking a predicted domain, containing a signal peptide and/or transmembrane domain, as well as the presence of orthologs in other apicomplexan parasites.
(F) Gene Ontology (GO) enrichment analysis for biological process and molecular function for significantly downregulated phosphoproteins in the CDPK5-KO with predicted GO terms (p < 0.05).
To identify the potential targets of CDPK5, we compared the abundance of these 69 phosphoproteins at 48 and 56 h time points to uncover proteins that are significantly hypophosphorylated in the CDPK5-KO strain (log2 fold change less than −1.5 and p < 0.05). Total peptide amounts for each sample were used for normalization, and protein abundances were calculated by summing the abundances of their respective peptides. We found 16 phosphoproteins to be significantly downregulated in the CDPK5-KO strain at 48 h (Figure 6B; Table S4). At the 56 h time point, 27 phosphoproteins were found to be significantly downregulated in the CDPK5-KO strain (Figure 6C; Table S4). A comparison of these significantly downregulated phosphoproteins revealed 11 proteins that were commonly downregulated at both time points (Figure 6D; Table S5).
We next wanted to gain an understanding of possible functions of these 32 significantly downregulated phosphoproteins in the CDPK5-KO strain at 48 and 56 h. Searching the CryptoDB, we found that 50% (16/32) of these downregulated phosphoproteins were “uncharacterized proteins” and contain no predicted functional domain (Figure 6E; Table S4). Around 47% (15/32) of these downregulated phosphoproteins have a predicted signal peptide and/or transmembrane domain, suggesting that these may be secreted (Figure 6E; Table S4). Only 13 out of 32 proteins had orthologs in other apicomplexan parasites, thus indicating that the signaling mechanism for gamete egress may be wired differently in Cryptosporidium (Figure 6E). We performed Gene Ontology (GO) term enrichment analysis of downregulated proteins and found that biological processes such as deoxyribonucleotide catabolism (GO: 0009264, 0009166, and 0009262) and nucleotide-sugar transmembrane transport (GO: 0015780, 0090480, and 0015783) were impacted. Molecular function analysis also suggested that nucleotide-sugar transporter (GO: 0005457, 0036080, 0005338, 0015932, and 1901505) and peptidase (GO: 0004177 and 0008238) (Figure 6F; Table S6) activities were affected. Interestingly, the downregulated proteins included a GDP fucose transporter (encoded by the cgd3_500 gene) and six uncharacterized proteins (encoded by genes cgd1_3780, cgd2_3090, cgd4_3530, cgd6_3050, cgd7_1340, and cgd8_5300). These seven genes show expression in the late male gene cluster (same as cdpk5) that corresponds with male gamete egress and in early/late female stages according to the single-cell transcriptomics data on the full Cryptosporidium life cycle available in the CryptoDB. Together, these findings identified putative substrates of C. parvum CDPK5 and provide an insight into the molecular pathways regulated by this kinase.
DISCUSSION
The single-host life cycle of Cryptosporidium is highly programmed, which ensures a timely coordinated progression from asexual stages toward sexual development.11,12 Signaling mechanisms that control the development of male and female sexual stages and details on how this process unfolds are poorly understood. Our knowledge about the male gametogenesis process is based on limited electron microscopy studies, with no information about the molecular players involved in this process. It is imperative to understand how male gametes develop and egress from the microgamont, since this is a critical step for the productive fertilization of macrogamonts and subsequent oocyst formation. Our study fulfills this gap in knowledge by providing insights into the molecular signaling mechanism mediated by the Cryptosporidium CDPK5 and its role in male gametogenesis, male gamete egress, and parasite virulence, thus identifying a potential target for blocking disease transmission.
Our results demonstrate that the C. parvum CDPK5 is expressed only in microgamonts and newly released male gametes but is absent in asexual stages. Utilizing super-resolution microscopy and 3D image reconstruction analysis, we found that the Cryptosporidium microgamont undergoes transformations in nuclear morphology with a dynamic localization of CDPK5 during this process, resulting in the egress of sixteen non-flagellated male gametes. Interestingly, this microgametogenesis process in Cryptosporidium is markedly different from the malaria parasite. In Plasmodium, the male gametocyte undergoes changes in shape from falciform to round, three mitotic divisions, elongation of axonemes, and the emergence of haploid microgametes by the process of exflagellation.17,18
In addition to the exclusive expression of C. parvum CDPK5 during the male sexual stage, its dispensability for parasite survival and its role in regulating male gamete egress, oocyst production, and disease burden in vivo altogether suggest its remarkably distinct function from its ortholog in other apicomplexan parasites. In Plasmodium, CDPK5 has been reported to be essential for parasite survival and to play critical roles in the egress of merozoites from red blood cells and liver-stage schizonts.24,30 The malarial CDPK4 and CDPK2, on the other hand, have been reported to play essential roles in the exflagellation process to yield eight motile haploid male gametes.19,20,23 Interestingly, we have previously demonstrated that the Cryptosporidium CDPK1 (ortholog of the malarial CDPK4) plays a key role in asexual proliferation, with no expression in the sexual stage.27 Thus, our findings and phylogenetic analysis support a greater evolutionary relatedness of Cryptosporidium CDPK5 to eugregarines. It appears that the function of this kinase is specifically adapted to fit the intestinal lifestyle and single-host but multistage life cycle of this parasite.
Our findings clearly demonstrate that loss of CDPK5 leads to an egress defect resulting in less male gametes released from the microgamont. We hypothesize that a decrease in male gamete numbers may impact the number of macrogamonts that could be fertilized, leading to overall decreased oocyst production and shedding in infected mice. Reduced oocyst production after fertilization can also influence the within-host re-infection cycle, and this may have resulted in overall less intestinal burden (as observed in intestinal histology) and virulence observed in IFN-γ KO mice. Alternatively, the lower parasite burden could also result from the CDPK5-KO male gametes being somehow defective in fertilizing the macrogamont, resulting in decreased oocyst production. However, it is technically challenging to test the ability of CDPK5-KO male gametes to fertilize female gamonts in vitro due to the block in gamete fusion and oocyst development in HCT-8 cell cultures.11 Moreover, we lack assays to directly quantify fertilization events in the intestinal organoids41 and air-liquid interface cultures derived from intestinal epithelial stem cells42 or mice intestine.
The punctate localization of Cryptosporidium CDPK5 on the cell periphery in the mature gamont and the increase in expression before egress are particularly intriguing. This intense signal before egress is strikingly similar to a phenomenon seen in T. gondii, where the micronemal proteins at the apical end of the parasite pulse just before egress due to calcium influx.43,44 Our findings using the calcium ionophore A23187 to induce male gamete egress suggest that raising the intracellular calcium leads to increased egress in the presence of a functional CDPK5. However, the events that lead to changes in calcium levels for the activation of CDPKs within the parasite still need to be discovered. Future biochemical studies can be designed to understand the enzymatic activity of CDPK5 and its activation upon direct binding to calcium. This would involve expressing and purifying recombinant CDPK5 protein and evaluating the effect of increasing Ca2+ concentrations on enzyme activity. Additionally, site-directed mutagenesis of the EF-hand binding domains and/or catalytic domain would provide insights into their role in kinase activation.
Our phosphoproteomics analysis reveals several uncharacterized secretory proteins as potential substrates of CDPK5 and the deregulation of nucleotide-sugar transmembrane transport and nucleotide catabolism processes. These targets can be further validated using an orthogonal approach such as proximity-based biotin labeling. Since many of these identified proteins have no predicted functional domain and most of them are present only in Cryptosporidium, characterizing their biological function will increase our understanding of male gametogenesis.
In conclusion, this study provides interesting cell biological in-sights into the male gametogenesis process in Cryptosporidium. We have discovered the important role of Cryptosporidium CDPK5 in male gamete egress, identified its potential substrates, and demonstrated its importance in reducing virulence and disease severity in an animal model of cryptosporidiosis. Future studies are required to understand the vaccine potential of this kinase or targeting this pathway for developing therapeutics to improve clinical outcome and block disease transmission.
Limitations of the study
Our results show that increasing calcium levels enhance male gamete egress in CDPK5-HA, but not in CDPK5-KO, parasites. We acknowledge that our study does not unequivocally demonstrate that CDPK5 activity is indeed regulated by calcium. Although it is beyond the scope of this study, future biochemical investigations on the regulation of this kinase upon calcium binding can be conducted. Regarding proteomics, we were limited by the parasite numbers that could be purified from the feces of infected mice. Furthermore, mice infected with the CDPK5-KO transgenic strain shed even lower number of oocysts. With no protocols available for specifically isolating Cryptosporidium microgamonts and male gametes in large quantity for phosphoproteomics, we could only use a mixed population (male and female stages) of infected cultures at 48 and 56 h. Nevertheless, the expression of CDPK5 only in male stages and not in the female (macrogamont) stage resulted in the detection of 296 phosphosites in 113 parasite proteins, leading to the identification of putative CDPK5 substrates and biological processes. Further studies are required to characterize the function of the predicted secretory proteins and dissect how CDPK5 regulates downstream metabolic and cellular processes during male gametogenesis. We hope that the development of new methods in the future will permit the purification of specific parasite stages in large quantities. This would help to increase the number of biological replicates, stringent data normalization, and identification of additional phosphoproteins.
STAR☆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sumiti Vinayak (sumiti@illinois.edu).
Materials availability
Parasite strains and plasmids generated in this study are available on request.
Data and code availability
Data reported in the paper will be shared by the lead contact upon request. The LC-MS data have been deposited with the ProteomeXchange Consortium via the jPOST partner repository (https://jpostdb.org) and are publicly available as of the date of publication The dataset identifier is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| anti-rat-HA | Roche | Clone 3F10; RRID: AB_390918 |
| anti-5E3 | (Wilke et al.)33 | N/A |
| anti-alpha tubulin | (Tandel et al.)11 | N/A |
| anti-tryptophan synthase B (trpB) | (Vinayak et al.)32 | N/A |
| anti-Histone 3 acetyl Lys9 | Millipore Sigma | Cat# 07–352 |
| CP23 | Lifespan Biosciences | Cat# LS-C137378–100; RRID: AB_10947007 |
| VVL-FITC | Vector Laboratories | Cat# FL1231 |
| Alexa Fluor®488 Goat Anti-Rat | Thermo Fisher Scientific | Cat# A11006; RRID: AB_2534074 |
| Alexa Fluor® 568 Goat Anti-Rat | Thermo Fisher Scientific | Cat# A11077; RRID: AB_2534121 |
| Alexa Fluor® 568 Goat Anti-Mouse | Thermo Fisher Scientific | Cat# A11004; RRID: AB_2534072 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| DH5α Competent Cells | ThermoFisher | FEREC0112 |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| A23187 | Millipore Sigma | Cat# C7522 |
| Paromomycin sulfate | Gemini Bio | Cat# P9297 |
| Sodium taurocholate hydrate | Sigma | Cat# 86339 |
| Bovine serum albumin | Sigma | Cat# A7030 |
| 2-mercaptoethanol | Millipore Sigma | Cat# 63689 |
| Triton X-100 | ThermoFisher | Cat# BP151–100 |
| ProLong™ Gold Antifade Reagent | ThermoFisher | Cat# P36930 |
| 16% Paraformaldehyde | Fisher | Cat# 50980487 |
| 10% neutral buffered formalin | Sigma | Cat# HT501128 |
| Ampicillin Sodium Salt | Fisher | Cat# BP1760–25 |
| Streptomycin Sulfate | Sigma | Cat# S6501–50G |
| Vancomycin Hydrochloride | GoldBio | Cat# V-200–25 |
| Sucrose | Fisher | Cat# S5–3 |
| Cesium chloride | Fisher | Cat# BP210–500 |
| Hoechst | Thermo Fisher | Cat# H3570 |
| BIPPO | Moritz Treeck and Jeffrey Dvorin’s laboratories | N/A |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Nano-Glo Luciferase Assay kit | Promega | Cat# N1130 |
| SF Cell Line 4D-Nucleofector X Kit L | Lonza | Cat# V4XC-2024 |
| Quick-DNA Fecal/Soil Microbe Kit | Zymo Research | Cat# D6012 |
| RNeasy Mini kit | Qiagen | Cat# 74104 |
| QIAshredder | Qiagen | Cat# 79654 |
| SuperScript IV First-Strand synthesis System | Life Technologies | Cat# 18091050 |
| Dnase I Amp | Life Technologies | Cat# 18068015 |
| RNase-Free DNase set | Qiagen | Cat# 79254 |
| PowerUp™ SYBR Green Master Mix | Applied Biosystems | Cat# A25742 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69504 |
|
| ||
| Deposited Data | ||
|
| ||
| LC-MS data deposited with the ProteomeXchange Consortium | jPOST partner repository (https://jpostdb.org) | PXD051082 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| HCT-8 | ATCC | CCL-244 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| IFN-gamma KO mice, B6.129S7-Ifngtm1Ts/J | Jackson laboratory | strain # 002287 |
| Cryptosporidium parvumAUCP-1 strain | Mark Kuhlenschmidt laboratory, UIUC | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Provided in Table S2 | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| C. parvum Cas9/guide vector | (Vinayak et al.)32 | N/A |
| CplicHA3-Eno-Nluc-Neo-Eno vector | (Choudhary et al.)27 | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| Prism | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| ImageJ | ImageJ | https://imagej.net/ij/ |
| OrthoMCL | OrthoMCL database | https://orthomcl.org/ |
| Imaris software | Imaris | https://imaris.oxinst.com |
| ZEN software | ZEN | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Proteome Discoverer | Byonic search engine (Protein Metrics) implemented in Proteome Discoverer software (Thermo) | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice strain and infection
Mice were housed in dedicated biosafety level 2 rooms maintained at a temperature of 68–73°F on a 12:12 h light-dark cycle in the College of Veterinary Medicine animal facility, University of Illinois at Urbana-Champaign (UIUC). Mice were provided irradiated rodent diet and water ad libitum. All mice procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of UIUC under protocol numbers 17188 and 20144. Breeder pairs for IFN-γ KO mice (B6.129S7-Ifngtm1Ts/J, strain # 002287), purchased from Jackson laboratory were used to maintain an in-house breeding colony. Female or male mice (4–6 weeks old) randomly assigned to groups (n = 4 or n = 5 mice per cage) were used for generating and passaging stable transgenic parasite. Prior to infection with C. parvum (oocysts, sporozoites or fecal slurry), 4–5-week-old mice were given water containing antibiotics (1 mg/ml ampicillin, 1 mg/mL streptomycin and 0.5 mg/mL vancomycin) for 7 days.
Parasite strain
Cryptosporidium parvum oocysts (AUCP-1 strain from Dr. Mark Kuhlenschmidt’s laboratory, University of Illinois atUrbana-Champaign) were used to create the stable transgenic strains. All stable transgenics generated in this study were propagated in IFN-γ KO mice.
Cell lines
Human ileocecal colorectal adenocarcinoma (HCT-8) (ATCC, CCL-244) cells were grown at 37°C, 5% CO2 conditions in RPMI medium with L-glutamine, 10% fetal bovine serum (FBS), 0.1 U/ml penicillin, 0.1 μg/mL streptomycin, 0.25 μg/mL amphotericin B and 1mM sodium pyruvate.
METHOD DETAILS
Construction of maximum likelihood phylogeny
The C. parvum cgd2_1300 protein sequence was used as a query to perform a BLAST search for identifying apicomplexan parasite homologs with e-value <1e-10. The sequences obtained were further filtered using the OrthoMCL database to select only core sequences, while peripheral hits were removed. Alignment was performed using MUSCLE45 and tree was created using Randomized Axelerated Maximum Likelihood (RaxML) algorithm using the GTR-GAMMA model with 1000 bootstrap replications.46
Sequence analysis of CDPK5
Illumina paired end sequence data of 158 C parvum and 96 C hominis samples were downloaded from the NCBI Short Read Archive (SRA) database. After initial quality assessment using fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the reads were assembled using SPAdes version 3.15.547 integrated in the shovill pipeline (https://github.com/tseemann/shovill). The cdpk5 ORF from all 254 assembled genomes was retrieved using BLAST search and were translated for protein sequence alignment.
Cloning of guide sequences into Cas9 vector and generation of repair DNA
Guide RNAs for the cdpk5 gene (cgd2_1300) or 3ʹ untranslated region (3ʹUTR) were designed and cloned into BbsI sites of the C. parvum Cas9/guide vector.32 Since the parasite lacks machinery for non-homologous end-joining, the Cas9-induced double-stranded breaks were repaired by providing DNA for homologous recombination.32,48,49 A linear DNA was generated by overhang PCR using the CplicHA3-Eno-Nluc-Neo-Eno vector as a template and 50 bp flanks on both sides for double homologous recombination. Primers used for guide cloning and overhang PCRs for generating tagged and knockout strains are provided in Table S2.
Generation of stable transgenic C. parvum strains
Cryptosporidium parvum AUCP-1 oocysts were subjected to excystation, and sporozoites were electroporated with the Cas9/guide plasmid (50 μg) and repair DNA template (50 μg) using Lonza Nucleofector 4D device.32,48,49 Transfected sporozoites were delivered to IFN-γ KO mice via oral gavage after buffering the acidic stomach with 8% sodium bicarbonate. The day after transfection, mice were provided paromomycin (16 mg/mL) in drinking water to select for neomycin-resistant transgenic parasites. Feces from mice were collected every three days for luciferase measurements. Oocysts were purified from fecal material from high shedding days (as measured by fecal luminescence) by sucrose flotation and cesium chloride purification protocols.32,48–50 To passage transgenic strains, fecal slurry or purified oocysts were used to infect new cages of 5–6-week-old IFN-γ KO mice.
Fecal luminescence assay to measure parasite burden
Pooled fecal material from mice cage were collected and stored at 4°C. and fecal luminescence assays was performed.32 20 mg of homogenized feces were measured in a tube, to which 10–15 glass beads and 1 mL of lysis buffer (50 mM Tris-HCl pH 7.6, 2 mM DTT, 2 mm EDTA, 10% v/v glycerol and 1% Triton X-100) was added. Contents of the tube were thoroughly mixed using an Eppendorf thermomixer C (2000 rpm, for 5 min) followed by centrifugation at 8000 rpm for 10 s. Aliquots of 100 μL of the supernatant were added to three wells of a 96-well white plate, followed by addition of 100 μL of Nanoluc substrate that was diluted in 1:50 Nanoluc lysis buffer (Promega, Madison, WI). Luminescence measurements were done on an Wallac VICTOR2 1420 multilabel counter (PerkinElmer Inc) or Biotek Synergy LX multimode (Agilent) luminescence reader.
Validation of transgenics using PCR
Fecal genomic DNA was extracted from 100 mg feces using the Zymo fecal/soil DNA Miniprep kit (Zymo Research, Irvine, CA) using manufacturer’s instructions. PCR was performed using this genomic DNA to confirm the correct 5ʹ and 3ʹ recombination (integration) events. Primers used for validating transgenics have been provided in Table S2.
Mice survival and virulence experiments
For infection with CDPK5-KO and CDPK5-HA, two cages of mice (n = 4 mice per cage) for each transgenic were used. The weight of each mouse was recorded before infection and daily after infection. Mice were infected with 5,000 transgenic oocysts by oral gavage, and paromomycin was provided in the drinking water. Pooled feces from cages were collected daily for luminescence assay.
Infection of HCT-8 host cell with C. parvum and immunofluorescence assay
HCT-8 (ATCC, CCL-244) cells were grown at 37°C, 5% CO2 conditions to 60–70% confluency on cover slips in 6-well plates.27,32 Host cell media was replaced with Cryptosporidium infection media (RPMI containing 2% FBS, 0.1 U/ml penicillin, 0.1 μg/mL streptomycin and 0.25 μg/mL amphotericin B) prior to infection. Cells were infected with transgenic oocysts for 24, 42, 48 or 56 h, fixed in 4% paraformaldehyde. For egress assays with BIPPO or ionophore A23187 (both compounds dissolved in DMSO), cells were infected with transgenic oocysts and 50 μM of the BIPPO or 4 μM A23187 was added at 47.5 h post infection. Permeabilization was performed using 0.25% Triton X-100 in PBS, followed by blocking with 3% bovine serum albumin (BSA) overnight, washings with PBS and incubation with primary antibody for 1 h.27 Primary antibodies used were anti-rat-HA (clone 3F10, Roche), mouse anti-5E3 (kind gift from Dr. David Sibley, Washington University School of Medicine), rabbit anti-Histone 3 acetyl Lys9 (H3K9ac, Millipore Sigma), mouse anti-alpha tubulin and rabbit anti-tryptophan synthase B (kind gift from Boris Striepen laboratory, University of Pennsylvania). Fluorophore-conjugated secondary antibodies (Alexa Fluor 488, Alexa Fluor 568) at a dilution of 1:500, and fluorescein labeled Vicia villosa lectin (VVL, from Vector Laboratories) at a 1:4000 dilution was used. Nuclear staining was done using Hoechst 33342 DNA stain (2 μg/mL). Coverslips were inverted and mounted on glass slides using Vectashield antifade mounting medium (Vector laboratories). Images were acquired on a Zeiss ELYRA S1 super-resolution structured illumination (SR-SIM) microscope and processed using ZEN software and ImageJ. 3D reconstruction of z stack images was done using Imaris software.
Western blotting
Wild type and transgenic CDPK5-HA oocysts (5x106) were subjected to bleach treatment, washed three times with ice-cold PBS, and used to infect HCT-8 cells grown in 24-well plates. Infected cells were harvested by scraping the monolayer after 48 h of infection. Cell pellet was suspended in laemmli sample buffer and boiled for 5 min, followed by centrifugation at 13,000 rpm for 3 min before loading on a 4 to 20% Tris-glycine-sodium dodecyl sulfate (SDS) Mini-PROTEAN precast gel (BioRad). After SDS-polyacrylamide gel electrophoresis (PAGE), proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. Membrane was blocked in 5% non-fat milk for 2 h and then incubated with 1:1,000 dilution of primary antibody (anti-rat HA, clone 3F10) overnight at 4°C on rocker. After three washes in PBS with 0.1% Tween 20 (PBST), the membrane was incubated with anti-rat horseradish peroxidase (HRP)-conjugated secondary antibody (1:20,000 dilution) for 1 h, followed by multiple washing of the blot with PBST. The HRP signal was detected by chemiluminescence using the SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific) on a FluorChem imager. The blot was stripped and reprobed with a 1:1000 dilution of primary mouse anti-CP23 (Lifespan Biosciences) and anti-mouse HRP-conjugated secondary antibody (1:20,000) for loading control analysis.
Intestinal histology
For histology, two cages of mice were orally infected with 5000 oocysts of CDPK5-HA and CDPK5-KO transgenics. Mice (n = 3 per cage) were euthanized at day 13 post infection and the small intestine was harvested and flushed with PBS. The distal section was fixed in 10% neutral buffered formalin (Sigma, St. Louis, MO) for 24 h and then transferred to a jar with 70% ethanol until processing. Sections were embedded in paraffin, sectioned, and stained with hematoxylin-eosin stain. Slides were imaged on an Olympus BX51 light microscope fitted with a DP70 camera. Villus length and crypt depth were measured (total of 30 measurements for each strain) using ImageJ to quantify intestinal damage.
Quantification of oocyst shedding
Genomic DNA was extracted from 100 mg of fecal material collected from IFN-γ KO mice infected with 5000 oocysts of CDPK5-HA and CDPK5-KO using the Zymo fecal/soil DNA Miniprep kit (Zymo Research, Irvine, CA). To generate a standard curve, feces from uninfected mice were spiked with different amounts (101 to 108) of wild type C. parvum oocysts and used for DNA extraction. PCR was done on extracted DNA using primers and fluorescent probe against the C. parvum 18S rRNA gene and Biorad Sso advanced Universal Probe Super mix (BioRad) on the ABI 7500 real-time PCR system51
Reverse transcription PCR
Wild type and transgenic oocysts were used to infect multiple wells of HCT-8 cells grown on 24-well plates for 48 h. Cells were scraped and collected for RNA isolation using the Qiagen Rneasy Minikit. QIAshredder columns (Qiagen) were used for efficient lysis, and on-column DNase I treatment was performed to remove genomic DNA. Removal of genomic DNA was confirmed by PCR using intron-specific primers, and additional DNaseI treatment of RNA was performed if required. 5 μg of RNA was used for cDNA preparation using SuperScriptIV First-Strand synthesis system (Invitrogen) following manufacturer’s instructions. PowerUp SYBR Green Master Mix (Applied Biosystems) was used for cDNA amplification on the QuantStudio 3 Real-time PCR system (Applied Biosystems). Sequences of the primers used to quantify expression of C. parvum cdpk5, as well as C. parvum 18S rRNA and human actin gene primers are provided in Table S2. Relative expression of the cdpk5 gene was calculated using the ΔΔCT method.
Proteomics and mass spectrometry data analysis
HCT-8 cells grown in 24-well plates were infected with 5x106 CDPK5-HA and CDPK5-KO oocysts (three wells for each strain) for 48 and 56 h. Cells were harvested from three wells and combined. Pellets were washed with ice-cold 1x PBS and suspended in lysis buffer containing 6M guanidine HCl (GuHCl) in 100 mM triethylammonium bicarbonate (TEAB) followed by heating for 5 min at 95°C. All samples were flash frozen in liquid nitrogen and stored at −80°C until proteomic analysis.
For proteomics, samples were lysed in 10 mM tris(2-carboxyethyl)phosphine HCl (TCEP) and 40 mM 2-chloroacetamide (CAA) followed by heating at 95°C for 10 min to promote reduction and alkylation of disulfide bonds. Samples were cleared of any remaining debris by centrifugation at 14000 g at 4°C for 10 min. Protein amount was calculated by BCA assay (Pierce) following manufacturer’s instructions. Proteins were digested with LysC (Fujifilm Wako Chemicals) for 4 h at 25°C using a ratio of 1:100 w/w enzyme:substrate, followed by overnight trypsin (Promega) digestion (1:50 w/w; enzyme: substrate) at 37°C. Samples were acidified with trifluoroacetic acid (TFA) to a final concentration of 0.5% TFA, desalted with 100 mg Sep-Pak C18 columns (Waters) and then dried in a vacuum centrifuge. Peptides were then resuspended in water with 0.1% formic acid (FA), and the protein amounts were re-normalized according to a colorimetric peptide BCA assay (Pierce). Phosphopeptides from 200 μg of starting peptides was enriched by iron-immobilized metal ion affinity chromatography (Fe-IMAC) in a microtip format52 before being desalted once more using StageTips.53 Dried phosphopeptide samples were resuspended in 15 μL of 0.1% formic acid (FA) in 5% acetonitrile (ACN), while the non-enriched peptide samples were diluted to a concentration of 250 ng/μL in 0.1% FA in 5% ACN. 1 μL from each sample was injected into an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Peptides were separated using a 50 cm Acclaim PepMap 100 C18 column (2 μm particle size, 75 μm ID) and mobile phases of 0.1% FA (A) and 0.1% FA in 80% ACN (B) at a flow rate of 300 nL/min. The gradient started at 2% B, increased to 5% B over 2 min, increased to 35% B over 110 min, and then increased to 50% B over 10 more minutes; this was followed by column washing and equilibration. The column was maintained at 60°C over the course of the run. Peptides were analyzed with a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) using data-dependent acquisition in positive mode. MS1 scans from 350 to 1500 m/z were acquired at 120k resolution (3e6 AGC; 50 ms max IT), followed by Higher-energy collisional dissociation (HCD) fragmentation (30 NCE) of the 15 most abundant ions. MS2 scans were acquired at 15,000 resolution with an isolation window of 1.0 m/z and a dynamic exclusion time of 45 s (6e3 minimum AGC; 30 ms max IT). Two replicate runs were conducted for each sample.
Raw liquid chromatography-mass spectrometry (LC-MS/MS) data was analyzed against the Uniprot Cryptosporidium parvum (3,805 entries) and Homo sapiens (82,202 entries) databases using the Byonic search engine v.5.3.5 (Protein Metrics) implemented in ProteomeDiscoverer v2.4.1.15 (Thermo Fisher). Tryptic digestion was specified with a maximum of two missed cleavages, while peptide and fragment mass tolerances were set to 10 ppm and 20 ppm, respectively. Variable modifications including oxidation of methionine, acetylation of protein N-termini, deamidation of glutamine and asparagine, and phosphorylation of serine, threonine, and tyrosine; a fixed modification to account for cysteine carbamidomethylation was also added to the search. The protein level false discovery rate (FDR) was set to 1% using a reversed decoy database strategy. Label-free quantitation using precursor intensities was also done in ProteomeDiscoverer using the Minora Feature Detector, Feature Mapper, and Precursor Ions Quantifier nodes. Chromatographic alignment was performed for feature mapping between files with a maximum retention time shift of 5 min and a precursor mass tolerance of 10 ppm. Protein abundance ratios were calculated as the median of the pairwise peptide ratios, and p values for these ratios were determined using t test.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis were performed using GraphPad Prism v10. Unpaired t test was used to measure difference in oocyst shedding between CDPK5-HA and CDPK5-KO transgenics. Mann-Whitney test, t test and two-way ANOVA were used to compare egress phenotypes in the cell biological phenotypic analysis, and intestinal damage. A two-tailed p < 0.05 was considered significant among all comparisons tested. Details of the statistical parameters have been described in the results or figure legends. This includes the number of technical or biological replicates and standard deviation for each experiment.
Supplementary Material
Highlights.
CDPK5 is a male-stage-specific kinase in Cryptosporidium
CDPK5 plays a role in egress of male gametes
Genetic ablation of CDPK5 reduces virulence, disease severity, and oocyst shedding in vivo
Phosphoproteomics reveals putative substrates of CDPK5
ACKNOWLEDGMENTS
We thank Dr. Boris Striepen and Dr. David Sibley for providing the C. parvum antibodies used in immunofluorescence assays. We are grateful to Dr. Moritz Treeck and Dr. Jeffrey Dvorin for providing the BIPPO compound. Figures S1 and 6A and the graphical abstract were created with BioRender.com. We thank the Institute for Genome Biology (IGB) microscopy core, the Proteomics Core Facility of the Roy J. Carver Biotechnology Center, the Department of Comparative Biosciences histology core, and the animal facilities at UIUC for their support. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under award number R01AI150961 (to S.V.) and start-up funds from the University of Illinois at Urbana-Champaign.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114263.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. 10.1016/s0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, Panchalingam S, Sur D, Zaidi AKM, Faruque ASG, et al. (2016). The Burden of Cryptosporidium Diarrheal Disease among Children < 24 Months of Age in Moderate/High Mortality Regions of Sub-Saharan Africa and South Asia, Utilizing Data from the Global Enteric Multicenter Study (GEMS). PLoS Neglected Trop. Dis 10, e0004729. 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, Sow SO, Sur D, Zaidi AKM, Faruque ASG, et al. (2019). The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Global Health 7, e568–e584. 10.1016/s2214-109x(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert IH, Vinayak S, Striepen B, Manjunatha UH, Khalil IA, and Van Voorhis WC Cryptosporidiosis Therapeutics Advocacy Group CTAG; Cryptosporidiosis Therapeutics Advocacy CTAG (2023). Safe and effective treatments are needed for cryptosporidiosis, a truly neglected tropical disease. BMJ Glob. Health 8, e012540. 10.1136/bmjgh-2023-012540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, Colombara DV, De Hostos EL, Engmann C, Guerrant RL, et al. (2018). Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Global Health 6, e758–e768. 10.1016/s2214-109x(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, et al. (2015). Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Global Health 3, e564–e575. 10.1016/s2214-109x(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santin M (2020). Cryptosporidium and Giardia in Ruminants. Vet. Clin. North Am. Food Anim. Pract 36, 223–238. 10.1016/j.cvfa.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Gharpure R, Perez A, Miller AD, Wikswo ME, Silver R, and Hlavsa MC (2019). Cryptosporidiosis Outbreaks - United States, 2009–2017. MMWR Morb. Mortal. Wkly. Rep 68, 568–572. 10.15585/mmwr.mm6825a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amadi B, Mwiya M, Sianongo S, Payne L, Watuka A, Katubulushi M, and Kelly P (2009). High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect. Dis 9, 195. 10.1186/1471-2334-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, and Kelly P (2002). Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet 360, 1375–1380. 10.1016/s0140-6736(02)11401-2. [DOI] [PubMed] [Google Scholar]
- 11.Tandel J, English ED, Sateriale A, Gullicksrud JA, Beiting DP, Sullivan MC, Pinkston B, and Striepen B (2019). Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol 4, 2226–2236. 10.1038/s41564-019-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.English ED, Guérin A, Tandel J, and Striepen B (2022). Live imaging of the Cryptosporidium parvum life cycle reveals direct development of male and female gametes from type I meronts. PLoS Biol 20, e3001604. 10.1371/journal.pbio.3001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumaine JE, Tandel J, and Striepen B (2020). Cryptosporidium parvum. Trends Parasitol 36, 485–486. 10.1016/j.pt.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Tandel J, Walzer KA, Byerly JH, Pinkston B, Beiting DP, and Striepen B (2023). Genetic Ablation of a Female-Specific Apetala 2 Transcription Factor Blocks Oocyst Shedding in Cryptosporidium parvum. mBio 14, e0326122–22. 10.1128/mbio.03261-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrovska K, and Paperna I (1990). Cryptosporidium sp. of the starred lizard Agama stellio: Ultrastructure and life cycle. Parasitol. Res 76, 712–720. 10.1007/bf00931092. [DOI] [Google Scholar]
- 16.Göebel E, and Brändler U (1982). Ultrastructure of microgametogenesis, microgametes and gametogony of Cryptosporidium sp. in the small intestine of mice. PROTISTOLOGICA 18, 331–344. [Google Scholar]
- 17.Sinden RE, Canning EU, and Spain B (1976). Gametogenesis and fertilization in Plasmodium yoelii nigeriensis: a transmission electron microscope study. Proc. R. Soc. Lond. B Biol. Sci 193, 55–76. 10.1098/rspb.1976.0031. [DOI] [PubMed] [Google Scholar]
- 18.Yahiya S, Jordan S, Smith HX, Gaboriau DCA, Famodimu MT, Dahalan FA, Churchyard A, Ashdown GW, and Baum J (2022). Live-cell fluorescence imaging of microgametogenesis in the human malaria parasite Plasmodium falciparum. PLoS Pathog 18, e1010276. 10.1371/journal.ppat.1010276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansal A, Molina-Cruz A, Brzostowski J, Mu J, and Miller LH (2017). Plasmodium falciparum calcium-dependent protein kinase 2 is critical for male gametocyte exflagellation but not essential for asexual proliferation. mBio 8, 016566–17–e1717. 10.1128/mbio.01656-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, and Brinkmann V (2004). Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117, 503–514. 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 21.Absalon S, Blomqvist K, Rudlaff RM, DeLano TJ, Pollastri MP, and Dvorin JD (2018). Calcium-Dependent Protein Kinase 5 Is Required for Release of Egress-Specific Organelles in Plasmodium falciparum. mBio 9, 001300–e218. 10.1128/mbio.00130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, and Sibley LD (2010). Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465, 359–362. 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ojo KK, Pfander C, Mueller NR, Burstroem C, Larson ET, Bryan CM, Fox AMW, Reid MC, Johnson SM, Murphy RC, et al. (2012). Transmission of malaria to mosquitoes blocked by bumped kinase inhibitors. J. Clin. Invest 122, 2301–2305. 10.1172/jci61822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, et al. (2010). A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328, 910–912. 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rueckert S, Betts EL, and Tsaousis AD (2019). The Symbiotic Spectrum: Where Do the Gregarines Fit? Trends Parasitol 35, 687–694. 10.1016/j.pt.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Boisard J, Duvernois-Berthet E, Duval L, Schrével J, Guillou L, Labat A, Le Panse S, Prensier G, Ponger L, and Florent I (2022). Marine gregarine genomes reveal the breadth of apicomplexan diversity with a partially conserved glideosome machinery. BMC Genom 23, 485–522. 10.1186/s12864-022-08700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhary HH, Nava MG, Gartlan BE, Rose S, and Vinayak S (2020). A conditional protein degradation system to study essential gene function in Cryptosporidium parvum. mBio 11, e01231–20. 10.1128/mbio.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Jarreta J, Amos B, Aurrecoechea C, Bah S, Barba M, Barreto A, Basenko EY, Belnap R, Blevins A, Böhme U, et al. (2024). VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center in 2023. Nucleic Acids Res 52, D808–D816. 10.1093/nar/gkad1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, and Rutherford MS (2012). The Cryptosporidium parvum transcriptome during in vitro development. PLoS One 7, 317155–e31814. 10.1371/journal.pone.0031715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Govindasamy K, and Bhanot P (2020). Overlapping and distinct roles of CDPK family members in the pre-erythrocytic stages of the rodent malaria parasite, Plasmodium berghei. PLoS Pathog 16, e1008131. 10.1371/journal.ppat.1008131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bansal A, Molina-Cruz A, Brzostowski J, Liu P, Luo Y, Gunalan K, Li Y, Ribeiro JMC, and Miller LH (2018). PfCDPK1 is critical for malaria parasite gametogenesis and mosquito infection. Proc. Natl. Acad. Sci. USA 115, 774–779. 10.1073/pnas.1715443115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, and Striepen B (2015). Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477–480. 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilke G, Ravindran S, Funkhouser-Jones L, Barks J, Wang Q, Van-Dussen KL, Stappenbeck TS, Kuhlenschmidt TB, Kuhlenschmidt MS, and Sibley LD (2018). Monoclonal antibodies to intracellular stages of Cryptosporidium parvum define life cycle progression in vitro. mSphere 3, 001244–e218. 10.1128/msphere.00124-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guérin A, Roy NH, Kugler EM, Berry L, Burkhardt JK, Shin J-B, and Striepen B (2021). Cryptosporidium rhoptry effector protein ROP1 injected during invasion targets the host cytoskeletal modulator LMO7. Cell Host Microbe, 1–46. 10.1016/j.chom.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McRobert L, Taylor CJ, Deng W, Fivelman QL, Cummings RM, Polley SD, Billker O, and Baker DA (2008). Gametogenesis in malaria parasites is mediated by the cGMP-dependent protein kinase. PLoS Biol 6, 1399–e210. 10.1371/journal.pbio.0060139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard BL, Harvey KL, Stewart RJ, Azevedo MF, Crabb BS, Jennings IG, Sanders PR, Manallack DT, Thompson PE, Tonkin CJ, and Gilson PR (2015). Identification of Potent Phosphodiesterase Inhibitors that Demonstrate Cyclic Nucleotide-Dependent Functions in Apicomplexan Parasites. ACS Chem. Biol 10, 1145–1154. 10.1021/cb501004q. [DOI] [PubMed] [Google Scholar]
- 37.Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, and Blackman MJ (2013). Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog 9, e1003344. 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balestra AC, Koussis K, Klages N, Howell SA, Flynn HR, Bantscheff M, Pasquarello C, Perrin AJ, Brusini L, Arboit P, et al. (2021). Ca2+ signals critical for egress and gametogenesis in malaria parasites depend on a multipass membrane protein that interacts with PKG. Sci. Adv 7, eabe5396. 10.1126/sciadv.abe5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nofal SD, Dominicus C, Broncel M, Katris NJ, Flynn HR, Arrizabalaga G, Botté CY, Invergo BM, and Treeck M (2022). A positive feedback loop mediates crosstalk between calcium, cyclic nucleotide and lipid signalling in calcium-induced Toxoplasma gondii egress. PLoS Pathog 18, e1010901. 10.1371/journal.ppat.1010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nava S, Sadiqova A, Castellanos-Gonzalez A, and White AC (2020). Cryptosporidium parvum cyclic GMP-dependent protein kinase (PKG): An essential mediator of merozoite egress. Mol. Biochem. Parasitol 237, 111277. 10.1016/j.molbiopara.2020.111277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol 3, 814–823. 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, Baldridge MT, VanDussen KL, Shen B, Kuhlenschmidt MS, et al. (2019). A stem-cell-derived platform enables complete Cryptosporidium development in-vitro and genetic tractability. Cell Host Microbe 26, 123–134.e8. 10.1016/j.chom.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carruthers VB, and Sibley LD (1999). Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol. Microbiol 31, 421–428. 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- 44.Borges-Pereira L, Budu A, McKnight CA, Moore CA, Vella SA, Hortua Triana MA, Liu J, Garcia CRS, Pace DA, and Moreno SNJ (2015). Calcium Signaling throughout the Toxoplasma gondii lytic cycle: A STUDY USING GENETICALLY ENCODED CALCIUM INDICATORS. J. Biol. Chem 290, 26914–26926. 10.1074/jbc.m115.652511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prjibelski A, Antipov D, Meleshko D, Lapidus A, and Korobeynikov A (2020). Using SPAdes De Novo Assembler. Curr Protoc Bioinform 70, e102. 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 48.Sateriale A, Pawlowic M, Vinayak S, Brooks C, and Striepen B (2020). Genetic manipulation of Cryptosporidium parvum with CRISPR/Cas9. Methods Mol. Biol 2052, 219–228. 10.1007/978-1-4939-9748-0_13. [DOI] [PubMed] [Google Scholar]
- 49.Pawlowic MC, Vinayak S, Sateriale A, Brooks CF, and Striepen B (2017). Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol 46, 20B.2.1–20B.2.32. 10.1002/cpmc.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upton SJ (1997). In Cryptosporidium and Cryptosporidiosis, Fayer R, ed. (Boca Raton. FL: CRC Press; ), pp. 181–207. [Google Scholar]
- 51.Manjunatha UH, Vinayak S, Zambriski JA, Chao AT, Sy T, Noble CG, Bonamy GMC, Kondreddi RR, Zou B, Gedeck P, et al. (2017). A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature 546, 376–380. 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai CF, Hsu CC, Hung JN, Wang YT, Choong WK, Zeng MY, Lin PY, Hong RW, Sung TY, and Chen YJ (2014). Sequential Phosphoproteomic Enrichment through Complementary Metal-Directed Immobilized Metal Ion Affinity Chromatography. Anal. Chem 86, 685–693. 10.1021/ac4031175. [DOI] [PubMed] [Google Scholar]
- 53.Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896–1906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in the paper will be shared by the lead contact upon request. The LC-MS data have been deposited with the ProteomeXchange Consortium via the jPOST partner repository (https://jpostdb.org) and are publicly available as of the date of publication The dataset identifier is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| anti-rat-HA | Roche | Clone 3F10; RRID: AB_390918 |
| anti-5E3 | (Wilke et al.)33 | N/A |
| anti-alpha tubulin | (Tandel et al.)11 | N/A |
| anti-tryptophan synthase B (trpB) | (Vinayak et al.)32 | N/A |
| anti-Histone 3 acetyl Lys9 | Millipore Sigma | Cat# 07–352 |
| CP23 | Lifespan Biosciences | Cat# LS-C137378–100; RRID: AB_10947007 |
| VVL-FITC | Vector Laboratories | Cat# FL1231 |
| Alexa Fluor®488 Goat Anti-Rat | Thermo Fisher Scientific | Cat# A11006; RRID: AB_2534074 |
| Alexa Fluor® 568 Goat Anti-Rat | Thermo Fisher Scientific | Cat# A11077; RRID: AB_2534121 |
| Alexa Fluor® 568 Goat Anti-Mouse | Thermo Fisher Scientific | Cat# A11004; RRID: AB_2534072 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| DH5α Competent Cells | ThermoFisher | FEREC0112 |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| A23187 | Millipore Sigma | Cat# C7522 |
| Paromomycin sulfate | Gemini Bio | Cat# P9297 |
| Sodium taurocholate hydrate | Sigma | Cat# 86339 |
| Bovine serum albumin | Sigma | Cat# A7030 |
| 2-mercaptoethanol | Millipore Sigma | Cat# 63689 |
| Triton X-100 | ThermoFisher | Cat# BP151–100 |
| ProLong™ Gold Antifade Reagent | ThermoFisher | Cat# P36930 |
| 16% Paraformaldehyde | Fisher | Cat# 50980487 |
| 10% neutral buffered formalin | Sigma | Cat# HT501128 |
| Ampicillin Sodium Salt | Fisher | Cat# BP1760–25 |
| Streptomycin Sulfate | Sigma | Cat# S6501–50G |
| Vancomycin Hydrochloride | GoldBio | Cat# V-200–25 |
| Sucrose | Fisher | Cat# S5–3 |
| Cesium chloride | Fisher | Cat# BP210–500 |
| Hoechst | Thermo Fisher | Cat# H3570 |
| BIPPO | Moritz Treeck and Jeffrey Dvorin’s laboratories | N/A |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Nano-Glo Luciferase Assay kit | Promega | Cat# N1130 |
| SF Cell Line 4D-Nucleofector X Kit L | Lonza | Cat# V4XC-2024 |
| Quick-DNA Fecal/Soil Microbe Kit | Zymo Research | Cat# D6012 |
| RNeasy Mini kit | Qiagen | Cat# 74104 |
| QIAshredder | Qiagen | Cat# 79654 |
| SuperScript IV First-Strand synthesis System | Life Technologies | Cat# 18091050 |
| Dnase I Amp | Life Technologies | Cat# 18068015 |
| RNase-Free DNase set | Qiagen | Cat# 79254 |
| PowerUp™ SYBR Green Master Mix | Applied Biosystems | Cat# A25742 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69504 |
|
| ||
| Deposited Data | ||
|
| ||
| LC-MS data deposited with the ProteomeXchange Consortium | jPOST partner repository (https://jpostdb.org) | PXD051082 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| HCT-8 | ATCC | CCL-244 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| IFN-gamma KO mice, B6.129S7-Ifngtm1Ts/J | Jackson laboratory | strain # 002287 |
| Cryptosporidium parvumAUCP-1 strain | Mark Kuhlenschmidt laboratory, UIUC | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Provided in Table S2 | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| C. parvum Cas9/guide vector | (Vinayak et al.)32 | N/A |
| CplicHA3-Eno-Nluc-Neo-Eno vector | (Choudhary et al.)27 | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| Prism | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| ImageJ | ImageJ | https://imagej.net/ij/ |
| OrthoMCL | OrthoMCL database | https://orthomcl.org/ |
| Imaris software | Imaris | https://imaris.oxinst.com |
| ZEN software | ZEN | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Proteome Discoverer | Byonic search engine (Protein Metrics) implemented in Proteome Discoverer software (Thermo) | N/A |


