Abstract
Poly(dA·dT) sequences (T-tracts) are abundant genomic DNA elements with unusual properties in vitro and an established role in transcriptional regulation of yeast genes. In vitro T-tracts are rigid, contribute to DNA bending, affect assembly in nucleosomes and generate a characteristic pattern of CPDs (cyclobutane pyrimidine dimers) upon irradiation with UV light (UV photofootprint). In eukaryotic cells, where DNA is packaged in chromatin, the DNA structure of T-tracts is unknown. Here we have used in vivo UV photofootprinting and DNA repair by photolyase to investigate the structure and accessibility of T-tracts in yeast promoters (HIS3, URA3 and ILV1). The same characteristic photofootprints were obtained in yeast and in naked DNA, demonstrating that the unusual T-tract structure exists in living cells. Rapid repair of CPDs in the T-tracts demonstrates that these T-tracts were not folded in nucleosomes. Moreover, neither datin, a T-tract binding protein, nor Gcn5p, a histone acetyltransferase involved in nucleosome remodelling, showed an influence on the structure and accessibility of T-tracts. The data support a contribution of this unusual DNA structure to transcriptional regulation.
INTRODUCTION
Poly(dA·dT) sequences (called T-tracts) have been extensively characterised in vitro. They adopt an unusual structure (here referred to as T-tract structure) that is distinct from typical B-type DNA. T-tracts are essentially straight and rigid (1–3) with a compressed minor goove (4) and a short helical repeat of 10 bp per turn, compared to 10.5 bp per turn determined for canonical B-DNA (5,6). The structure of T-tracts is not sensitive to changes in cations or salt concentration (7), but folding of T-tract DNA in nucleosomes can disrupt the T-tract structure, indicating that the structural constraints in nucleosomes dominate over those of the T-tracts (8,9). Since the formation of DNA lesions depends on the DNA structure (10,11), the unusual T-tract structure generates a characteristic pattern of cyclobutane pyrimidine dimers (CPDs) upon irradiation with UV light (UV photofootprint). Tn-tracts in plasmid DNA with n ≥ 4 give high yields of CPDs at the 3′-end, low yields in the middle and variable yields towards the 5′-end, depending on the flanking base (12).
Because of their unusual structure, T-tracts influence the conformation of longer stretches of free DNA and they interfere with the formation and arrangement of nucleosomes. Phased runs of short T-tracts cause DNA bending (see for example 13,14) and bending is abolished when the T-tract structure is disrupted by a CPD lesion (15). Long poly(dA·dT) sequences exclude nucleosome formation during in vitro reconstitution (16–18) except at high temperatures (19). Short T-tracts (∼20 bp) form nucleosomes (8,20–23), but appear to be excluded from the centre (20,24). Whether T-tracts form nucleosomes apparently depends on their length, the flanking sequences and the reconstitution conditions. Thus, nucleosomes with different T-tracts may have differential stability with respect to disruption and nucleosome positioning.
The unusual structure of T-tracts and their potential to destabilise or exclude nucleosomes has implications for their functional role in eukaryotic cells. In the yeast Saccharomyces cerevisiae T-tracts >10 bp occur at frequencies higher than expected in the genome (25). In the HIS3, DED1 and URA3 genes, as well as in many other genes, T-tracts act as upstream promoter elements necessary for wild-type levels of transcription (26–28). Since transcription activation depends on the length of the T-tract and T-tracts can be replaced by poly(dG·dC), another rigid structure, it was suggested that the T-tracts operate not by recruitment of specific transcription factors, but rather by their intrinsic DNA structure. The T-tracts might affect nucleosome stability and enhance accessibility for binding of factors to nearby sequences (28). This hypothesis is consistent with the observations that several T-tract promoters of yeast genes (including URA3, HIS3 and DED1) are not packaged in stable nucleosomes as judged by enhanced nuclease sensitivity (22,29–31) and by rapid repair of UV lesions by photolyase (32). There are, however, examples where T-tracts were reported in nucleosomal regions (33,34), indicating that T-tracts alone are not sufficient to disrupt nucleosomes, but they might lead to a local distortion in the nucleosome (35). From these observations it is not clear whether and how T-tracts contribute to chromatin organisation and whether additional factors are required to disrupt nucleosomes. The only T-tract binding protein identified so far in yeast is datin. It recognises T-tracts >9–11 bp in vitro (36,37), but an effect of datin on nucleosome formation is not known. It was reported that Gcn5p, which is a histone acetyltransferase of the yeast SAGA complex, acts as a co-activator of HIS3 transcription (38) by acetylation of histones in the promoter region (39). This implies that nucleosome remodelling is involved in structural organisation of the HIS3 promoter.
The central questions addressed here are whether T-tracts in yeast promoters adopt the unusual rigid structure in vivo and whether those T-tracts are incorporated in nucleosomes.
MATERIALS AND METHODS
Yeast strains
JMY1 [MATa his3-Δ1 trp1-289 rad1-Δ ura3-52 YRpTRURAP (URA3 ARS1)] and FTY117 [MATa his3-Δ1 trp1-289 rad1-Δ ura3-52 YRpCS1 (HIS3 TRP1 ARS1)] have been described (32). GSY5 (MATa ade2-1 ura3-1,15 trp1-1 leu2-3, 112 can1-100 rad1Δ gcn5Δ::LEU2) and GSY2 (MATa ade2-1 ura3-1,15 trp1-1 leu2-3, 112 can1-100 rad1Δ) were generated from W303-1a by restoration of the genomic HIS3 gene using a 1.6 kb BamHI fragment with the pet56-HIS3-ded1 sequence of YRpCS1 for gene replacement and by disruption of RAD1 and GCN5, respectively. EWY1002C-1 [MATa his3-Δ200 leu2-3-112 ura3-52 lys2-801(am) trp1-1(am)] and EWY1002C-2 [MATa his3-Δ200 leu2-3-112 ura3-52 lys2-801(am) trp1-1(am) dat1Δ::LEU2] (36) were provided by Dr E. Winter and transformed with YRpCS1 (HIS3 TRP1 ARS1) to yield BSY1 and BSY2, respectively.
UV irradiation of yeast cells
UV irradiation and photoreactivation were as described (32). Yeast strains were grown at 30°C in minimal medium (2% dextrose, 0.67% Yeast Nitrogen Base without amino acids) supplemented with the required amino acids to a final density of ∼1 × 107 cells/ml. GSY2 and GSY5 were grown for 6 h in the presence of 3-aminotriazol to induce HIS3 transcription. Cells were harvested and resuspended in minimal medium to yield ∼3 × 107 cells/ml. The cell suspension was transferred to plastic trays to form thin layers and irradiated under four Sylvania G15T8 germicidal lamps (predominantly 254 nm as measured by an UVX radiometer; UVP Inc., CA). After irradiation the media were supplemented with the appropriate amino acids and 3-aminotriazol (GSY2 and GSY5). Photoreactivation of the cell suspension was done in plastic trays using Sylvania Type F15 T8/BLB bulbs (peak emission at 375 nm) at ∼1.5 mW/cm2 at 22–26°C. After different exposure times aliquots were collected and chilled on ice. One UV-irradiated sample was incubated at 23°C in the dark. DNA isolation was either done by phenol extraction or using Qiagen Genomic-Tips. DNA was dissolved in 10 mM Tris, 1 mM EDTA, pH 8.0 (TE). All post-irradiation steps were carried out under yellow light to prevent photoreactivation.
UV irradiation and photoreactivation of DNA
Purified DNA was exposed in droplets to UV light. For photoreactivation in vitro UV-irradiated DNA was mixed with Escherichia coli photolyase (0.1 µg/µl) to a final concentration of 0.1 µg photolyase/µg DNA and exposed in droplets to photoreactivating light for 50 min at 2 mW/cm2.
Primer extension analysis
DNA was digested with EcoRI, purified and dissolved in TE. Primer labelling and primer extension were done as described (40,41) with modifications for detection of CPDs in genomic DNA (42). The reaction products were analysed on 8% polyacrylamide gels. DNA damage at individual sites or at clusters of sites was calculated from PhosphorImager scans as a fraction of the total amount of signal per lane. The signal of non-irradiated DNA was subtracted to yield total damage [CPDs and pyrimidine (6–4) pyrimidone photoproducts (6–4PPs)]. The signal for DNA treated with E.coli photolyase displays non-CPD lesions. For repair analysis, initial damage at each site was set to 0% repair. The sequences of the primers were 5′-TGTCATCTTTGCCTTCGTTTATCTTGC (HIS3 gene, top strand), 5′-TGATTTATCTTCGTTTCCTGCA (URA3 gene, top strand) and 5′-TATTATCTCTGCTATTTTGTGACG (ILV1 gene, bottom strand).
RESULTS
UV photofootprinting was applied in yeast and to naked DNA to identify the structures of T-tracts based on their characteristic formation of CPD lesions (12). We focused on the promoter regions of the yeast URA3 and HIS3 genes, since the T-tracts of both genes act as transcriptional elements in vivo (26,27) and are located in nuclease-sensitive regions (22,29).
Poly(dA·dT) sequences located in open promoter regions adopt T-tract structure in vivo
The URA3 promoter was investigated in strain JMY1 harbouring the URA3 gene in a minichromosome (YRpTRURAP; Fig. 1). For damage induction, protein-free DNA and yeast cells in suspension were irradiated with UV light (predominantly 254 nm) which generates preferentially cis-syn CPDs and, to a much lesser extent, 6–4PPs (43). DNA was purified and DNA lesions were assessed by primer extension with Taq polymerase, which is efficiently blocked in front of the 3′-end of CPDs and 6–4PPs (40,41). The intensities of the bands reflect the yields of UV lesions. Irradiation conditions were chosen that generated <0.1 CPD/molecule in the promoter region.
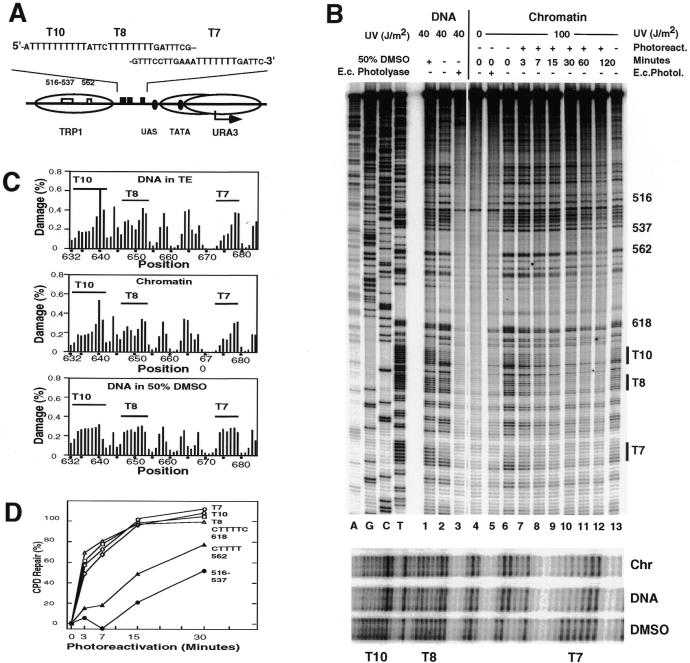
Figure 1.
UV photofootprints and repair analysis of T-tracts in the URA3 promoter of strain JMY1 [rad1Δ; YRpTRURAP (URA3, ARS1)]. (A) Schematic drawing of the chromatin structure and DNA elements in the top strand including nucleosome positions (ovals; overlapping positions at the TATA box) and T-tracts (T10, T8 and T7) (29,30). (B) Primer extension products showing damage formation and repair. UV damage generated by UV light (preferentially CPDs and 6–4PPs) in naked DNA: lane 1, in 50% DMSO; lane 2, in TE buffer; lane 6, in living cells (chromatin). Lanes 7–12, UV photoproducts after photoreactivation for 3–120 min in vivo. Lanes 3 and 5, non-CPDs (preferentially 6–4PPs) revealed by treatment with E.coli photolyase. Lane 4, DNA of non-irradiated cells. A, G, C and T, sequencing lanes. The T-tract regions of lanes 1, 2 and 6 are enlarged in the bottom panel. (C) Quantitative analysis. The graphs show the percentage of total DNA damage generated at individual dipyrimidine sequences. (D) DNA repair curves. Repair of CPDs is plotted versus incubation time (min) in the presence of photoreactivating light. The pyrimidine clusters 562 and 516–537 are located in the flanking nucleosome.
Damage induction is shown for DNA irradiated in TE buffer and in cellular chromatin (Fig. 1B, lanes 2 and 6). These lesions were predominantly CPDs, since they could be removed by incubation with E.coli photolyase in vitro (lanes 3 and 5) and by photoreactivation in vivo (lanes 7–12; see below).
The URA3 promoter contains a T10-, a T8- and a T7-tract in the top strand. In free DNA, damage induction in the T10- and T7-tracts was highest at the 3′-end and lower towards the 5′-end (Fig. 1B, lane 2 and enlargement, and C). Damage in the T8-tract was also highest at the 3′-end, decreased towards the middle of the tract, but slightly increased again towards the 5′-end. Both, the T10- and T7-tracts are flanked by adenines at their 5′-ends, whereas the T8-tract has a cytosine at this position (Fig. 1A). This damage pattern observed in the T-tracts of free DNA is characteristic for the unusual structure of T-tracts (12). Dimethyl sulphoxide (DMSO) unwinds DNA by dehydration (44), perturbs the unusual T-tract structure and generates a more even CPD pattern upon UV irradiation (12). When naked DNA was irradiated in 50% DMSO the damage yields were similar at all positions within the T-tracts, demonstrating a loss of the T-tract structure (Fig. 1B and C). We therefore conclude that the T7-, T8- and T10-tracts of the natural URA3 promoter adopt typical T-tract structures in aqueous solution.
When DNA damage was generated in yeast cells, the CPD patterns in all T-tracts were very similar to those induced in free DNA (Fig. 1B, lanes 6 and 2, and C). These data are direct evidence that the T-tracts of the URA3 promoter adopt the same rigid DNA structure in chromatin in vivo as in free DNA.
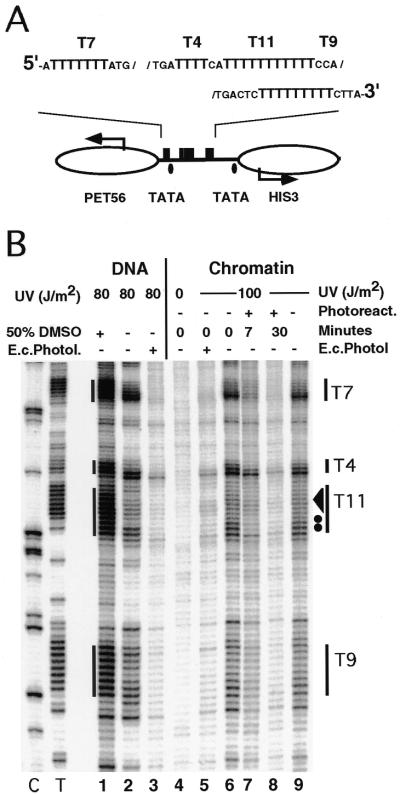
To test whether this observation also applies to other T-tracts, we analysed the HIS3 promoter region in the minichromosome YRpCS1 of strain FTY117. This region, which includes the divergent HIS3 and PET56 promoters (45), is nuclease sensitive and flanked by positioned nucleosomes (Fig. 2A). All T-tracts in free DNA as well as in chromatin showed a CPD pattern characteristic of T-tract structures, with enhanced damage formation at the 3′-end and disruption of the structure upon incubation in DMSO (Fig. 2B, lanes 1, 2 and 6). The enhanced yields of lesions at the 5′-end of the T9-tract are attributable to the flanking cytosine, as in the T8-tract of URA3. Thus, the HIS3 promoter in the minichromosome is a second example where T-tracts in vivo maintain the same rigid structure as in isolated DNA.
Figure 2.
UV photofootprints and repair analysis of T-tracts in the extrachromosomal HIS3 promoter of strain FTY117 [rad1Δ; YRpCS1 (HIS3 TRP1 ARS1)]. (A) Schematic drawing of chromatin structure and DNA elements (top strand; according to 22). (B) Primer extension products. For space reasons, only the lower part of the gel is shown.
T-tract structure of poly(dA·dT) sequences in genomic chromatin
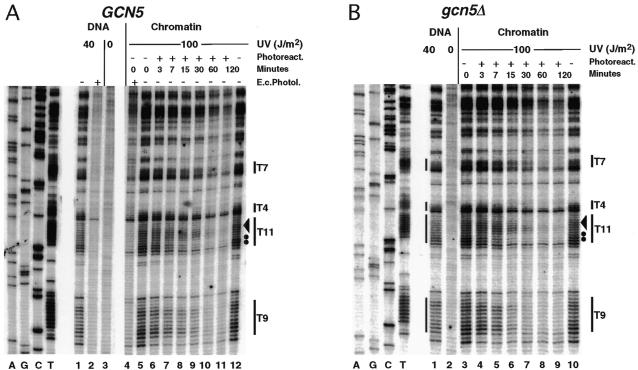
Chromatin folding involves several levels of DNA packaging from nucleosomes to higher order structures (46). It has been demonstrated that the arrangement of nucleosomes on the DNA sequence (nucleosome positioning) depends on a combination of DNA sequence, protein–DNA interactions, boundaries and chromatin folding (47). It is therefore conceivable that the location of a gene in chromosomes or small circular minichromosomes affects the structure of T-tracts due to local constraints imposed by packaging of DNA in chromatin. We therefore tested whether the T-tract structure also occurs in the genomic HIS3 gene. The HIS3 promoter has the same chromatin structure in the genome as in the minichromosome, namely with the T-tracts located in a nuclease-sensitive region (not shown). The UV damage pattern in chromatin was very similar to that observed in free DNA (Fig. 3A, lanes 1 and 5) and no difference was observed between the genomic copy and the HIS3 gene in the minichromosome. We conclude that the T-tract structure is maintained in the genome and that there are no constraints on the T-tracts in genomic chromatin which would affect their structure.
Figure 3.
UV photofootprints and repair analysis in the genomic HIS3 promoter in the presence and absence of Gcn5p. (A) Primer extension products of strain GSY2 (rad1Δ GCN5). (B) Primer extension products of strain GSY5 (rad1Δ gcn5Δ). Dots and the arrowhead indicate fast and more slowly repaired sites within T11, respectively.
Efficient CPD repair in poly(dA·dT) sequences by photolyase
It is essential to establish whether T-tracts are incorporated in nucleosomes, modified nucleosomes or complexed with other proteins. We used CPD repair by photolyase (photoreactivation) as a direct in vivo approach, in contrast to chromatin digestion with nucleases, which is an in vitro assay. Photoreactivation is a major pathway to remove UV-induced CPDs and it is modulated by nucleosomes and proteins bound to DNA (32,48–50). Nucleotide excision repair, which is the alternative pathway to remove CPDs, was inactivated by disruption of the RAD1 gene (rad1Δ). Immediately after damage induction the cell suspension was exposed to photoreactivating light for different times, the DNA was extracted and the remaining lesions were analysed.
In a low resolution repair study we previously showed that linker DNA between nucleosomes and whole open promoter regions of the URA3 and HIS3 genes were repaired in 15–30 min, while repair of nucleosomes required ∼2 h (32). Thus, photolyase is a new tool to measure whether a DNA lesion is in a nucleosome in vivo. The high resolution data presented here show rapid repair of all T-tracts in the URA3 and HIS3 promoters in minichromosomes (Figs 1B, lanes 7–12, and 2B, lanes 7 and 8) and in the genome (Fig. 3A, lanes 6–11). After 30 min >90% of CPDs were removed at all positions within the different T-tracts (T7, T10, T8 and CTTTC 618 in Fig. 1A and D), while DNA lesions which map in the flanking nucleosome were repaired at 40–50% (CTTT 562 and 516–537). Repair was exclusively by photolyase. No repair occurred in the absence of light (Figs 1B, lane 13, 2, lane 9, 3A, lane 12, and 3B, lane 10). This efficient repair demonstrates that these T-tracts were not folded in nucleosomes.
We noticed a subtle repair heterogeneity within T-tracts. While all sites in the T9-tract of HIS3 were repaired at similar rates, CPDs at the 5′-end of the T11-tract (excluding the TC dimer) were more rapidly repaired than CPDs towards the 5′-end. This was observed in the minichromosome and in the genome and might indicate unknown factors which interfere with repair (dots and arrowhead in Figs 2 and 3).
The T-tract structures in the HIS3 promoter are independent of Gcn5p
Gcn5p is a histone acetyltransferase of the SAGA complex and contributes to transcriptional activity of the HIS3 promoter (38), possibly by nucleosome remodelling. Thus, it is conceivable that Gcn5p activity is required to destabilise the nucleosomes in the promoter and thereby allow T-tracts to adopt their rigid structure. Figure 3B shows the results obtained in a strain with a deleted GCN5 gene (gcn5Δ). The photofootprints were identical to those obtained in the GCN5 wild-type strain GSY2. Thus, the T-tract structure in the HIS3 promoter is independent of Gcn5p. Moreover, there was no obvious difference in photoreactivation of T-tracts, indicating that the T-tracts remained accessible to photolyase in the absence of Gcn5p. Thus, both the photofootprints and rapid repair data argue against Gcn5p-dependent nucleosome remodelling in the T-tract region.
T-tract structures in vivo are independent of datin
Datin is the only known yeast protein that binds poly(dA·dT) sequences of 9–11 bp (36,37). Datin behaves as a repressor and activator through poly(dA·dT) sequences of the HIS3 and ILV1 promoters, respectively (28,51). To investigate whether datin affects the structure of T-tracts in vivo, yeast strains containing the wild-type gene (DAT1) or a disruption (dat1Δ) were transformed with YRpCS1 and the UV photofootprints in the HIS3 promoter were analysed (Fig. 4). There were no obvious differences in the CPD patterns of the T-tracts between the dat1Δ and wild-type (DAT1) strains and the patterns described above (Figs 2 and 3). Similarly, the promoter of the genomic ILV1 gene showed typical T-tract patterns that were not affected by deletion of the datin gene. We conclude that the T-tract structure as it occurs in chromatin in vivo is independent of datin.
Figure 4.
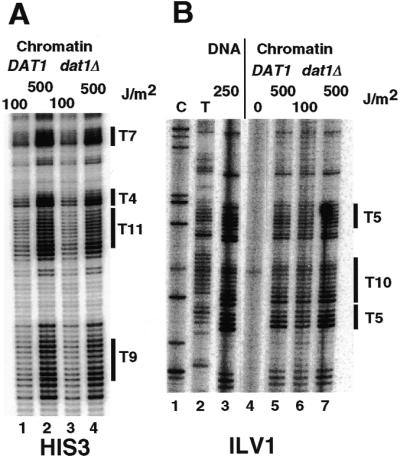
T-tract structure is independent of datin. (A) Primer extension analysis of UV photoproducts in the HIS3 promoter of strains BSY1 (DAT1, YRpCS1) and BSY2 (dat1Δ, YRpCS1). (B) Primer extension analysis of UV photoproducts in the ILV1 promoter of the same strains.
DISCUSSION
A wide variety of experiments established that T-tract DNA adopts an unusual rigid B-form structure in vitro and it was inferred that this particular structure serves a functional role in the genome. Here we show direct evidence that this unusual structure exists in chromatin of living cells. We identified these structures in promoters which are known to be transcriptionally regulated by T-tracts. Moreover, we show that these T-tracts are not folded in nucleosomes nor affected by datin or Gcn5p. This suggests that the T-tract structure per se is involved in transcriptional regulation of these genes.
Very recent work reported T-tract structures in artificial minichromosome constructs (52). Here we show T-tract structures in natural promoters. The T-tract structures did not depend on whether the gene (HIS3) was located in a small circular minichromosome (3.2 kb) or in the genome, demonstrating that there were no structural constraints in chromosomes or minichromosomes which would compromise these structures. Moreover, T-tract structure was observed in tracts which varied in length from 4 to 11 bp. We also observed T-tract structures in the nuclease-sensitive 3′-end of the URA3 gene and in the DED1 promoter (not shown). Therefore, the unusual structure of T-tracts appears to be a dominant feature in yeast chromosomes and further suggests a functional role of T-tract structures. It was estimated that there are ∼1500 yeast genes whose promoters contain T-tracts >10 bp (28). It is reasonable to extrapolate that many of these promoters will reveal T-tract structures when analysed.
Our results shed light on how chromatin is organised under conditions where T-tract structures were observed in the HIS3 and URA3 promoters. Nuclease digestion of isolated chromatin in vitro previously established that the T-tracts are located in nuclease-sensitive regions flanked by positioned nucleosomes (22,29,30). However, another report proposed that a ‘perturbed’ nucleosome includes the T-tracts of a modified HIS3 promoter (28). Here we observed that the UV damage patterns of the promoter regions were identical in vivo and in naked DNA. These in vivo photofootprints argue against a nucleosome, since examples are known where T-tract structure was lost upon incorporation in nucleosomes (8,9). In addition, we found that all T-tracts were repaired within <30 min and repair was much faster than repair of CPDs in nucleosomes (32). Thus, three lines of evidence (nuclease digestion, photofootprinting and DNA repair data) are consistent with the absence of nucleosomes in the T-tract regions of the HIS3 and URA3 promoters.
A difficult question to address is whether the T-tracts structures in vivo are stabilised by proteins or whether they are maintained by their intrinsic properties. Since T-tract size varied between T4 and T11, one would have to propose a class of different T-tract stabilising proteins. All the photofootprints were very similar in naked DNA and in chromatin, in small and long T-tracts, in minichromosomes and chromosomes and in the presence and absence of datin and Gcn5p. With the exception of the mild repair delay in the T11-tract of HIS3, there was no indication for proteins in these regions. If proteins bind to T-tracts in vivo, they do not disturb the T-tract structure and they do not inhibit repair or they are displaced by damage induction. All those observations together make it more likely that T-tracts are not tightly associated with proteins and therefore participate in transcription regulation by virtue of their intrinsic DNA structure, as previously proposed (28).
We still lack a clear explanation as to why these T-tract regions are not packaged in nucleosomes. Based on controversial results on how T-tracts can be incorporated in nucleosomes and that T-tracts were found in nucleosomal as well as in non-nucleosomal regions (see Introduction), T-tracts per se appear not to be sufficient to exclude nucleosome formation in vivo. However, under some conditions they may destabilise nucleosomes or affect their positions, as reported for long T-tracts artificially inserted in minichromosome constructs (52). To establish a non-nucleosomal region, additional activities might be required. A sequence-specific protein, Abf1, and a T-rich element appear to be required to create a nucleosome-free region in the yeast RPS28A gene (31). Other obvious candidates are nucleosome remodelling activities which contain histone acetyltransferases, are brought to promoter regions by transcription factors and destabilise nucleosomes by acetylation of histones (53). Gcn5p is a histone acetyltransferase of the SAGA complex and contributes to transcriptional activity of the HIS3 promoter (38). Disruption of the GCN5 gene was reported to lead to invasion of the HIS3 promoter by a nucleosome (54). Our photofootprints and repair results do not support that observation. Using a chromatin immunoprecipitation approach it was reported that the promoter region contains acetylated histones (39). The probe used in that study was 0.6 kb long, which, according to our data, includes the nucleosomal regions flanking the promoters of the HIS3 and PET56 genes (22). Therefore, the observation of acetylated histones is compatible with non-nucleosomal T-tracts and modified nucleosomes in the flanking regions.
With respect to DNA repair, we also need to consider that chromatin structures are dynamic and might react on damage induction, e.g. by rearranging nucleosome positions. If the T-tract structure affects the stability or positions of nucleosomes, one would expect that disruption of the T-tract structure by CPD formation (15) would allow regeneration of nucleosomes in these regions. The rapid repair by photolyase shown here is strong evidence against such a chromatin rearrangement after damage formation.
Having shown that T-tract structure exists in T-tracts of different length in vivo, it will be interesting to use UV photofootprinting and photolyase to understand the structure and role of T-tracts in other regions of the genome and in other organisms. Of particular interest is the characterisation of T-tracts in nucleosomal arrays as well as phased T-tracts that induce DNA bending in vitro.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr E. Winter for yeast strains and Dr U. Suter for continuous support. This work was supported by grants from the Swiss National Science Foundation and by the Swiss Federal Institute of Technology (ETH) (grants to F.T.)
REFERENCES
- 1.Nelson H.C., Finch,J.T., Luisi,B.F. and Klug,A. (1987) Nature, 330, 221–226. [DOI] [PubMed] [Google Scholar]
- 2.DiGabriele A.D., Sanderson,M.R. and Steitz,T.A. (1989) Proc. Natl Acad. Sci. USA, 86, 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leroy J.L., Charretier,E., Kochoyan,M. and Gueron,M. (1988) Biochemistry, 27, 8894–8898. [DOI] [PubMed] [Google Scholar]
- 4.Alexeev D.G., Lipanov,A.A. and Skuratovskii,I. (1987) Nature, 325, 821–823. [DOI] [PubMed] [Google Scholar]
- 5.Peck L.J. and Wang,J.C. (1981) Nature, 292, 375–378. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes D. and Klug,A. (1980) Nature, 286, 573–578. [DOI] [PubMed] [Google Scholar]
- 7.Leslie A.G., Arnott,S., Chandrasekaran,R. and Ratliff,R.L. (1980) J. Mol. Biol., 143, 49–72. [DOI] [PubMed] [Google Scholar]
- 8.Schieferstein U. and Thoma,F. (1996) Biochemistry, 35, 7705–7714. [DOI] [PubMed] [Google Scholar]
- 9.Hayes J.J., Bashkin,J., Tullius,T.D. and Wolffe,A.P. (1991) Biochemistry, 30, 8434–8440. [DOI] [PubMed] [Google Scholar]
- 10.Becker M.M. and Wang,J.C. (1984) Nature, 309, 682–687. [DOI] [PubMed] [Google Scholar]
- 11.Becker M.M. and Wang,Z. (1989) J. Mol. Biol., 210, 429–438. [DOI] [PubMed] [Google Scholar]
- 12.Lyamichev V. (1991) Nucleic Acids Res., 19, 4491–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koo H.S., Wu,H.M. and Crothers,D.M. (1986) Nature, 320, 501–506. [DOI] [PubMed] [Google Scholar]
- 14.Wu H.M. and Crothers,D.M. (1984) Nature, 308, 509–513. [DOI] [PubMed] [Google Scholar]
- 15.Wang C.I. and Taylor,J.S. (1991) Proc. Natl Acad. Sci. USA, 88, 9072–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson R.T. and Kunzler,P. (1979) Nucleic Acids Res., 6, 1387–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel G.R. and Martinson,H.G. (1981) Nucleic Acids Res., 9, 6869–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes D. (1979) Nucleic Acids Res., 6, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puhl H.L. and Behe,M.J. (1995) J. Mol. Biol., 245, 559–567. [DOI] [PubMed] [Google Scholar]
- 20.Prunell A. (1982) EMBO J., 1, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puhl H.L., Gudibande,S.R. and Behe,M.J. (1991) J. Mol. Biol., 222, 1149–1160. [DOI] [PubMed] [Google Scholar]
- 22.Losa R., Omari,S. and Thoma,F. (1990) Nucleic Acids Res., 18, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schieferstein U. and Thoma,F. (1998) EMBO J., 17, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satchwell S.C., Drew,H.R. and Travers,A.A. (1986) J. Mol. Biol., 191, 659–675. [DOI] [PubMed] [Google Scholar]
- 25.Dechering K.J., Cuelenaere,K., Konings,R. and Leunissen,J. (1998) Nucleic Acids Res., 26, 4056–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Struhl K. (1985) Proc. Natl Acad. Sci. USA, 82, 8419–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy A., Exinger,F. and Losson,R. (1990) Mol. Cell. Biol., 10, 5257–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer V. and Struhl,K. (1995) EMBO J., 14, 2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoma F. (1986) J. Mol. Biol., 190, 177–190. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka S., Livingstone-Zatchej,M. and Thoma,F. (1996) J. Mol. Biol., 257, 919–934. [DOI] [PubMed] [Google Scholar]
- 31.Lascaris R.F., Groot,E., Hoen,P.B., Mager,W.H. and Planta,R.J. (2000) Nucleic Acids Res., 28, 1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suter B., Livingstone-Zatchej,M. and Thoma,F. (1997) EMBO J., 16, 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdone L., Camilloni,G., Di Mauro,E. and Caserta,M. (1996) Mol. Cell. Biol., 16, 1978–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubbi L., Camilloni,G., Caserta,M., Di Mauro,E. and Venditti,S. (1997) Biochem. J., 328, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z. and Thiele,D.J. (1996) Cell, 87, 459–470. [DOI] [PubMed] [Google Scholar]
- 36.Winter E. and Varshavsky,A. (1989) EMBO J., 8, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reardon B.J., Gordon,D., Ballard,M.J. and Winter,E. (1995) Nucleic Acids Res., 23, 4900–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterner D.E., Grant,P.A., Roberts,S.M., Duggan,L.J., Belotserkovskaya,R., Pacella,L.A., Winston,F., Workman,J.L. and Berger,S.L. (1999) Mol. Cell. Biol., 19, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo M.H., Zhou,J., Jambeck,P., Churchill,M.E. and Allis,C.D. (1998) Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wellinger R.E. and Thoma,F. (1997) EMBO J., 16, 5046–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wellinger R.E. and Thoma,F. (1996) Nucleic Acids Res., 24, 1578–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aboussekhra A. and Thoma,F. (1998) Genes Dev., 12, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA-Repair and Mutagenesis. ASM Press, Washington, DC.
- 44.Lee C.H., Mizusawa,H. and Kakefuda,T. (1981) Proc. Natl Acad. Sci. USA, 78, 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struhl K. (1985) Nucleic Acids Res., 13, 8587–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornberg R.D. and Lorch,Y. (1999) Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- 47.Thoma F. (1992) Biochim. Biophys. Acta, 1130, 1–19. [DOI] [PubMed] [Google Scholar]
- 48.Aboussekhra A. and Thoma,F. (1999) EMBO J., 18, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suter B., Wellinger,R.E. and Thoma,F. (2000) Nucleic Acids Res., 28, 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoma F. (1999) EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreira J.M., Remacle,J.E., Kielland-Brandt,M.C. and Holmberg,S. (1998) Mol. Gen. Genet., 258, 95–103. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu M., Mori,T., Sakurai,T. and Shindo,H. (2000) EMBO J., 19, 3358–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman J.L. and Kingston,R.E. (1998) Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 54.Filetici P., Aranda,C., Gonzalez,A. and Ballario,P. (1998) Biochem. Biophys. Res. Commun., 242, 84–87. [DOI] [PubMed] [Google Scholar]