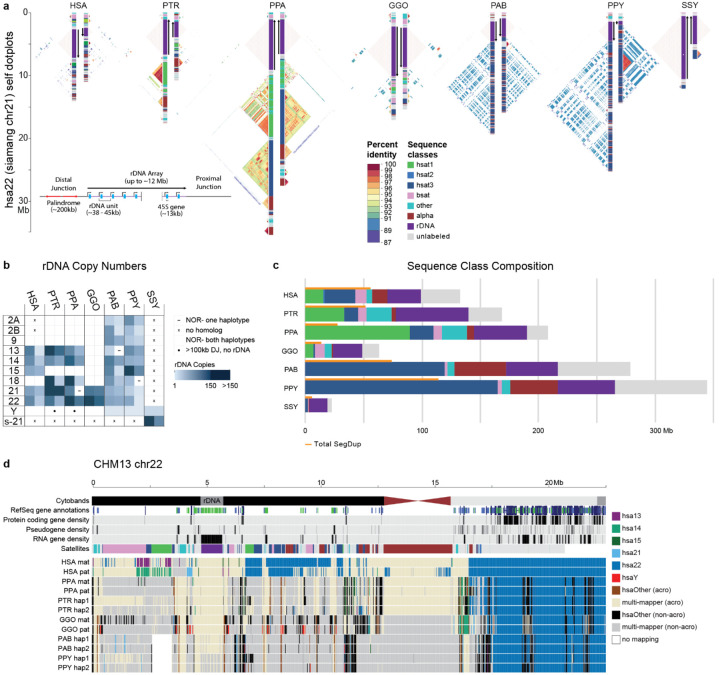
Figure 6. Organization and sequence composition of the ape acrocentric chromosomes.
a) Sequence identity heatmaps and satellite annotations for the NOR+ short arms of both HSA22 haplotypes across all the great apes, and siamang chr21 (the only NOR+ chromosome in siamang) drawn with ModDotPlot82. The short arm telomere is oriented at the top of the plot, with the entirety of the short arm drawn to scale up to but not including the centromeric α-satellite. Heatmap colors indicate self-similarity within the chromosome, and large blocks indicate tandem repeat arrays (rDNA and satellites) with their corresponding annotations given in between. Human is represented by the diploid HG002 genome. b) Estimated number of rDNA units per haplotype (hap) for each species. HSA numbers are given in the first column, with the exception of “s-21” for siamang chr21, which is NOR+ but has no single human homolog. c) Sum of satellite and rDNA sequence across all NOR+ short arms in each species. “unlabeled” indicates sequences without a satellite annotation, which mostly comprise SDs. Total SD bases are given for comparison, with some overlap between regions annotated as SDs and satellites. d) Top tracks: chr22 in the T2T-CHM13v2.0 reference genome displaying various gene annotation metrics and the satellite annotation. Bottom tracks: For each primate haplotype, including the human HG002 genome, the chromosome that best matches each 10 kbp window of T2T-CHM13 chr22 is color coded, as determined by MashMap83. On the right side of the centromere (towards the long arm), HSA22 is syntenic across all species; however, on the short arm synteny quickly degrades, with very few regions mapping uniquely to a single chromosome, reflective of extensive duplication and recombination on the short arms. Even the human HG002 genome does not consistently align to T2T-CHM13 chr22 in the most distal (left-most) regions.