Abstract
We have determined the crystal structure of the RNA octamer duplex r(guguuuac)/r(guaggcac) with a tandem wobble pair, G·G/U·U (motif III), to compare it with U·G/G·U (motif I) and G·U/U·G (motif II) and to better understand their relative stabilities. The crystal belongs to the rhombohedral space group R3. The hexagonal unit cell dimensions are a = b = 41.92 Å, c = 56.41 Å, and γ = 120°, with one duplex in the asymmetric unit. The structure was solved by the molecular replacement method at 1.9 Å resolution and refined to a final R factor of 19.9% and Rfree of 23.3% for 2862 reflections in the resolution range 10.0–1.9 Å with F ≥ 2σ(F). The final model contains 335 atoms for the RNA duplex and 30 water molecules. The A-RNA stacks in the familiar head-to-tail fashion forming a pseudo-continuous helix. The uridine bases of the tandem U·G pairs have slipped towards the minor groove relative to the guanine bases and the uridine O2 atoms form bifurcated hydrogen bonds with the N1 and N2 of guanines. The N2 of guanine and O2 of uridine do not bridge the ‘locked’ water molecule in the minor groove, as in motifs I and II, but are bridged by water molecules in the major groove. A comparison of base stacking stabilities of motif III with motifs I and II confirms the result of thermodynamic studies, motif I > motif III > motif II.
INTRODUCTION
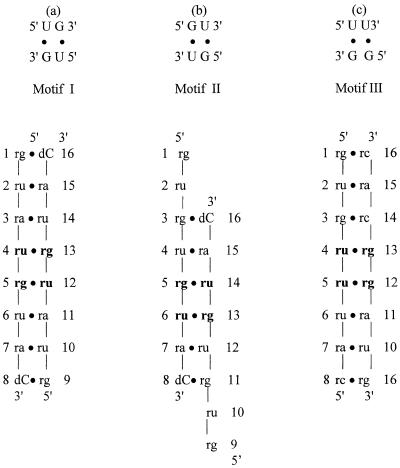
Base pairs in DNA are important due to the crucial role of complementary DNA Watson–Crick base pairs in genetic information transfer from one generation to the next. Mispairs in DNA are usually corrected by repair enzymes in order to maintain the fidelity of the genetic code (1,2). RNAs have very substantial differences in biological functions than DNA (3). Mispaired base pairings are frequently observed in natural RNA molecules (4,5) and seem to be essential in biological functions such as RNA processing and RNA–protein interactions. Among mispairs, the wobble base pair (G·U) is the most frequent non-canonical structure conserved at certain sites in rRNA (6,7). It is often involved in functional interactions, particularly in RNA–protein recognition (8–12). Indeed, G·U pairs in tRNA stems, e.g. alanyl-tRNA, are recognized by its cognate synthetase (8–10) and are also crucial for splice site selection in group I introns (13–15). Adjacent or tandem G·U pairs have three motifs (Fig. 1a; 16). Studies of the conservation of tandem G·U pairs in 16S and 23S rRNAs showed that motif I (5′-UG-3′/3′-GU-5′) occurs most frequently. The next most frequent occurrence is motif III (5′-UU-3′/3′-GG-5′), and motif II (5′-GU-3′/3′-UG-5′) is the least frequently observed. The order of occurrence of motif I > motif III > motif II is in agreement with thermodynamic studies (16–18). The crystal structures of motifs I and II were reported by this laboratory earlier (19,20). While the present motif III sequence was being crystallized, the crystal structure of the catalytic domain of the group I intron was determined (21). While this paper was being written another 14 bp RNA oligonucleotide containing motif III was reported (22). Here we report a simple octamer double helix of motif III as in motifs I and II and compare their base stacking stabilities.
Figure 1.
Schematic representations of base pairings of the three tandem G·U pairs: (a) motif I with the sequence r(guaugua)dC; (b) motif II with the sequence r(gugugua)dC; (c) motif III with the sequence r(guguuuac)/r(guaggcac).
MATERIALS AND METHODS
Synthesis and purification
The non-self-complementary RNA octamer r(guguuuac)/r(guaggcac) was synthesized by the phosphoramidite method using an Applied Biosystem DNA synthesizer 391 and cleaved from the solid support using 3:1 (v/v) ammonia/ethanol and incubated overnight in the same solution at 55°C for deprotection. The 2′-hydroxyl groups were deprotected by overnight incubation with 0.8 ml of 1 M tetrabutylammonium fluoride. An equal amount (0.8 ml) of 0.1 M triethylamine acetate was added and the mixture was evaporated in a lyophilizer. The RNA fragment was subsequently precipitated with ammonium acetate/ethanol. The sample was further purified by ion exchange FPLC, using lithium chloride for the eluting gradient.
Crystallization and data collection
The crystallization experiments were performed by the hanging drop vapor diffusion method at room temperature (293 K). The best crystals were obtained with 40 mM sodium cacodylate buffer pH 7.0, 50 mM magnesium chloride, 1.0 mM cobalt hexamine chloride and 5% (v/v) methyl 2,4-pentanediol (MPD) against 55% MPD in the reservoir. Crystals of dimensions 0.3 × 0.2 × 0.4 mm were obtained in 2 weeks. Data were collected from a single crystal at room temperature on a R-AXIS IIc imaging plate and a 50 kV/100 mA graphite monochromated CuKα (1.5418 Å) X-ray beam. A total of 2862 independent reflections corresponding to 98.5% of the theoretically possible data were collected to 1.9 Å resolution with an Rmerge of 5.5%. The data completeness for the last resolution bin, 1.9–1.97 Å, was 97.4%. The data were processed using the programs DENZO and SCALEPACK (23). The crystals belonged to the rhombohedral space group R3 with hexagonal settings a = b = 41.92 Å, c = 56.41 Å, α = β = 90.0° and γ = 120°. The asymmetric unit contains one duplex with a volume/bp of 1192 Å3. Crystal data and refinement statistics are summarized in Table 1.
Table 1. Crystal data and refinement statistics for r(guguuuac)/r(guaggcac).
| Crystal data | |
| Crystal system | Rhombohedral |
| Space group | R3 |
| Cell parameters (hexagonal setting) | |
| a = b (Å) | 41.92 |
| c (Å) | 56.41 |
| γ (°) | 120 |
| Asymmetric unit | 1 duplex |
| Volume/base pair (Å3) | 1192 |
| Resolution (Å) | 1.9 |
| No. of unique reflections | 2862 |
| Data completeness (%) | 98.5 |
| Rsym (%) | 5.5 |
| Refinement | |
| Resolution range used (Å) | 10.0–1.9 |
| No. of reflections used [≥2σ(F)] | 2862 |
| Rwork/Rfree(%) | 19.9/23.3 |
| r.m.s. deviation from target values | |
| Parameter file | param_nd.dna |
| Bond length (Å) | 0.013 |
| Bond angle (°) | 1.68 |
Structure solution and refinement
The structure was solved by the molecular replacement method, using the atomic coordinates of the self-complementary duplex r(guauaua)dC (24) as the starting model in the hexagonal unit cell: a = b = 43.07 Å and c = 59.36 Å. Refinement was performed using the program CNS (25): 10% of the reflections were selected at random for the Rfree calculation. The starting model gave a Rwork/Rfree of 0.363/0.425. The structure was then refined as a rigid body using 2396 reflections between 10.0 and 2.0 Å resolution followed by a positional refinement. The Rwork/Rfree dropped to 0.319/0.357. The model was then annealed by heating the system to 4000 K and slow cooling to room temperature at 0.5 fs sampling intervals. This dropped the Rwork/Rfree to 0.265/0.314. The bases were then corrected by omit │Fo│ – │Fc│ maps calculated by removing one base pair at a time. The model was subsequently refined by positional and individual B factor refinement with reflections from 10.0 to 1.9 Å. Thirty water molecules located in the difference density maps were added. The final Rwork/Rfree was 0.199/0.233. The model contains 335 nucleic acid atoms and 30 water molecules. The r.m.s. deviation of the final model from ideal geometry was 0.013 Å in bond length and 1.68° in bond angle, respectively. The atomic coordinates and structure factors have been deposited with the Nucleic Acid Database (26), accession code AR0022.
RESULTS AND DISCUSSION
Overall molecular structure
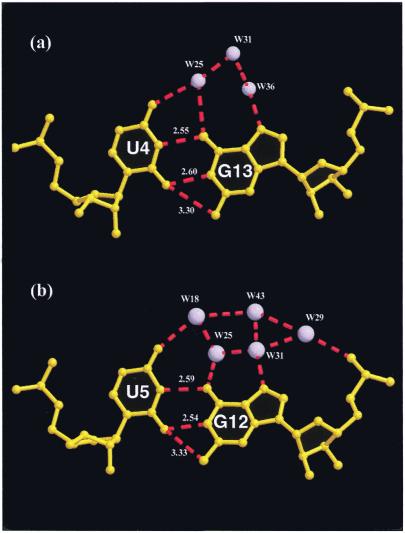
Figure 1 gives a schematic diagram of the sequences. The non-self-complementary octamer r(guguuuac)/r(guaggcac) forms a right-handed A-RNA duplex with 10.6 residues/turn, exhibits a narrow major groove (deep groove, 3.54 Å) and a wide minor groove (shallow groove, 11.27 Å). The duplexes stack in columns forming a pseudo-continuous infinite RNA helix. The base pairs at the junction of the two duplexes are at a very low twist angle of 5°. All ribose sugars are in the characteristic C3′-endo conformation. The average base pair displacement is –3.45 Å. The bases are anti and the backbone bond conformations C4′–C5′ (α) and P–O5′ (γ) are g– and g+, respectively. The average intra-strand phosphate–phosphate distance is 5.71 Å, typical of chains containing C3′-endo sugars. The uridine bases of the tandem wobble U·G/U·G pairs move into the minor groove such that the O2 atoms of the U residues are close enough to form bifurcated hydrogen bonds with N1 and N2 of the guanines (Fig. 2). In motifs I and II the tandem wobble base pairs form normal hydrogen bonds and the N2 atoms of G residues are at non-hydrogen bonding distances (>3.7 Å) from the O2 atoms of the U residues and lock a water molecule in the minor groove, which is not observed in motif III. However, in the crystal structure of a 14mer RNA containing motif III the tandem wobble base pairs lock a water molecule in the minor groove (22).
Figure 2.
Hydration network of the G·U wobble pairs; the water molecules are labeled. Note that the major grooves are hydrated. (a) The first G·U wobble pair; (b) the second G·U pair.
Hydration and crystal packing
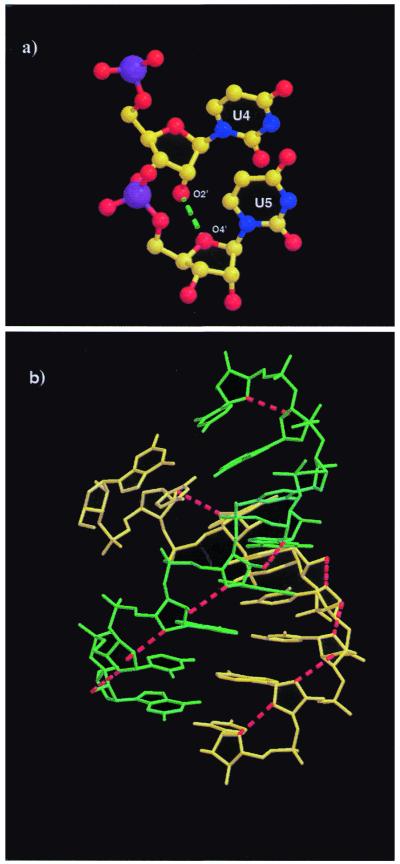
A total of 30 ordered solvent molecules were found in the current structure. The majority of the water molecules are located in the major groove and are especially concentrated in the central part of the helix where the G·U wobbles occur. The N7 atoms of most of the purines in the major groove are hydrogen bonded with water molecules. In fact, the N7 atoms of residues A7 and A15 are linked to the respective O1P groups via water bridges. The N7 atom of G3 and the O4 atom on U4 of the adjacent base are connected by a water molecule. A similar pattern (N7–N4) is found between the adjacent bases G13 and C14. Unlike motifs I and II, the helix backbone of motif III is not highly hydrated. Some O2′H groups on the sugars are linked to water molecules. Water molecules are found to be hydrogen bonded to O2′ atoms of G1, U4 and G9 of strand I and G12, G13 and G15 of strand II. Most of the O2′ atoms form intra-molecular hydrogen bonds to the adjacent O4′ atoms of the sugar on the 3′-side, thus stabilizing the double helix (Fig. 3). The tandem wobble pairs are extensively hydrated in the major groove while no water molecules are found in the minor groove (Fig. 2), maybe because of slippage of the uridine bases towards the minor groove. The acceptor O4 (U4) and N7 (G13) atoms in the major groove are linked by three water-mediated hydrogen bonds (W25, W31 and W36), while O4 (U5) and O6 (G12) are linked by two water molecules, W18 and W25. The acceptors O4 (U5), O6 (G12), N7 (G12) and O1P (G12) in the major groove are linked to each other via a network of five water molecules, W18, W25, W29, W31 and W43 (Fig. 2). These water bridges in the major groove side could play an important role in stabilizing the G·U wobble pairs in the RNA helix and also in RNA–protein interactions. In all three motifs the major grooves in the center of the double helices are highly hydrated. The high negative potential, particularly the presence of O4 atoms in the major groove, makes the G·U wobbles potential metal binding sites.
Figure 3.
(a) Close view of the O2′–O4′ interaction between residues U4 and U5. (b) The intramolecular hydrogen bonds between O2′ hydroxyl groups and O4′ sugar ring atoms in motif III are shown by broken red lines.
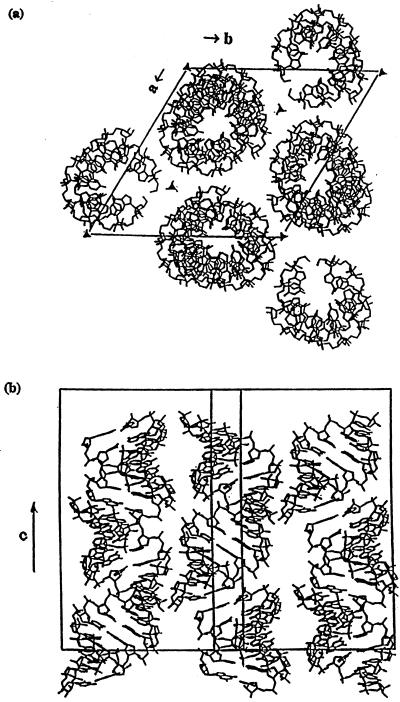
The stacked duplexes are closely packed around the 31-screw axis in a pseudo-continuous column parallel to the crystallographic c-axis (Fig. 4). The inter-molecular contact mainly occurs by interaction between the 31-screw axis related molecules through the donor 2′-OH groups and the N4 atoms in the major groove. Several CH–O contacts also occur in the minor groove (Table 2), for instance C5′(G3)H–O4′(U4) = 3.19 Å. The 3-fold rotation axis related molecules are further apart and do not have any direct inter-molecular interactions.
Figure 4.
(a) View of the crystal packing in the hexagonal unit cell down the c-axis. (b) View of the crystal packing in the unit cell perpendicular to the c-axis.
Table 2. Inter-molecular hydrogen bond contacts <3.3 Å.
| Atom | Atom | Duplex no.a | Distance (Å) |
|---|---|---|---|
| G1(N7) | C8(N4) | (I) | 3.29 |
| G1(O2′) | A15(C5′) | (II) | 3.14 |
| U2(O4′) | A15(C4′) | (II) | 3.19 |
| G3(O2P) | U4(O2′) | (II) | 2.56 |
| G3(C5′) | U4(O4′) | (II) | 3.19 |
| U4(O4′) | G3(C5′) | (III) | 3.19 |
| U4(O2′) | G3(O2P) | (III) | 2.56 |
| C8(N4) | G1(N7) | (IV) | 3.29 |
| G9(N7) | C16(N4) | (IV) | 3.26 |
| A15(C5′) | G1(O2′) | (III) | 3.14 |
| A15(C4′) | U2(O4′) | (III) | 3.19 |
| C16(N4) | G9(N7) | (I) | 3.26 |
| C16(C4′) | C16(O2′) | (III) | 3.14 |
| C16(O2′) | C16(O2′) | (III) | 2.54 |
| C16(O3′) | C16(O2′) | (III) | 3.25 |
| C16(O2′) | C16(C4′) | (II) | 3.14 |
| C16(O2′) | C16(O2′) | (II) | 2.54 |
| C16(O2′) | C16(O3′) | (II) | 3.25 |
aThe roman numeral in parentheses represents the duplex number with the following symmetry: I, y – x + 1, –x + 1, z; II, y – x + 2, –x + 1, z; III, –y + 1, x – y – 1, z; IV, –y + 1, x – y – 1, z + 1.
Base stacking of the wobble and flanking Watson–Crick base pairs in motifs I, II and III
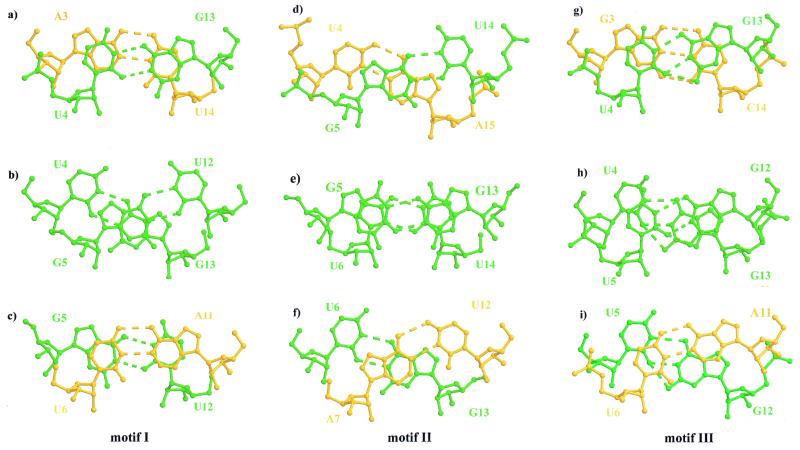
Base stacking is crucial for stabilizing adjacent base pairs. The structures of motifs I, II and III do not tell us about thermodynamic stabilities, but they can provide a rationale. Thermodynamic studies have shown that the stabilities of the tandem wobble pairs in these three motifs (U·G/G·U, G·U/U·G and G·G/U·U) are in the order motif I > motif III > motif II (17,18), which is the same order as their occurrence in nature. The helical parameters of motifs I and II have been published in earlier papers (19,20). The helical parameters of motif III have been calculated with the program NEWHEL92 (27) and the twist angles between adjacent G·U wobble pairs and the flanking Watson–Crick pairs have been tabulated (Table 3). Adjacent G·U wobble pairs have a twist angle of 32.2°, which is less than the average of the duplex (34.2°). The 3′-side flanking Watson–Crick base pair closest to a wobble pair shows a high twist angle (37.2°) compared to the average twist. In motif I the twist angle between the tandem G·U wobble pairs is high (38.13°), while twist angles are low at both flanking Watson–Crick ends (∼26.5°, Table 3). In motif II the twist angle between the tandem wobble pairs is very low (23.8°), while average values are seen at flanking Watson–Crick ends (Table 3). The twist angles mainly affect base pair stacking (Fig. 5). In motif I strong intra-strand stacking occurs between G·U wobble base pairs and their flanking Watson–Crick pairs on both the 5′- and the 3′-sides (Fig. 5a and c); guanine bases exhibit cross-strand stacking (Fig. 5b) and uridine bases are apparently not stacked. Motif II has a reversed base pair stacking pattern compared to motif I. The adjacent G·U wobble pairs are stacked with slight slippage (Fig. 5e), but the flanking Watson–Crick pairs exhibit cross-strand stacking between the purines (Fig. 5d and f), while uridines have no stacking interactions. In motif III the uridine bases in the same strand have only partial stacking (Fig. 5h). The six-membered purine and pyrimidine rings of the 5′-side wobble pair with its flanking Watson–Crick base pair exhibit a slightly slipped stacking (Fig. 5g), whereas the stacking of the wobble pair with the 3′-side flanking Watson–Crick pair is considerably less (Fig. 5i).
Table 3. Twist angles for the tandem wobble pairs and their flanking Watson–Crick base pairs in motif I, r(guaugua)dC, motif II, r(gugugua)dC, and motif III, r(guguuuac)/r(guaggcac).
| Motif I | Motif II | Motif III | |||
|---|---|---|---|---|---|
| Base step | Twist (°) | Base step | Twist (°) | Base step | Twist (°) |
| A3·U14 | U4·A15 | G3·C14 | |||
| 26.72 | 30.16 | 33.93 | |||
| U4·G13 | G5·U14 | U4·G13 | |||
| 38.13 | 23.82 | 32.20 | |||
| G5·U12 | U6·G13 | U5·G12 | |||
| 26.34 | 34.04 | 37.23 | |||
| U6·A11 | A7·U12 | U6·A11 | |||
| Average | 30.40 | 29.34 | 34.15 | ||
Figure 5.
Base pair stacking in motifs I (a–c), II (d–f) and III (g–i). All the G·U wobbles are shown in green and all the flanking Watson–Crick pairs are yellow. (a) Stacking of the first G·U wobble with its 5′ flanking Watson–Crick pair with a twist angle of 26.7°. (b) Stacking of the tandem G·U wobbles with a high twist angle of 38.1°. (c) Stacking of the second G·U wobble with its 3′ flanking Watson–Crick pair with a low twist angle of 26.3°. (d) Stacking of the first G·U wobble with its 5′ flanking Watson–Crick pair with a twist angle of 30°. (e) Stacking of the tandem G·U wobbles with a low twist angle of 24°. (f) Stacking of the second G·U wobble with its 3′ flanking Watson–Crick pair with a high twist angle of 34°. (g) Stacking of the U4·G13 wobble pair with its 5′ flanking Watson–Crick pair with a twist angle of 33.93°. (h) Stacking of the tandem wobble pairs in motif III, U·U/G·G, with a twist angle of 32.20°. (i) Stacking of the 3′ side G·U wobble with its 3′ flanking Watson–Crick pair with a high twist angle of 37.23°.
The three stacking steps taken together are the main determinants of the stability of the various motifs of the tandem wobbles. In motif I intra-strand slippage occurs between the adjacent G·U wobbles (Fig. 5b) while good intra-strand base stacking is retained at both flanking Watson–Crick ends (Fig. 5a and c) without much distortion of the A-type helix. It seems that base stacking in motif I is comparatively the strongest. In motif II the tandem G·U/U·G wobbles stack with slightly slipped bases and on both the 5′- and 3′-sides only cross-strand stacking occurs with the flanking Watson–Crick pairs (Fig. 5d–f). Thus, motif II has the least stacked tandem base pairs because it has two weak cross-strand stackings in which the uridines are unstacked. The base pair stacking in motif III is intermediate between those of motifs I and II (Fig. 5g–i).
When motif III is compared to the crystal structures of the group I intron at 2.8 Å resolution (21) and the 14 bp RNA at 2.1 Å resolution (22), where the flanking Watson–Crick base sequence is different, the base stacking pattern is nevertheless similar (base step twist angles are given in Table 4). In the 14 bp RNA crystal structure the 3′-side U·G wobble and its Watson–Crick pair (G·C) show less intra-strand stacking and increased cross-strand stacking (22). Although we do not have the structure of the motif III sequence with a pyrimidine on the 5′-side and a purine on the 3′-side, the stacking pattern is probably the same as above. Thermodynamic studies have shown that motif I is the most stable, independent of the flanking Watson–Crick sequence (17). In general, motif II is destabilizing and motif III is slightly more stable than motif II (16–18). Considering all the possibilities for the four flanking Watson–Crick base pairs, there are 16 different possible combinations. We have compared the base stacking in motif III of three crystal structures (the current structure, the group I intron and 14 bp RNA), however, we do not have crystal structures for motifs I and II with combinations of flanking Watson–Crick base pairs. Nevertheless, based on the results of the available crystal structures for motif III, we have shown that the stacking stabilities can be rationalized as motif I > motif III > motif II, which is consistent with their thermodynamic stabilities.
Table 4. Twist angles for the tandem wobble pairs and their flanking Watson–Crick base pairs in motif III in the group I intron and 14 bp RNA duplex.
| Group I intron | 14 bp RNA | ||
|---|---|---|---|
| Base step | Twist (°) | Base step | Twist (°) |
| A146·U157 | A4·U25 | ||
| 38.42 | 28.26 | ||
| G147·U156 | U5·G24 | ||
| 35.52 | 34.47 | ||
| G148·U155 | U6·G23 | ||
| 24.74 | 36.23 | ||
| G149·C154 | G7·C22 | ||
| Average | 34.6 | 33.0 | |
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge support of this work by grant GM-17378 from the National Institutes of Health. We thank the Board of Regents of Ohio for an Eminent Scholar Chair and Endowment to M.S. We acknowledge the Hayes Investment Fund of the Ohio Regents for partial support towards the purchase of an R-axis II C imaging plate system.
REFERENCES
- 1.Modrich P. (1987) Annu. Rev. Biochem., 56, 435. [DOI] [PubMed] [Google Scholar]
- 2.Pearl L.H. and Savva,R. (1995) Trends Biochem. Sci., 20, 421. [DOI] [PubMed] [Google Scholar]
- 3.Varani G. and Pardi,A. (1994) In Nagai,K. and Mattaj,I.W. (eds), RNA–Protein Interactions. Oxford University Press, Oxford, UK, pp. 1–24.
- 4.Varani G., Wimberly,B.and Tinoco,I.,Jr (1989) Biochemistry, 28, 7760. [DOI] [PubMed] [Google Scholar]
- 5.Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) Biochemistry, 32, 1078. [DOI] [PubMed] [Google Scholar]
- 6.Woese C.R., Gutell,R.R., Gupta,R. and Noller,H.F. (1983) Microbiol. Rev., 47, 621–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutell R.R., Larsen,N. and Woese,C.R. (1994) Microbiol. Rev., 58, 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClain W.H. and Floss,K. (1988) Science, 240, 793–796. [DOI] [PubMed] [Google Scholar]
- 9.McClain W.H. and Floss,K. (1988) J. Mol. Biol., 202, 697–709. [DOI] [PubMed] [Google Scholar]
- 10.Hou Y.M. and Schimmel,P. (1988) Nature, 333, 140–145. [DOI] [PubMed] [Google Scholar]
- 11.McClain W.H., Gabriel,K. and Scheneider,J. (1996) RNA, 2, 105–109. [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuno H. and Sundaralingam,M. (1978) Nucleic Acids Res., 5, 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cech T.R. (1987) Sciences, 236, 1532–1539. [DOI] [PubMed] [Google Scholar]
- 14.Barford E.T. and Cech,T.R. (1989) Mol. Cell. Biol., 9, 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strobel A.A. and Cech,T.R. (1995) Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- 16.Gautheret D., Konings,D. and Gutell,R. (1995) RNA, 1, 807–814. [PMC free article] [PubMed] [Google Scholar]
- 17.He L., Kierzek,R., SantaLucia,J.J., Walter,A.E. and Turner,D.H. (1991) Biochemistry, 30, 11124–11132. [DOI] [PubMed] [Google Scholar]
- 18.Wu M., McDowell,J.A. and Turner,D.H. (1995) Biochemistry, 34, 3204–3211. [DOI] [PubMed] [Google Scholar]
- 19.Biswas R., Wahl,M.C., Ban,C. and Sundaralingam,M. (1997) J. Mol. Biol., 267, 1149–1156. [DOI] [PubMed] [Google Scholar]
- 20.Biswas R. and Sundaralingam,M. (1997) J. Mol. Biol., 270, 511–519. [DOI] [PubMed] [Google Scholar]
- 21.Cate J.H. and Doudna,J.A. (1996) Structure, 4, 1221–1229. [DOI] [PubMed] [Google Scholar]
- 22.Trikha J., Filman,D.J. and Hogle,J.M. (1999) Nucleic Acids Res., 27, 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z. and Minor,W. (1997) Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Wahl M.C., Ban,C., Sekharudu,C., Ramakrishnan,B. and Sundaralingam,M. (1996) Acta Crystallogr., 52D, 655–667. [DOI] [PubMed] [Google Scholar]
- 25.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J., Kuszewski,J., Niges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Acta Crystallogr., 54D, 905–921. [DOI] [PubMed] [Google Scholar]
- 26.Berman H.M., Olson,W.K., Berveridge,D.L., Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.H., Srinivasan,A.R. and Schneider,B. (1992) Biophys. J., 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fratini A.V., Kopka,M.L., Drew,H.R. and Dickerson,R.E. (1982) J. Biol. Chem., 257, 14686–14707. [PubMed] [Google Scholar]